Ballistic Labeling of Pyramidal Neurons in Brain Slices and in Primary Cell Culture
Summary
We present a protocol to label and analyze pyramidal neurons, which is critical for evaluating potential morphological alterations in neurons and dendritic spines that may underlie neurochemical and behavioral abnormalities.
Abstract
It has been reported that the size and shape of dendritic spines is related to their structural plasticity. To identify the morphological structure of pyramidal neurons and dendritic spines, a ballistic labeling technique can be utilized. In the present protocol, pyramidal neurons are labeled with DilC18(3) dye and analyzed using neuronal reconstruction software to assess neuronal morphology and dendritic spines. To investigate neuronal structure, dendritic branching analysis and Sholl analysis are performed, allowing researchers to draw inferences about dendritic branching complexity and neuronal arbor complexity, respectively. The evaluation of dendritic spines is conducted using an automatic assisted classification algorithm integral to the reconstruction software, which classifies spines into four categories (i.e., thin, mushroom, stubby, filopodia). Furthermore, an additional three parameters (i.e., length, head diameter, and volume) are also chosen to assess alterations in dendritic spine morphology. To validate the potential of wide application of the ballistic labeling technique, pyramidal neurons from in vitro cell culture were successfully labeled. Overall, the ballistic labeling method is unique and useful for visualizing neurons in different brain regions in rats, which in combination with sophisticated reconstruction software, allows researchers to elucidate the possible mechanisms underlying neurocognitive dysfunction.
Introduction
In 2000, Gan et al. described a rapid labeling technique for individual neurons and glia in the nervous system that combined various lipophilic dyes, allowing for the simultaneous labeling of many brain cells with different colors1,2. More recently, a ballistic labeling technique was described by Seabold et al.3 that introduced fluorescent dyes (Dil) into the neurons of brain slices. A versatile staining technique, ballistic labeling is appreciated for its ability to be utilized in multiple animal species and across a wide range of ages. Furthermore, it can be combined with immunostaining to identify subpopulations of brain cells3. Compared with traditional techniques (e.g., Golgi-Cox silver impregnation, microinjection)4, ballistic labeling affords an opportunity to more clearly distinguish morphological characteristics, including dendritic spines, a feature that is critical for drawing inferences about neuronal complexity and synaptic connectivity5.
Excitatory pyramidal neurons are characterized by a single, large apical dendrite, multiple shorter basal dendrites, and thousands of dendritic spines6. Pyramidal neurons are found in multiple brain regions related to higher order cognitive processing, including the prefrontal cortex (PFC) and hippocampus. In the PFC, pyramidal neurons are observed in layers II/III and layer V, with each exhibiting unique morphology. Specifically, pyramidal neurons in layer II/III of the PFC have a shorter apical dendrite and less branching than pyramidal neurons in layer V6. Within the hippocampus, pyramidal neurons are located in both the CA1 and CA3 regions, with each displaying distinct morphologies. Specifically, pyramidal neurons in the CA1 region exhibit a more distinctive apical dendrite, with branching occurring farther from the soma, relative to the CA3 region6.
Dendritic spines on pyramidal neurons in both the PFC and hippocampus are the primary site of excitatory synapses7. Morphological characteristics of dendritic spines, which are classically characterized into three primary categories (i.e., thin, stubby, or mushroom8), have been related to the size of the excitatory synapse9. Thin spines, characterized by a long, thin neck, small bulbous head, and smaller postsynaptic densities, are more unstable and develop weaker connections. However, mushroom spines, which have a larger dendritic spine head, are recognized for forming stronger synaptic connections, an effect resulting from their larger size. In sharp contrast, stubby spines are devoid of a spine neck, exhibiting an approximately equal head and neck volume ratio8. Within the hippocampus, branched spines may also be observed, whereby the spine has multiple heads that emerge from the same dendritic spine neck10. Therefore, the morphological changes of dendritic spines could reflect functionality and structural capacity. Furthermore, studies have demonstrated that the size and shape of dendritic spines relates to their structural plasticity, leading to the idea that small spines are involved in learning and attention, whereas larger, more stable spines, are involved in long-term processes, including memory11. Additionally, the distribution of dendritic spines along the dendrite may be associated with synaptic connectivity5,12.
Thus, the present methodological paper has three goals: 1) Present our protocol for ballistic labeling, which has been utilized with a success rate (i.e., neurons meeting selection criteria and appropriate for analysis) of 83.3%5,12,13 and across multiple brain regions (i.e., PFC, nucleus accumbens, hippocampus); 2) Demonstrate the generalizability of the technique and its application to neurons grown in vitro; 3) Detail the methodology utilized in neuronal reconstruction software and the inferences that can be drawn from such data.
Protocol
All animal protocols were reviewed and approved by the Animal Care and Use Committee at the University of South Carolina (federal assurance number: D16-00028).
1. Preparation of DiI/Tungsten bead tubing
- Dissolve 100 mg of polyvinylpyrrolidone (PVP) with 10 mL of ddH2O. Vortex the PVP solution lightly.
- Fill the tubing with the PVP solution (see Table of Materials) and leave it for 20 min. Then, expel the PVP solution through the other end of the tubing using a 10 mL syringe.
- Combine 170 mg of tungsten microcarrier beads with 250 µL of methylene chloride. Vortex the tungsten bead suspension thoroughly.
- Combine 6 mg of lipophilic DilC18(3) dye with 300 µL of methylene chloride. Vortex the DilC18(3) dye solution thoroughly.
NOTE: Perform steps 1.3–1.4 in a fume hood. - Pipette 250 µL of the tungsten bead suspension onto a glass slide. Wait for the suspension to air dry (~3 min).
- Add 300 µL of DilC18(3) dye solution on the top of the tungsten bead suspension layer. Mix the DiIC18(3) dye solution and tungsten bead suspension thoroughly with a pipette tip and allow the mixture to air dry (~3 min).
- Once dried, use a razor blade to split the mixture into two 1.5 mL centrifuge tubes. Fill the tubes with water.
- Sonicate in a water bath (Amp I, 100%) until homogenous (~5 min). Ensure that the tip of the sonicator lies directly on the tubes with the bead suspension.
- Combine the two 1.5 mL of homogenized mixture into a 15 mL conical tube. Sonicate the mixture for another 3 min.
- Draw the tungsten beads-DilC18(3) dye mixture into the PVP-coated tubing using a 10 mL syringe. Feed the tubing into the preparation station (see Table of Materials).
- Rotate for 1 min on the preparation station. Carefully remove all the water from the tubing using a 10 mL syringe.
- Turn on the nitrogen gas and adjust the nitrogen flow to approximately 0.5 Liters per minute (LPM), rotate the tubing in the preparation station, and dry with nitrogen for 30 min.
- Remove the tubing from the preparation station and cut into 13 mm lengths (matching the loading size of the cartridge) using a tubing cutter. Keep the 13 mm lengths in scintillation vials in the dark.
2. Preparation of brain sections
NOTE: Adult male F344/N rats were pair housed in a controlled environment under a 12/12 light:dark cycle with ad libitum access to food and water. All animals were cared for using guidelines established by the National Institutes of Health in the Guide for the Care and Use of Laboratory Animals.
- Deeply anesthetize rats using 5% sevoflurane.
- Proceed to the following step when the rats are not responsive to noxious stimuli and reflexes are absent.
- Secure the rat in a supine position inside a chemical fume hood.
- Make an incision through the skin along the thoracic midline. Separate the diaphragm and open the chest with scissors. Insert a 20 G × 25 mm needle into the left ventricle.
- Immediately cut the right atrium with scissors. Perfuse 50 mL of 100 mM PBS with 5 mL/min of flow rate. Perfuse 100 mL of 4% paraformaldehyde buffered in 100 mM PBS.
- Remove the entire rat brain right after perfusion.
- Postfix the entire brain for 10 min with 4% paraformaldehyde.
NOTE: Do not postfix in 4% paraformaldehyde for more than 30 min, because it will affect the labeling. - Cut 500 µm thick coronal sections using a rat brain matrix (see Table of Materials). Make the first cut and keep the blade in place. Make a second cut using a second blade and vertically remove the first blade, keeping the tissue on the blade surface.
- Place the brain slices in a 24 well plate with 1 mL of 100 mM PBS in each well. Repeat this process until all slices have been cut.
3. Ballistic labeling and visualization of brain sections
- Remove the PBS from each targeted well.
- Load the cartridge with a piece of DiI/Tungsten bead tubing and place it into the applicator.
- Put a piece of filter paper between two mesh screens. Connect the applicator to the helium hose. Adjust the output pressure of helium to 90 pounds per square inch (psi).
- Place the applicator vertically by hand on the center of the targeted well at a distance of 1.5 cm between the sample and the mesh screen. Fire the DiI/Tungsten beads tubing.
NOTE: Be sure to remove all PBS from the targeted wells before shooting. - Load the cartridge with the next DiI/Tungsten bead tubing. Continuously fire the DiI/Tungsten beads from the tubing on the remaining slices.
- Fill the 24 well plate with 100 mM of PBS. Wash with 500 µL of fresh 100 mM of PBS 3x. Do not let the slices flip over while washing with PBS.
- Add 500 µL of fresh 100 mM PBS and keep the slices at 4 °C in the dark for 3 h.
- Transfer the brain slices onto glass slides using a fine brush.
NOTE: Three brain sections can be transferred onto each glass slide. - Immediately add 1 mL of antifade mounting medium onto each section. Place a 22 mm x 50 mm coverslip over the brain sections. Dry the glass slides in the dark for 2 days.
- Turn on the confocal microscope system and switch to a 60× objective.
- Set the confocal microscope system to have a magnification of 60× (A/1.4, oil) and a Z-plane interval of 0.15 μm (pinhole size 30 μm, back-projected pinhole radius 167 nm). Use a 543 nm wavelength to acquire images of the neurons of interest.
- Obtain Z-stack images for the targeted neuron type based on brain region boundaries and morphological characteristics of neurons.
NOTE: Acquire at least three images fromm each animal.
4. Use of the methodology with cell culture
- Isolate primary cortical neurons from F344/N rats at postnatal day one using previously reported methodology14.
- Culture primary cortical neurons in a 35 mm glass bottom dish for one week. Change half of the culture medium with fresh neuron growth medium at third day after isolation. Wash the glass bottom dish 2x with 1 mL of 100 mM PBS.
- Fix with 4% paraformaldehyde for 15 min at room temperature. Repeat steps 3.2–3.6 to ballistically label the cells.
- Wash with 1 mL of 100 mM PBS 3x. Add 500 µL of fresh 100 mM of PBS and keep it at 4 °C in the dark for 3 h.
- Add 200 µL of antifade mounting medium.
- Obtain Z-stack images for each targeted neuron using the parameters in step 3.10.
5. Neuronal analysis and dendritic spine quantification
- Blind neurons using code numbers to prevent experimenter bias.
- Establish selection criteria for neurons based on the brain region of interest.
NOTE: Selection criteria for neurons include continuous dendritic staining, low background, no dye clusters inside of the cells, minimal diffusion of the DiI dye into the extracellular space, correct morphology of pyramidal neurons (Figure 1). - Open neuronal reconstruction software (See the attached Video 1).
- Load an image file by clicking on the “File Folder” image in the upper left-hand corner.
- Click “Soma” and mark the soma of the neurons on the image.
- Click “Tree” and select “User-Guided”.
- Trace all dendritic branches of interest.
NOTE: For pyramidal neurons, which are characterized by one apical dendrite and multiple basilar dendrites, only the apical dendrite is traced. Make sure all connecting branches are attached to one another. - Click “Spine”.
- Define Detection Parameters and click “Detect All”.
NOTE: For brain slices, the consistent parameters utilized across brain regions are: 2.0 (Outer Range), 0.3 (Minimum Height), 100% (Detector Sensitivity), and 10 (Minimum Count). For cell culture, it is necessary to increase the Detector Sensitivity and decrease the Minimum Count. - Classify spines by selecting “Classify All”.
NOTE: Dendritic spines are classified using an algorithm integral to the neuronal reconstruction software15. - Save tracing by selecting the Disk Image in the upper left-hand corner.
- Perform neuronal and dendritic spine morphological analyses.
- Open neuronal reconstruction quantitative analysis software (See attached Video 2).
- Load image by clicking “File” and “Open Data File”.
- Click “Analysis” and “Branched Structure Analysis” to analyze neuronal morphology and dendritic spine morphology.
- For neuronal morphology, click “Tree Totals” and select the box for “Dendrite Totals”.
- For dendritic spine morphology, click “Spines” and then select the box for “Spine Details”.
- Save output as a text (.txt) file by right-clicking the output table and selecting “Save to File”.
- Click “Analyze” and “Sholl Analysis”.
- Set the Starting Radius to 10 μm and the Radius Increment to 10 μm.
- Click the box “Dendrites” and click “Display”.
- Save output as a text (.txt) file by right-clicking the Output Table and selecting “Save to File”.
6. Data analysis
- Analyze the neuronal morphology (i.e., dendritic branching complexity) data.
- Add the number of dendrites at each branch order and divide by the total number of dendrites. Multiply by 100 to calculate the relative frequencies for the number of dendrites at each branch order.
- Analyze the Sholl analysis data to examine neuronal arbor complexity and dendritic spine connectivity.
- Calculate the mean and standard error of the mean for the number of intersections at each radius.
- Sum the number of spines dependent upon spine type (i.e., thin, stubby, mushroom) at each radius and divide by the total number of spines for that spine type. Multiply by 100 to calculate the relative frequencies for the number of spines at each radius.
Representative Results
In Figure 2A, the typical pyramidal neurons in the hippocampal region in the rat brain sections were identified by ballistic labeling technology, characterized by one large apical dendrite and several smaller basal dendrites around the soma. Figure 2B shows the neuron in the neuronal reconstruction quantitative analysis software after the soma was detected, dendritic branches were traced, and spines were detected. Subsequently, the data were analyzed using neuronal reconstruction quantitative analysis software, which provided an opportunity to assess the dendritic branching complexity (Figure 2C) and neuronal arbor complexity (Figure 2D).
In Figure 2C, we utilized the centrifugal branch ordering method, collected from the “Tree Totals” output, to count the number of segments traversed along each dendrite and assigned branch order. The relative frequency of segments at each branch order was examined for branch orders 1–15. When shifts in the distribution of dendritic branches were observed between groups, alterations in dendritic branching complexity could be inferred. Furthermore, a Sholl analysis was conducted as a complementary measure of neuronal arbor complexity, whereby the number of dendritic intersections occurring every 10 µm from the soma was quantified in each sample section (Figure 2D). When shifts in the number of dendritic intersections were observed between groups, alterations in neuronal arbor complexity could be inferred.
Morphological changes in dendritic spines could be assessed using length (µm), head diameter (µm), and volume (µm3), as seen in Figure 3A–B. Furthermore, spines were classified using the automatic assisted classification system in the neuronal reconstruction software. The relative frequency of the number of spines between each radius was examined for thin, mushroom, and stubby spines. Given our understanding of which spine types form stronger synaptic connections (i.e., mushroom relative to stubby) and neurotransmitter afferents, shifts in the distribution of spines along the neuron can indicate synaptic connectivity.
Furthermore, we validated the utility of the ballistic labeling technique on a primary pyramidal neuron in cell culture. First, we cultured primary hippocampal neurons at postnatal D1 (day 1) on a cell culture plate coated with poly-L-lysine for 2 weeks or until approximately 70% confluency. Then the samples were fixed with 4% PFA for 15 min and washed 2x with PBS. Beginning at step 3.1 of the present protocol, we ballistically labeled and imaged the primary pyramidal neurons from the hippocampus. Data showed stabilized labeling and identified pyramidal neurons based on the triangle shape of the soma and large apical dendrite (Figure 4A). Utilization of neuronal reconstruction software to investigate dendritic spines of primary pyramidal neurons grown in cell culture offers opportunities similar to those in the rat brain. Example outcomes are illustrated for the distribution of thin dendritic spines (Figure 4B) and dendritic spine length measured in µm (Figure 4C). However, it is noteworthy that the primary pyramidal neurons grown in cell culture had less dendritic branching, precluding the assessment, at least in this example, of dendritic branching complexity and neuronal arbor complexity.
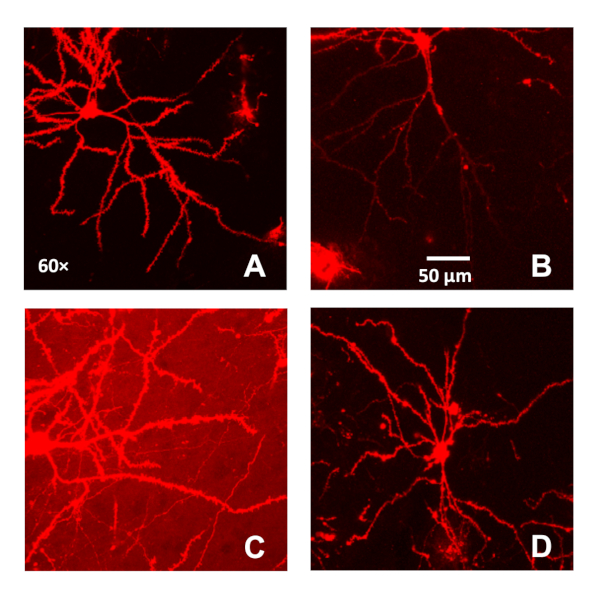
Figure 1: Selection criteria utilized for pyramidal neurons in the medial prefrontal cortex labeled using ballistic labeling technology. (A) A representative confocal image (60x) of a well-labeled pyramidal neuron from the medial prefrontal cortex. A single pyramidal neuron with a clear soma and apical dendrite included continuous, bright dendritic staining with low background. (B–C) A representative confocal image (60x) of a pyramidal neuron from the medial prefrontal cortex with light staining at the more distal branches (B) and high background (C). (D) A representative confocal image (60x) of a labeled neuron from the medial prefrontal cortex (based on Bregma coordinates) that has flawed morphological characteristics. Please click here to view a larger version of this figure.
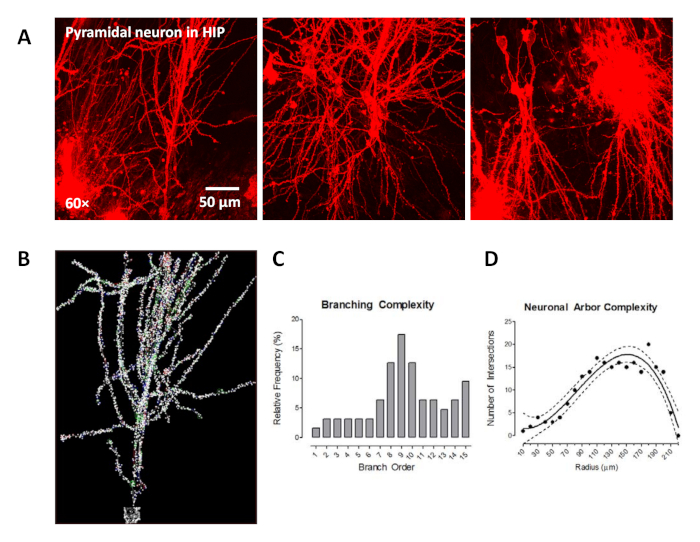
Figure 2: The labeling of pyramidal neurons in the hippocampus (HIP) using ballistic labeling technology and neuroanatomic assessments. (A) Three representative confocal images (60x) of pyramidal neurons labeled by ballistic tungsten beads. (B) The assessment of neuronal morphology: dendritic branch order analysis and Sholl analysis. The traced image of the dendritic spine in which the spine morphology was also identified using dendritic spine analysis software. (C) Branch order analyses utilized to examine the relative frequency of dendritic branches at different branch orders. (D) The numbers of dendritic intersections every 10 µm from the soma were assessed using Sholl analysis as a measure of neuronal arbor complexity. Data are described as relative frequencies of the entire dataset (C) or fit with 95% confidence intervals (D). Please click here to view a larger version of this figure.
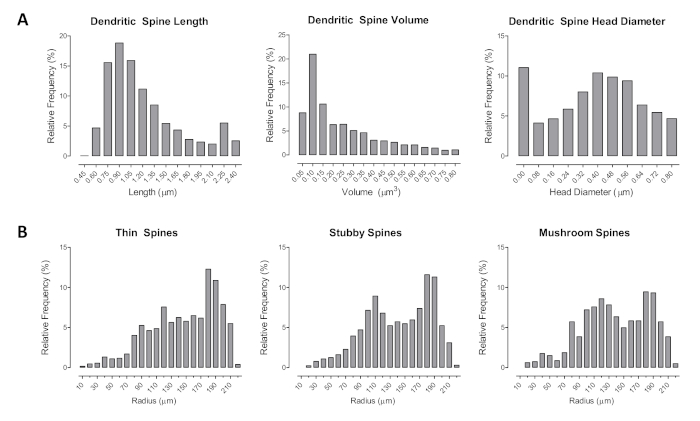
Figure 3. The assessment of dendritic spine morphology. (A–B) The distribution of dendritic spines illustrated as a function of spine type (i.e., thin, stubby, mushroom). Additional dendritic spine parameters were also analyzed (i.e., length, volume, head diameter) as an assessment of dendritic spine morphology. Data are illustrated as relative frequencies of the entire dataset. Please click here to view a larger version of this figure.
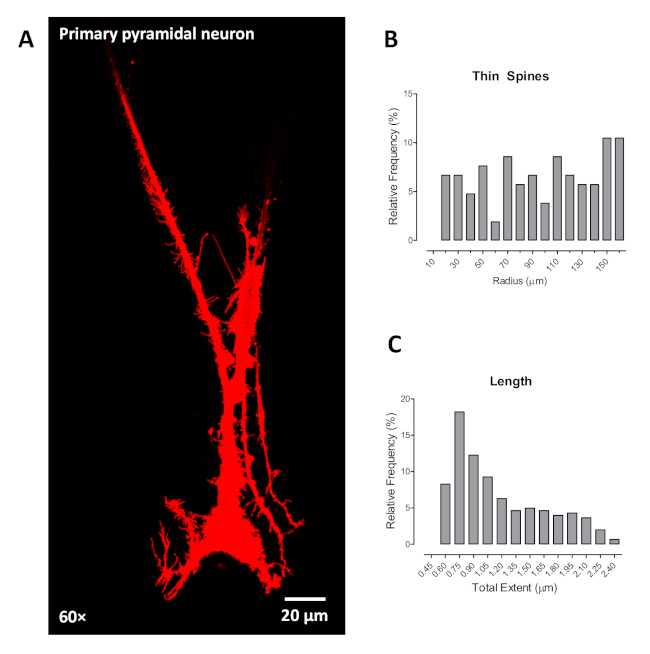
Figure 4. The labeling of primary cortical neuron in vitro using ballistic labeling technology. (A) Representative confocal images (60x) of primary cortical neurons labeled by ballistic tungsten beads in vitro. Example results similar to those acquired from ballistic labeling in brain slices are shown for the distribution of thin dendritic spines (B) and length (C). Data are illustrated as relative frequencies of the entire dataset. Please click here to view a larger version of this figure.
Video 1: Procedures of neuronal tracing and dendritic spine detection. Please click here to view this video. (Right-click to download.)
Video 2: Procedures of data collection and output for quantitative analysis. Please click here to view this video. (Right-click to download.)
Discussion
In this protocol, we describe a versatile labeling technique for neurons from both rat brain and those grown in vitro. Furthermore, we report the methodology for utilizing neuronal reconstruction software and neuronal reconstruction quantitative analysis software to assess neuronal morphology and dendritic spines. The assessment of neuronal morphology and dendritic spines provides an opportunity to determine alterations in dendritic branching complexity, neuronal arbor complexity, dendritic spine morphology, and synaptic connectivity.
When conducting the protocol, researchers should pay special attention to a few steps. First, post-fixing in 4% PFA for too long will damage the integrity of the lipophilic membrane and cause the dye to leak outside of the cells. Second, compared to the specificity of ballistic labeling in brain slices, which targets only neurons, labeling of primary cortical neurons in vitro introduces nonspecific labeling of the glia because ballistic labeling in brain slices is not specific for a type of neuron (i.e., pyramidal neuron, medium spiny neuron, granule cell). Thus, Bregma coordinates, morphological assessments, or specific cell markers should be combined with the ballistic labeling method. Third, the thickness of the brain slices can be between 200–500 µm; for best results it should be optimized. Fourth, the efficiency of labeling and dye penetration is related to many factors, such as helium pressure, incubating times after ballistic application, the DiI/Tungsten beads, the distance between the mesh screen and the surface of brain slices, etc. The protocol should be optimized for each study. Fifth, large clumps or clusters of ballistic dye coated tungsten beads during preparation must be avoided, because clumps would not allow individual neurons to be distinguished. We also determined that DiI diffuses throughout individual neurons more completely than DiO in this ballistic methodology.
Nevertheless, compared with traditional labeling methods4, the ballistic labeling technique makes high resolution confocal imaging possible, allowing for the assessment of neuronal and dendritic spine morphology. Furthermore, neuronal reconstruction software utilizes an algorithm for automatic assisted classification of dendritic spines (i.e., thin, mushroom, stubby), branch ordering measurements, classical Sholl analysis, and measurement of morphological features of dendritic spines, such as length (µm), head diameter (µm), and volume (µm3). The quantification of multiple neuronal parameters affords an opportunity to better understand the mechanisms underlying neurocognitive dysfunction.
Overall, the ballistic labeling method allows visualization of neuronal structures in different brain regions of the rat and in cell culture, which is important to elucidate the possible mechanisms underlying neurocognitive dysfunction. In the present study, we present a method to label pyramidal neurons by a ballistic labeling technique. Furthermore, combined with neuronal reconstruction software, we demonstrated the ability to examine neuronal and dendritic spine morphology in hippocampal pyramidal neurons. Group differences in neuronal and/or dendritic spine morphology provide an opportunity to understand the mechanisms underlying neurocognitive dysfunction.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was funded by NIH grants HD043680, MH106392, DA013137, and NS100624.
Materials
| 20Gx25mm PrecisionGlide needle | BD | 305175 | |
| 24-well cell culture plate | Costar | 3562 | |
| 35 mm Glass Bottom Dishes | MatTek Corporation | P35G-1.5-20-C | |
| Antibiotic-Antimycotic solution | Cellgro | 30004CI | 100X |
| B-27 supplement | Life Technologies | 17504-044 | 50X |
| Barrel liner | BIO-RAD | 165-2417 | |
| Borax | Sigma | B9876 | |
| Boric acid | Sigma | B0252 | |
| Cartridge holder | BIO-RAD | 165-2426 | |
| Confocal imaging software | Nikon | EZ-C1 | version 3.81b |
| Confocal microscope | Nikon | TE-2000E | |
| Cover glass | VWR | 637-137 | |
| DilC18(3) | Fisher Scientific | D282 | |
| DMEM/F12 medium | Life Technologies | 10565-018 | |
| Dumont #5 Forceps | World Precision Instruments | 14095 | |
| Dumont #7 Forceps | World Precision Instruments | 14097 | |
| F344 rat | (Harlan Laboratories, Indianapolis, IN) | ||
| Glucose | VWR | 101174Y | |
| GlutaMax | Life Technologies | 35050-061 | 100X |
| HBSS | Sigma | H4641 | 10X |
| Helios diffusion screens | BIO-RAD | 165-2475 | |
| Helios gene gun kit | BIO-RAD | 165-2411 | |
| Helios gene gun system | BIO-RAD | 165-2431 | |
| Helium hose assembly | BIO-RAD | 165-2412 | |
| Iris Forceps | World Precision Instruments | 15914 | |
| Iris Scissors | World Precision Instruments | 500216 | |
| Methylene chloride | Fisher Scientific | D150-1 | |
| Neurobasal medium | Life Technologies | 21103-049 | |
| Neurolucida 360 software | mbf bioscience | dendritic spine analysis | |
| Paraformaldehyde | Sigma-Aldrich | 158127-500G | |
| Paraformaldehyde | Sigma | P6148 | |
| Poly-L-Lysine | Sigma | P9155 | |
| Polyvinylpyrrolidone | Fisher Scientific | 5295 | |
| ProLong Gold antifade reagent | Fisher Scientific | P36930 | mounting medium |
| Rat brain matrix, 300 – 600g, Coronal, 0.5mm | Ted Pella | 15047 | |
| Sevoflurane | Merritt Veterinary Supply | 347075 | |
| Sodium Bicarbonate | Life Technologies | 25080 | |
| SuperFrost Plus Slides | Fisher Scientific | 12-550-154% | |
| Syringe kit | BIO-RAD | 165-2421 | |
| Tefzel tubing | BIO-RAD | 165-2441 | |
| Trypsin-EDTA | Life Technologies | 15400-054 | |
| Tubing cutter | BIO-RAD | 165-2422 | |
| Tubing Prep station | BIO-RAD | 165-2418 | |
| Tungsten M-25 Microcarrier 1.7 µm | BIO-RAD | 165-2269 | |
| Vannas Scissors | World Precision Instruments | 500086 |
Referências
- Gan, W. B., Grutzendler, J., Wong, W. T., Wong, R. O., Lichtman, J. W. Multicolor “DiOlistic” labeling of the nervous system using lipophilic dye combinations. Neuron. 27, 219-225 (2000).
- Gan, W. B., Grutzendler, J., Wong, R. O., Lichtman, J. W. Ballistic delivery of dyes for structural and functional studies of the nervous system. Cold Spring Harbor Protocol. 2009 (4), 5202 (2009).
- Seabold, G. K., Daunais, J. B., Rau, A., Grant, K. A., Alvarez, V. A. DiOLISTIC labeling of neurons from rodent and non-human primate brain slices. Journal of Visualized Experiments. (41), (2010).
- Spacek, J. Dynamics of the Golgi method: a time-lapse study of the early stages of impregnation in single sections. Journal of Neurocytology. 18 (1), 27-38 (1989).
- McLaurin, K. A., Li, H., Booze, R. M., Mactutus, C. F. Disruption of Timing: NeuroHIV Progression in the Post-cART Era. Science Reports. 9 (1), 827 (2019).
- Spruston, N. Pyramidal neurons: dendritic structure and synaptic integration. Nature Reviews Neurosciences. 9 (3), 206-221 (2008).
- Megias, M., Emri, Z., Freund, T. F., Gulyas, A. I. Total number and distribution of inhibitory and excitatory synapses on hippocampal CA1 pyramidal cells. Neurociência. 102, 527-540 (2001).
- Peters, A., Kaiserman-Abramof, I. R. The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. American Journal of Anatomy. 127, 321-355 (1970).
- Harris, K. M., Sultan, P. Variation in the number, location, and size of synaptic vesicles provides an anatomical basis for the nonuniform probability of release at hippocampal CA1 synapses. Neuropharmacology. 34, 1387-1395 (1995).
- Sorra, K. E., Fiala, J. C., Harris, K. M. Critical assessment of the involvement of perforations, spinules, and spine branching in hippocampal synapse formation. Journal of Comparative Neurology. 398, 225-240 (1998).
- Mancuso, J. J., Chen, Y., Li, X., Xue, Z., Wong, S. T. C. Methods of dendritic spine detection: from Golgi to high-resolution optical imaging. Neurociência. 251, 129-140 (2012).
- McLaurin, K. A., et al. Synaptic connectivity in medium spiny neurons of the nucleus accumbens: A sex-dependent mechanism underlying apathy in the HIV-1 transgenic rat. Frontiers in Behavior Neurosciences. 12, 285 (2018).
- Roscoe, R. F., Mactutus, C. F., Booze, R. M. HIV-1 transgenic female rat: synaptodendritic alterations of medium spiny neurons in the nucleus accumbens. Journal of Neuroimmune Pharmacology. 9 (5), 642-653 (2014).
- Li, H., Aksenova, M., Bertrand, S. J., Mactutus, C. F., Booze, R. Quantification of Filamentous Actin (F-actin) Puncta in Rat Cortical Neurons. Journal of Visualized Experiments. (108), e53697 (2016).
- Rodriguez, A., Ehlenberger, D. B., Dickstein, D. L., Hof, P. R., Wearne, S. L. Automated Three-Dimensional Detection and Shape Classification of Dendritic Spines from Fluorescence Microscopy Images. PLoS ONE. 3 (10), 1371 (2008).

