Rapid and Cost-Effective RNA Extraction of Rat Pancreatic Tissue
Summary
The purity and integrity of the isolated RNA is a vital step in RNA dependent assays. Here, we present a practical, rapid, and inexpensive method to extract RNA from a small quantity of undamaged pancreatic tissue.
Abstract
Regardless of the extraction method, optimized RNA extraction of tissues and cell lines are carried out in four stages: 1) homogenization, 2) effective denaturation of proteins from RNA, 3) ribonuclease inactivation, and 4) removal of contamination from DNA, proteins, and carbohydrates. However, it is very laborious to maintain the integrity of RNA when there are high levels of RNase in the tissue. Spontaneous autolysis makes it very difficult to extract RNA from pancreatic tissue without damaging it. Thus, a practical RNA extraction method is needed to maintain the integrity of pancreatic tissues during the extraction process. An experimental and comparative study of existing protocols was carried out by obtaining 20-30 mg of rat pancreatic tissues in less than 2 minutes and extracting the RNA. The results were assessed by electrophoresis. The experiments were carried out three times for generalization of the results. Immersing pancreatic tissue in RNA stabilization reagent at -80 °C for 24 h yielded high integrity RNA, when the RNA extraction reagent was used as the reagent. The results obtained were comparable to the results obtained from commercial kits with spin column bindings.
Introduction
Structural gene data can be transcribed to a functional product through gene expression. RNA analysis is used to discover differences in gene expression across different conditions. There are a number of methods to extract nucleic acids as follows: guanidinium thiocyanate, extraction via phenol-chloroform, cellulose-based chromatography, extraction by silica matrices, and anion-exchange1,2.
Proper detection of gene expression is influenced by the integrity of RNA isolated from tissues; therefore, it is vital to evaluate the integrity of RNA isolated from tissues before further tests are carried out because complementary molecular tests on low-quality RNA may jeopardize diagnostic application results. Thus, high integrity RNA is needed for molecular biological tests with different diagnostic applications: quantitative RT-PCR, micro-arrays, ribonuclease protection assay, northern blot analysis, RNA mapping, and cDNA library construction3,4.
RNA becomes rather unstable after being kept for a long time. Long mRNA fragments over 10 kb are particularly susceptible to degradation5,6. Thus, researchers must consider various factors that influence the integrity of purified RNA. The purity of RNA must be protected against RNases, proteins, genomic DNA, and enzymatic inhibitor contamination. In addition, the best and acceptable absorption ratio of RNA to UV (260/280) must be within the range of 1.8-2.0 with minimum fragmentation over electrophoresis. Recently developed laboratory techniques have enabled scientists to evaluate the integrity of molecular analysis sample more practically7,8.
It is much more difficult to extract undamaged RNA from pancreatic tissue than other types of tissues because of the high quantity of ribonucleases (RNases). However, existing extraction methods, namely the rapid ejection of the pancreatic tissue from the abdominal cavity and homogenization at low temperatures to impede RNases, have proven ineffective7,8,9,10,11,12,13,14.
The purpose of the present comparative experimental study is to modify and compare existing methods to determine the most efficient methods. To that end, various protocols of RNA extraction were modified and compared. It was specifically aimed at determining the least expensive method requiring a minimum amount of pancreatic tissue.
Protocol
Ethical approval for this study was obtained from Shiraz University of Medical Sciences (Approval number: 93-01-01-71783-07-2014).
NOTE: Use male Sprague–Dawley rats weighing 250 g. Place the vial containing a sliver of pancreatic tissue immersed in RNA stabilizing reagent in a liquid nitrogen tank at -80 °C and use RNA extraction reagent solution to maintain the integrity of RNA.
1. Removal of the rat pancreatic tissue
- Prepare the operating room and place all required materials under the hood.
- Sterilize all the surgical instruments (Dressing forceps, Brown-Adson forceps, Iris scissors, and Littler scissors) in an oven for at least 4 h at 240 °C to inactivate RNases15. Sterilize the surface of the surgery place with 70% alcohol under the hood.
- Inject ketamine/xylazine intraperitoneally using an insulin syringe [80/8 mg/kg]. Check the depth of anesthesia by pinching the rat’s toes for a lack of response.
- Add 1 mL of RNA stabilization reagent in the proper microtube (2 mL) to inhibit activation of endonucleases in excised tissues.
- Immediately place the rat on the surgical board (head away from the surgeon) for a mid-line incision.
- Sterilize the entire abdominal surface with 70% alcohol and remove rat hair using a hair clipper with #40 blades.
- Make a V-shape incision to open the abdomen from the pubic area to the front legs with Dressing forceps, Brown-Adson forceps, Iris scissors, and Littler scissors.
- Flip the abdominal organs to the left side to expose the pancreas. Carefully find the pancreatic tissue, which spreads in the abdominal cavity. Locate the area under the spleen to find the pancreas without being confused with the fat tissue.
- Immediately remove 20-30 mg of pancreas from the abdominal cavity in less than 2 min. Put the removed tissue in the 2 mL microtube quickly and cut it with a sterile cutter to penetrate the separated tissues with RNA stabilization reagent.
- Place the microtube in a nitrogen tank to freeze immediately. Keep the microtube in a -80 °C freezer for 24 h16.
- After 24 h, transfer the tiny pieces of pancreatic tissues to an ice container.
- Inject potassium chloride (KCl) intracardially for euthanasia of rats after surgery.
2. RNA extraction
- Sterilize the surface under the hood with 70% alcohol.
- Put 1 mL of the RNA extraction reagent, which contains guanidinium thiocyanate to inhibit RNase, in the sterile microtube.
- Transfer the separated pancreatic tissue to the microtube. Transfer the microtube to the liquid nitrogen to freeze immediately.
- Homogenize the tissue with the micro tip probe sonicator at 4 °C set to level 20 for 60 s on and 5 s off.
- Incubate each homogenized sample for 5 min at 4 °C using crushed ice to allow the nucleoprotein complexes to be completely dissociated.
- Enrich the samples with 0.2 mL of chloroform for each 1 mL of RNA extraction reagent. Cap the tube firmly and shake it forcefully for 15 s.
- Incubate the tube for 15 min on the crushed ice (4 °C) to separate the reagent in three phases.
- Run the centrifuge at 12,000 x g for 15 min at +2 to +8 °C. Transfer the colorless aqueous phase to a new microtube.
- Add 0.5 mL of 100% isopropanol to the aqueous phase. Close the tube and then invert it at least 3 times to mix the RNA thoroughly.
- Incubate the sample for 5-10 min on a cold box (4 °C) to allow RNA precipitation.
- Run the centrifuge at 12,000 x g for 15 min at +2 to +8 °C. Discard the supernatant.
- Enrich each of the centrifuge tubes with 1 mL of 75% ethanol.
- Wash the RNA pellet by inverting the tube at least 3 times.
- Run the centrifuge at 7,500 x g at +2 to +8 °C for 5 min. Discard the supernatant to remove the excess ethanol from the RNA pellet by air-drying.
- Re-suspend the RNA pellet in diethylpyrocarbonate (DEPC)-treated RNase-free water.
- Pass the solution through a pipette tip several times to dissolve the RNA pellet. Then, incubate the solution for 10-15 min at +65 °C.
3. Evaluating RNA Integrity with denaturation electrophoresis
- Prepare gel running buffer: 50 mM NaAc (DEPC treated) and 0.5 M EDTA (pH 8.0) in DEPC-treated H2O.
- Prepare 5x formaldehyde gel-running buffer (MOPS running buffer): 0.1 M MOPS (pH 7.0), 40 mM NaAc, 5 mM EDTA (pH 8.0).
- Dissolve 20.6 g of MOPS in 800 mL of DEPC treated 50 mM sodium acetate. Adjust the pH to 7.0 with 2 N NaOH. Add 10 mL of DEPC-treated 0.5 M EDTA (pH 8.0). Adjust the volume of the solution to 1 L with DEPC-treated water.
- Filter the solution through a 0.2 µm filter and keep it at room temperature away from light for the sake of sterilization. The buffer becomes yellow with time if it is exposed to light or is autoclaved. A straw-colored buffer works well, but darker ones do not17.
- Prepare a 1.5% agarose gel18. For 50 mL, add 0.75 g of agarose and 31 mL of H2O. Microwave the solution in the microwave for 1 min. Add 9 mL of formaldehyde and 10 mL of 5x MOPS running buffer.
- Prepare the samples for the gel. Mix the following in a sterile microfuge tube:
X µL RNA (up to 30 µg)
2 µL of 5x gel-loading buffer
10 µL of formamide
4 µL of MOPS running buffer
1 µL of 0.1 mg/mL EtBr
3 µL of formaldehyde 5-x µL of DEPC-H2O - Incubate the samples at 65 °C for 15 min and cool them on ice. Centrifuge for 5 s until all the fluid is deposited.
- Pre-run the gel at 5 V/cm for 5 minutes.
- Load the samples into the lanes of the gel immediately and then submerge the gel in 1x formaldehyde gel-running buffer. Run at 3-4 V/cm19.
Representative Results
Evaluation of the integrity of RNA in the RNA extraction reagent according to a routine and modified surgical protocol without RNA stabilization reagent
Unacceptable bands were observed after the extraction of RNA with the RNA extraction reagent from a routine surgical protocol. Lane 1 shows RNA from the liver as a control. Lane 2 shows the degraded status of 28S/18S rRNA bands in total RNA obtained from a routine surgical protocol. When the quantity of pancreatic tissue was reduced to 50 mg (lane 3) or 20-30 mg (lane 4) and the surgery was performed immediately (modified protocol) without the RNA stabilization reagent, RNA separation was less successful than in the liver tissue control and unspecific bands were observed.
Evaluation of the integrity of RNA samples according to a modified surgical protocol immersed in RNA stabilization reagent
The integrity of RNAs produced with the RNA extraction reagent depends on the preservation time and temperature (lane 5-8). In comparison with the control liver tissue, RNA separation was not successful when the amount of pancreatic tissue was 50 mg (lane 5) or 20-30 mg (lane 6). RNA was extracted immediately after the tissue was immersed in RNA stabilization reagent. No specific band was observed when 20-30 mg of tissue was submerged in RNA stabilization reagent at -80 °C for 48 h and RNA was extracted based on the protocol. According to the electrophoresis results, the RNA was completely degraded (lane 7). As depicted in lane 8, acceptable bands (28S/18S rRNA) were observed after submerging 20-30 mg of pancreatic tissue in RNA stabilization reagent at -80 °C for 24 h, and then RNA was extracted.
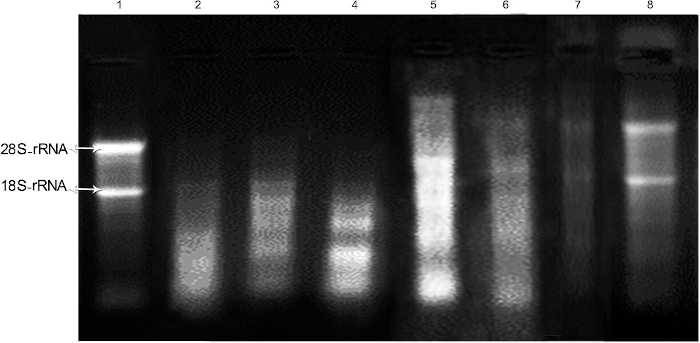
Figure 1: Assessment of the integrity of RNA isolated from rat pancreatic tissues using RNA extraction reagent according to the protocols under the investigation. Lane 1 depicts the integrity of RNA obtained from the liver as a control. Lane 2 represents the status of 28S/18S rRNA bands in total RNA obtained from a routine surgical protocol. Lane 3 represents the status of 28S/18S rRNA bands in total RNA obtained from a modified surgical protocol and 50 mg of tissue. Lane 4 represents the status of 28S/18S rRNA bands in total RNA obtained from a modified surgical protocol and 20-30 mg of tissue. Lane 5 represents the status of 28S/18S rRNA bands in total RNA obtained from a modified surgical protocol and 50 mg of tissue extracted immediately after being immersed in RNA stabilization reagent. Lane 6 shows the integrity of RNA obtained from 20-30 mg of pancreatic tissue from a modified surgical protocol extracted immediately after being immersed in RNA stabilization reagent. Lane 7 depicts the integrity of RNA obtained from 20-30 mg of tissue from a modified surgical protocol after 48 h of immersion in an RNA stabilization reagent at -80 °C. Lane 8 depicts the integrity of RNA obtained from 20-30 mg of tissue from a modified surgical protocol after 24 h of immersion in an RNA stabilization reagent at -80 °C. Please click here to view a larger version of this figure.
Discussion
In molecular biology it is vital to obtain high-quality RNA. The presence of the ribonuclease enzymes in cells and tissues quickly degrades RNA and makes the extraction complex. RNases are stable enzymes functioning without any co-factors. Small amounts of RNase are adequate to destroy RNA. When the rat pancreatic tissue is removed from the abdominal cavity, it is necessary to disinfect the surgical instruments by strong detergents, rinse them thoroughly and put them in an oven for at least 4 h at 240 °C to inactivate RNases before surgery. Given the fact that the RNase level is extremely high in the pancreas, the place of surgery is sterilized with NaOH and mild bleach to deactivate the RNases. While pancreatic tissue is being removed during dissection, the RNA would degrade. To increase efficiency, it is necessary for the dissection to be completed as quickly as possible20,21,22,23,24.
The pancreas is a critical tissue for the body’s homeostatic mechanisms. Hence, improved pancreatic RNA extraction procedures help researchers better understand active pathways. The present protocol proposed a model for an efficient, simple, and optimized method for RNA extraction from the pancreas. Different common RNA extraction methods from pancreatic tissue were evaluated. It was concentrated on the effect of frozen storage and RNase inhibition strategies influencing RNA quality. The two most significant factors influencing the integrity of RNA are surgery duration and the amount of collected pancreatic tissue. Recent studies have revealed that there is a positive correlation between RNA degradation and the amount of pancreatic tissue8,13,20.
In this protocol, 20-30 mg of pancreatic tissue was obtained in less than 2 min from the anesthetized rats. Lengthy surgical steps may lead to activation of endogenous endonucleases in the pancreas and degrade the RNA quickly. In this study, RNA was isolated from different samples with guanidinium thiocyanate, and phenol-chloroform extraction techniques used liquid nitrogen to impede RNA activity. The method achieved three objectives: rapid permeation of RNA stabilization reagent in pancreatic tissues, protection of cellular RNA, and increased preservation time. The results were optimal when the samples containing RNA stabilization reagent were kept at -80 °C for 24 h.
However, RNA integrity increased significantly when RNA stabilization reagent was introduced. Moreover, this process was reproducible. A small section of the pancreas (20-30 mg) was dissected during surgery from anesthetized rats and submerged in 1 mL of RNA stabilization reagent at -80°C for 1-2 days. As seen in lane 8, storage for 24 h was the optimal time.
The aforementioned methods allowed a smaller quantity of RNA stabilization reagent to penetrate into the organ. Furthermore, the degradation process discontinued shortly after the experiments because the size of the pieces dissected was small. In this protocol, the vital step is cutting the immersed tissue in RNA stabilization reagent to very small pieces as soon as possible until it penetrates the cells and suppresses activation of RNase. In the homogenizing step, it is very crucial to prevent bubble production in the RNA extraction reagent and perform all steps at 4 °C. It is crucial to separate the aqua phase (the phase containing RNA) very carefully to avoid DNA contamination. Although autolysis and the presence of endogenous RNases compromise intact RNA isolation from the rat pancreas, the integrity of RNA was maintained in the proposed pancreas perfusion method. Thus, the proposed method is a straightforward, reproducible, and inexpensive procedure that requires smaller quantities of RNA stabilization reagent than the other existing methods.
Like any study, this protocol has some limitations. First, the integrity and yield of obtained RNA is less than using whole pancreatic tissue as only 20-30 mg of tissue is used. Second, a large number of samples cannot be taken in one day because RNA extraction and also cDNA synthesis tests must be done fast and consecutively to decrease RNA degradation. Third, to perform research projects with different rat groups, it is essential to take exactly 20-30 mg of tissue from the same surgical area to decrease variation of data because rat pancreatic tissue diffuses completely in the peritoneal cavity.
To conclude, using RNA extraction reagent solution after RNA stabilization reagent perfusion is a good alternative to expensive and column-based RNA extraction kits.
Declarações
The authors have nothing to disclose.
Acknowledgements
The present study was financially supported by Shiraz University of Medical Sciences (Grant No. 93-01-01-71783-07-2014). We thank Mr. Zomorodian and Mr. Rostami at the Department of e-Learning in Medical Sciences, Virtual School and Center of Excellence in e-Learning, Shiraz University of Medical Sciences for editing the video.
Materials
| Agarose | Merck | 116801 | Germany |
| Atoclave | Teb Zaim | Iran | |
| Centrifuge | Sigma | Germany | |
| Chloroform | Merck | 107024 | Germany |
| Diethylpyrocarbonate (DEPC)-treated water | Sigma | Germany | |
| EDTA | sigma | 60-00-4 | Germany |
| Electrophoresis tank | Payapajoohesh | Iran | |
| Eppendorf microTube | Extragene | Taiwan | |
| EtBr | sigma | E 8751 | Germany |
| Ethanol | Merck | 81870 | Germany |
| Falcon Tube | Extragene | Taiwan | |
| Formaldehyde | Merck | 344198 | Germany |
| Formamide | Merck | 344206 | Germany |
| Homogenizer-sunicator | Microson XL 2000 | USA | |
| Isopropanol | sigma | 19516 | Germany |
| Ketamine hydrochloride | sigma | 1867-66-9 | Germany |
| Laminar Flow Hood | Jal Tajhiz | Iran | |
| Mgnetic stirrer | Labrotechnik | USA | |
| Microcentrifuge | Eppendorf | Germany | |
| Micropipette Tips | Extragene | Taiwan | |
| MOPS | sigma | 85022106 | Germany |
| Na AC | Merck | 567422 | Germany |
| NaOH | Merck | 109137 | Germany |
| Oven | Teb Zaim | Iran | |
| PH meter | Knick | Germany | |
| RNA Later/RNA stabilization reagent | Qiagen | 76104 | USA |
| Surgical instrument | Agn Thos | German made | |
| Syringes | AvaPezeshk | Iran | |
| TriPure reagent/RNA extraction reagent | Roche | 11667157001 | USA |
| Vortex | Labinco | Netherland | |
| Water bath | Memmert | Germany | |
| zylazine | sigma | 7361-61-7 | Germany |
Referências
- McCarthy, B., Hoyer, B. Identity of DNA and diversity of messenger RNA molecules in normal mouse tissues. Proceedings of the National Academy of Sciences. 52 (4), 915-922 (1964).
- Tan, S. C., Yiap, B. C. DNA, RNA, and protein extraction: the past and the present. BioMed Research International. 2009, (2009).
- Peirson, S. N., Butler, J. N. RNA extraction from mammalian tissues. Circadian Rhythms: Methods and Protocols. , 315-327 (2007).
- Skidmore, A. F., Beebee, T. J. Characterization and use of the potent ribonuclease inhibitor aurintricarboxylic acid for the isolation of RNA from animal tissues. Biochemical Journal. 263 (1), 73-80 (1989).
- Mukhopadhyay, T., Roth, J. A. Isolation of total RNA from tissues or cell lines: visualization in gel. RNA Isolation and Characterization Protocols. , 55-59 (1998).
- Raeymaekers, L. Quantitative PCR: theoretical considerations with practical implications. Analytical Biochemistry. 214 (2), 582-585 (1993).
- Sparmann, G., Jäschke, A., Loehr, M., Liebe, S., Emmrich, J. Tissue homogenization as a key step in extracting RNA from human and rat pancreatic tissue. Biotechniques. 22 (3), 408 (1997).
- Kiba, T., et al. High-quality RNA extraction from rat pancreas for microarray analysis. Pancreas. 35 (1), 98-100 (2007).
- Gill, S. S., Aubin, R. A., Bura, C. A., Curran, I. H., Matula, T. I. Ensuring recovery of intact RNA from rat pancreas. Molecular Biotechnology. 6 (3), 359-362 (1996).
- Hernandez, G. E., Mondala, T. S., Head, S. R. Assessing a novel room temperature RNA storage medium for compatibility in microarray gene expression analysis. Biotechniques. 47 (2), 667 (2009).
- Mullin, A. E., Soukatcheva, G., Verchere, C. B., Chantler, J. K. Application of in situ ductal perfusion to facilitate isolation of high-quality RNA from mouse pancreas. Biotechniques. 40 (5), 617 (2006).
- Li, D., et al. A modified method using TRIzol reagent and liquid nitrogen produces high-quality RNA from rat pancreas. Applied Biochemistry and Biotechnology. 158 (2), 253-261 (2009).
- Griffin, M., Abu-El-Haija, M., Abu-El-Haija, M., Rokhlina, T., Uc, A. A simplified and versatile method for obtaining high quality rna from pancreas. Biotechniques. 52 (5), 332 (2012).
- Jun, E., et al. Method optimization for extracting high-quality RNA from the human pancreas tissue. Translation Oncology. 11 (3), 800-807 (2018).
- Green, M. R., Sambrook, J. J. How to win the battle with RNase. Cold Spring Harbor Protocols. (2), 101857 (2019).
- Li, D. -. S., Yuan, Y. -. H., Tu, H. -. J., Dai, L. -. j. A protocol for islet isolation from mouse pancreas. Nature Protocols. 4 (11), 1649 (2009).
- Armstrong, J. A., Schulz, J. R. J. Agarose gel electrophoresis. Current Protocol: Essential Laboratory Techniques. (1), 1-20 (2008).
- Aranda, P. S., LaJoie, D. M., Jorcyk, C. Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis. 33 (2), 366-369 (2012).
- Sambrook, J., Fritsch, E. F., Maniatis, T. Molecular cloning: a laboratory manual. Cold spring harbor laboratory press. , (1989).
- Potenza, N., et al. Hybridase activity of human ribonuclease-1 revealed by a real-time fluorometric assay. Nucleic Acids Research. 34 (10), 2906-2913 (2006).
- Jackson, D., Lewis, F., Taylor, G., Boylston, A., Quirke, P. Tissue extraction of DNA and RNA and analysis by the polymerase chain reaction. Journal of Clinical Pathology. 43 (6), 499-504 (1990).
- Quesada, I., Tudurí, E., Ripoll, C., Nadal, &. #. 1. 9. 3. ;. Physiology of the pancreatic α-cell and glucagon secretion: role in glucose homeostasis and diabetes. Journal of Endocrinology. 199 (1), 5-19 (2008).
- Quertinmont, E., Nicaise, C., Gustot, T., Deviere, J. Tissue Homogenization with the MagNA Lyser Instrument for Total RNA Extraction Using the TriPure Reagent. Liver (mg). 100 (100), 100 (2004).
- Dastgheib, S., Irajie, C., Assaei, R., Koohpeima, F., Mokarram, P. Optimization of RNA extraction from rat pancreatic tissue. Iranian Journal of Medical Sciences. 39 (3), 282 (2014).