Visualization of SARS-CoV-2 using Immuno RNA-Fluorescence In Situ Hybridization
Summary
Here, we describe a simple method that combines RNA fluorescence in situ hybridization (RNA-FISH) with immunofluorescence to visualize severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA. This protocol may increase understanding of the molecular characteristics of SARS-CoV-2 RNA-host interactions at a single-cell level.
Abstract
This manuscript provides a protocol for in situ hybridization chain reaction (HCR) coupled with immunofluorescence to visualize severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in cell line and three-dimensional (3D) cultures of human airway epithelium. The method allows highly specific and sensitive visualization of viral RNA by relying on HCR initiated by probe localization. Split-initiator probes help amplify the signal by fluorescently labeled amplifiers, resulting in negligible background fluorescence in confocal microscopy. Labeling amplifiers with different fluorescent dyes facilitates the simultaneous recognition of various targets. This, in turn, allows the mapping of the infection in tissues to better understand viral pathogenesis and replication at the single-cell level. Coupling this method with immunofluorescence may facilitate better understanding of host-virus interactions, including alternation of the host epigenome and immune response pathways. Owing to sensitive and specific HCR technology, this protocol can also be used as a diagnostic tool. It is also important to remember that the technique may be modified easily to enable detection of any RNA, including non-coding RNAs and RNA viruses that may emerge in the future.
Introduction
SARS-CoV-2 is a novel human betacoronavirus that emerged at the end of 2019, causing an unprecedented pandemic a few months later. Because the virus is new to science, much of its biology and its impact on host cells remain unknown. Therefore, mapping the virus-cell and -tissue tropism during infection is important if its basic biological characteristics and its effects on the host are to be understood. Several techniques are used to examine virus-host interplay including biochemical, biological, and physical assays. In situ hybridization is a common method that employs labeled complementary DNA, RNA, or modified nucleic acid probes, which localize to specific DNA or RNA sequences in a cell or tissue.
A new RNA fluorescent in situ hybridization (RNA-FISH) method has been developed that incorporates modifications to increase sensitivity by amplifying the signal-to-noise ratio via an HCR1. HCR allows the study of RNA localization at a single-cell level. Owing to its high specificity, sensitivity, and resolution, this method is useful not only for basic science studies, but also for applicatory projects, e.g., diagnostics. Recently, the feasibility of this method was demonstrated for detecting SARS-CoV-2 RNA localized to ciliated cells within fully differentiated 3D human airway epithelium (HAE) cultures2. HAE cultures constitute one of the most advanced tools used to study viral infection in the context of the "natural infection" microenvironment3,4.
Several reports on human coronaviruses (HCoV), including SARS-CoV-2, highlight the importance of epigenetic modifications with respect to HCoV infection and pathophysiology [reviewed in 5], e.g., the methylation pattern of the gene encoding the angiotensin-converting enzyme 2 (ACE-2) receptor6,7. Interestingly, mass-spectrometric screening identified several epigenetic factors that interact with the SARS-CoV-2 proteome8. More specifically, nonstructural protein 5 (NSP5) binds to the epigenetic regulator, histone deacetylase 2, and the catalytically inactive NSP5 (C145A) interacts with tRNA methyltransferase 1 (24). Additionally, NSP16 methyltransferase activity is blocked by the methyltransferase inhibitor, sinefungin9. However, the exact role of these epigenetic factors in COVID-19 remains unclear. Replication of HCoV takes place in the cytoplasm of the infected cell, and triggers inflammatory responses that are regulated by epigenetic modifications10.
For instance, HCoV-229E fine-tunes nuclear factor-kappa B signaling and profoundly reprograms the host cellular chromatin landscape by increasing acetylation of H3K36 and H4K5 in certain regions11. The Middle East respiratory syndrome-related coronavirus infection increases levels of H3K27me3 and depletes H3K4me3 at the promoter regions of subsets of specific interferon-sensitive genes12. Additionally, viral RNA triggers cell immune responses, as demonstrated for flaviviruses13, retroviruses14,15, and coronaviruses16. The epigenetic markers on viral RNA may play a role in recognition by cellular sensors, as shown for m7A methylation of human immunodeficiency virus-1 RNA17. However, questions remain: What is the impact of SARS-CoV-2 RNA on the immune response, and are epigenetic marks involved?
Here, an optimized RNA-FISH method coupled with immunofluorescence analysis of cell lines and 3D tissues (fully differentiated HAE) has been described. Although cytological methods, such as FISH and immunofluorescence, are used widely, this new-generation in situ hybridization method based on HCR has never been used for virus detection (except in a recent publication)2. In general, immunostaining and FISH require the following steps: permeabilization to enable penetration of probes or antibodies; fixation in which cellular material is fixed and preserved; detection in which antibodies or nucleic acid probes are applied; and finally, mounting of the samples for visualization.
Although existing protocols share these general features, they vary markedly with respect to the parameters involved. Here, an optimized, simple, immuno-RNA-FISH protocol has been described to detect SARS-CoV-2 RNA in HAE cultures and Vero cells. The technique comprises the following steps: (1) fixation of cells with paraformaldehyde; (2) permeabilization with detergent or methanol (MeOH); (3) rehydration through a graded series of MeOH solutions (HAE cultures only); (4) detection; (5) amplification using HCR technology to detect SARS-CoV-2 RNA; (6) immunostaining; and (7) imaging under a confocal microscope.
Protocol
1. Buffer preparation
- For 500 mL of 2x PHEM buffer, combine 18.14 g of piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 6.5 g of 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES), 3.8 g of ethylene glycol tetraacetic acid (EGTA), and 0.99 g of magnesium sulfate (MgSO4). Make the volume up to ~400 mL with distilled water (dH2O), stir, and adjust the pH to 7.0 using 10 M potassium hydroxide (KOH) or sodium hydroxide (NaOH). Make the final volume up to 500 mL, and then split into 50 mL aliquots. Store at -20 °C until required.
NOTE: The buffer will not be clear until the pH reaches 7.0. - Prepare a stock solution of 37% w/v paraformaldehyde (PFA). For 50 mL, mix 18.5 g of PFA and 35 mL of dH2O in a glass bottle. Place the bottle on a magnetic stirrer with heating. Add 900 µL of 1 M KOH or NaOH, and stir until the solution becomes clear. Allow to cool and transfer to a 50 mL conical centrifuge tube; top up to 50 mL with dH2O. The solution can be stored at -20 °C until required.
NOTE: Formaldehyde should be handled in a fume hood while wearing protective gloves and a lab coat. - Prepare fixation buffer (3.7% PFA buffered with PHEM). For 50 mL, combine 5 mL of 37% PFA solution, 25 mL of 2x PHEM buffer, and 20 mL of dH2O in a 50 mL conical centrifuge tube. Store at -20 °C until required.
NOTE: After thawing, the buffer can be stored at 4 °C for up to 3 months. - Prepare PBST (0.1% Tween-20 in 1x phosphate-buffered saline [PBS]). For 50 mL, add 50 µL of 100% Tween-20 to 50 mL of 1x PBS and mix well.
NOTE: The solution can be stored at room temperature (RT). - Prepare rehydration buffers by combining MeOH/PBST in ratios of 3:1, 1:1, and 1:3 (final volume of 100 mL of each; see Table 1).
NOTE: MeOH is very toxic and flammable. As it can damage the optic nerve, it should be handled in a fume hood avoiding any open flames. As mixing MeOH and PBST generates an exothermic reaction, combine the solutions on ice. - For 1000 mL of 20x SSC, combine 175.3 g of sodium chloride (NaCl) and 88.2 g of sodium citrate in a beaker, and fill up with 800 mL of dH2O. Stir until dissolved, and adjust the pH to 7.2 using NaOH. Add dH2O to a total volume of 1000 mL, and autoclave or filter through a 0.22 µm filter into an autoclaved bottle.
- For 50 mL of 5x SSCT, combine 12.5 mL of 20x SSC buffer with 37.5 mL of dH2O, and add 50 µL of 100% Tween-20. Mix well.
- For 50 mL of 50% 5x SSCT/50% PBST, combine 25 mL of 5x SSCT with 25 mL of PBST.
- For 50 mL of 2x SSC, combine 5 mL of 10x SSC buffer with 45 mL of dH2O. Mix well.
NOTE: All SSC buffers should be stored at RT in the dark.
2. Target definition, probes, and amplifiers
- Use the tools available on the manufacturer's website to design amplifiers and probes. Ensure that the probes are complementary to the reverse DNA strand of the SARS-CoV-2 N gene (Supplementary Figure 1).
- Determine the best probe set size (a set of 20 probes is sufficient for visualization).
- Set the amplifiers used for target RNA detection.
- Use HCR amplifier B1 labeled with Alexa Fluor 647.
NOTE: An HCR amplifier comprises metastable HCR hairpins h1 and h2. Multiplexed experiments can be designed using this protocol. If this is planned, choose a different HCR amplifier (B1, B2, …) for each target RNA to be imaged within the same sample (e.g., amplifier B1 for target 1, amplifier B2 for target 2, …).
- Use HCR amplifier B1 labeled with Alexa Fluor 647.
3. Cell culture and infection with SARS-CoV-2
- Culture Vero (monkey kidney epithelial cells) cells in Dulbecco's modified Eagle medium containing 5% fetal bovine serum.
- Seed 50,000 cells onto coverslips (No. 1, 15 × 15 mm) in a 12-well plate.
- Prepare fully differentiated HAE cultures as described18 on permeable Transwell insert supports with a membrane (diameter = 6.5 mm) and culture in bronchial epithelial growth medium until fully confluent.
- Virus infection
- Inoculate cells with SARS-CoV-2 at 1000x median tissue culture infectious dose (TCID50) per mL
- Incubate cells for 2 h at 37 °C.
- Wash cells twice with PBS to remove unbound virus.
- Culture cells for 48 h.
4. SARS-CoV-2 RNA-FISH in Vero cells cultured on coverslips
DAY 1
- Fixing and permeabilizing cells
- Fix infected cells with 3.7% w/v PFA solution for 1 h at RT.
- Aspirate the 3.7% PFA solution, and wash the cells twice using 1x PBS.
- Permeabilize the cells with PBST solution for 10 min at RT with agitation.
- Aspirate the PBST, and wash the cells twice with 1x PBS.
- Detection
- Aspirate the 1x PBS solution, and wash the cells twice with 2x SSC at RT.
- Aspirate the 2x SSC solution, and prehybridize the samples by adding at least 300 µL of probe hybridization buffer. Cover the wells containing the cells, and incubate at 37 °C for 30 min.
- Prepare the probe solution by adding 1.2 pmol of probe mixture to the probe hybridization buffer.
- Use 1.2 µL of the 1 µM probe stock to prepare 300 µL of working stock.
- Remove the prehybridization solution, and transfer the coverslips to a humidified chamber.
- Pipette 30-50 µL of probe solution onto parafilm to form individual droplets.
- Incubate the samples overnight (12-18 h) at 37 °C.
DAY 2 - Transfer the coverslips back into a 12-well plate, and remove excess probe solution by washing for 4 x 5 min with 400 µL of probe wash buffer at 37 °C.
NOTE: As an alternative to placing 30-50 µl aliquots onto parafilm and under coverslips for incubation, add 300 µL of probe solution directly to coverslips in a 12-well plate. This procedure is simpler, but requires substantial amounts of reagents. Heat the probe wash buffer to 37 °C before use. Calculate the amount of buffer needed, and transfer it to a 15 mL conical centrifuge tube. - Wash samples for 2 x 5 min with 5x SSCT at RT.
- Replace the 5x SSCT solution with 1x PBS, and store the samples at 4 °C until amplification.
- Amplification
- Remove the 1x PBS solution from the wells, and add at least 300 µL of amplification buffer to each well. Incubate the samples for 30 min at RT.
- Prepare each HCR hairpin (h1 and h2) by snap-cooling the desired volume in separate tubes.
- To prepare 300 µL of amplification solution, use 18 pmol of each hairpin (e.g., for 300 µL, use 6 µL of a 3 µM stock hairpin solution).
- Transfer the hairpin solution into tubes.
- Incubate at 95 °C for 90 s.
- Cool to RT for 30 min in the dark.
- Prepare a hairpin mixture by adding the snap-cooled "h1" and "h2" hairpins to the amplification buffer.
- Place drops of 30-50 µL of hairpin mixture onto parafilm.
- Incubate the samples overnight (12-18 h) in the dark at RT.
DAY 3 - Transfer the coverslips back into the 12 well plate, and remove excess hairpins by washing for 5 x 5 min with 5x SSCT at RT with agitation.
- Aspirate the 5x SSCT buffer, and replace it with 1x PBS.
NOTE: If required, use the RNA-FISH samples in a standard immunofluorescence assay, followed by staining of nuclei (see section 5).
- Nuclear staining and slide mounting
- Aspirate the 1x PBS solution, and replace it with 4′,6-diamidino-2-phenylindole (DAPI, 0.2 µg/mL) in 1x PBS solution.
- Incubate the samples for 10 min at RT in the dark.
- Aspirate the DAPI solution, and wash the cells twice with 1x PBS.
- Place two drops of 10 µL each of mounting medium; ensure that the drops are separated sufficiently to allow two coverslips to be placed on a single slide.
- Remove excess liquid by tapping the coverslips on a clean towel, and then place them in antifade mounting medium with the cells facing down.
- Place the mounted samples on a dry, flat surface in the dark and let them cure.
- Following curing, seal the edges of the coverslips with VALAP sealant or nail polish to prevent the samples from drying out.
5. SARS-CoV-2 RNA-FISH in HAE cultures
DAY 1
- Fixing and permeabilizing the HAE culture
- Aspirate the medium, and fix the infected cells using 3.7% PFA solution for 1 h at RT.
- Aspirate the 3.7% PFA solution, and wash the cells twice with 1x PBS.
- Replace the 3.7% PFA solution with 1x PBS under a Transwell insert.
- Discard the PBS, and dehydrate the samples using 2 x 5 min washes with 100% MeOH prechilled to -20 °C.
- After the second wash, replace the buffer with fresh, chilled MeOH for permeabilization under the Transwell insert. Store overnight at -20 °C.
DAY 2
- Rehydration
Rehydrate the samples through a graded series of MeOH/PBST solutions (each for 5 min) on ice: 75% MeOH/25% PBST, 50% MeOH/50% PBST, 25% MeOH/75% PBTS, and 100% PBST (twice).- Wash the cells for 5 min on ice with 50% 5x SSCT/50% PBST.
- Wash cells for 5 min on ice with 5x SSCT.
- Replace the 5x SSCT buffer with 1x PBS.
- Detection
- Incubate the cells (inside the Transwell insert) for 5 min on ice with 100 µL of probe hybridization buffer. Next, transfer the plate to incubator for 30 min at 37 °C (prehybridization).
NOTE: The probe hybridization buffer must be pre-heated to 37 °C before use. Calculate the required volume: 100 µL is needed for a single Transwell insert. - Prepare the probe solution. As 1 mL of probe solution requires 4 pmol of probe, add 4 µL of 1 µM probe stock solution to 1 mL of probe hybridization buffer, and mix well.
NOTE: For RNA detection, use 100 µL of probe solution per Transwell insert. Leave the probe solution on ice until the end of the prehybridization step. - Remove the prehybridization solution, and add the probe solution.
- Incubate the cells overnight (12-18 h) at 37 °C.
DAY 3 - Remove excess probe by washing for 4 x 15 min with 100 µL of probe wash buffer at 37 °C.
- Wash the samples for 2 x 5 min with 5x SSCT at RT.
- Replace the 5x SSCT with 1x PBS, and store the samples at 4 °C until amplification.
- Incubate the cells (inside the Transwell insert) for 5 min on ice with 100 µL of probe hybridization buffer. Next, transfer the plate to incubator for 30 min at 37 °C (prehybridization).
- Amplification
- Preamplify the samples by incubating them with amplification buffer for 30 min at RT.
- Prepare each hairpin by snap-cooling the desired volumes in separate tubes.
- To prepare 500 µL of amplification solution, use 30 pmol of each hairpin (e.g., for 500 µL, use 10 µL of 3 µM stock hairpin solution).
- Transfer the hairpin solution to the tubes.
- Incubate at 95 °C for 90 s.
- Cool to RT for 30 min in the dark.
- Prepare the hairpin solution by adding all snap-cooled hairpins to 500 µL of amplification buffer at RT.
- Remove the preamplification solution, and add the complete hairpin solution.
- Incubate the samples overnight (12-18 h) at RT in the dark.
- Remove excess hairpins by washing with 5x SSCT at RT as follows: 2 x 5 min, 2 x 15 min, and 1 x 5 min.
- Replace the 5x SSCT solution with 1x PBS, and store at 4 °C for not more than 2-3 days or proceed directly to nuclear staining.
NOTE: If required, use the RNA-FISH samples in a standard immunofluorescence assay, followed by nuclear staining (see section 6).
- Nuclear staining and slide mounting
- Aspirate the 1x PBS solution, and replace it with DAPI solution (0.2 µg/mL) in 1x PBS.
- Incubate the samples for 10 min at RT in the dark.
- Aspirate the DAPI solution, and wash the cells twice with 1x PBS.
- Place the cut-out membrane from the Transwell inserts onto 10 µL of antifade mounting medium with the cells facing up, and add extra mounting medium (5 µL) to the membrane.
- Cover the membranes with coverslips.
- Cure the mounted samples on a dry, flat surface in the dark.
- Following curing, seal the edges of the coverslips with VALAP sealant or nail polish to prevent the samples from drying out.
6. Immunofluorescence analysis of Vero cells and HAE cultures
NOTE: Perform the immunofluorescence assay on day 3 for cell lines or day 4 for HAE cultures. Use a different approach for each model. All differences are indicated clearly.
- Aspirate the 1x PBS solution from the wells.
- Block the samples by incubation with 1% w/v bovine serum albumin in PBST solution for 30 min at 37 °C.
- Prepare primary antibodies by preparing appropriate dilutions in blocking solution, and incubate.
- For Vero cells on coverslips:
- Place drops (30-50 µL) of antibody solution onto parafilm in a humidity chamber.
- Place coverslips onto the antibody drops with the cells facing down.
- For HAE, replace the blocking agent in the inserts with antibody solution (100 µL), and incubate the samples in a humidified chamber.
NOTE: Adjust the time and temperature for each incubation with primary antibody. Typical parameters are 2 h at RT or overnight at 4 °C.
- For Vero cells on coverslips:
- Wash the samples for 3 x 5 min with PBST.
- For cells on coverslips, transfer to a 12-well plate and add PBST solution.
- For HAE cultures, replace the antibody solution with PBST solution.
- Check the light sources and filters available for the confocal microscope.
- Choose secondary antibodies according to the host species. Choose the fluorophore parameters (excitation and emission wavelengths, spectrum width, and excitation efficiency according to the available light source).
NOTE: Spectral parameters can be modeled using online tools (see Table of Materials). - Prepare appropriate secondary antibody solutions by diluting them with blocking solution.
- Incubate with secondary antibodies (as in 6.3), but incubate for 1 h at 37 °C.
- Wash the samples as in step 6.4.
- Nuclear staining and slide mounting
- For cells on coverslips
- Aspirate the PBST, and replace it with DAPI (0.2 µg/mL) in 1x PBS solution.
- Incubate the samples for 10 min at RT in the dark.
- Aspirate the DAPI solution, and wash the cells twice with 1x PBS.
- Place the coverslips onto drops of 10 µL of mounting medium with the cells facing down.
- Seal the coverslips with nail polish.
- For HAE cultures
- Aspirate the 1x PBST, and replace it with DAPI (0.2 µg/mL) in 1x PBS solution.
- Incubate the samples for 10 min at RT in the dark.
- Aspirate the DAPI solution, and wash the cells twice with 1x PBS.
- Place the cut-out membranes onto drops of 10 µL of mounting medium with the cells facing up, and add extra (5 µL) mounting medium to the membrane.
- Cover the membranes with coverslips.
- Seal the coverslips with nail polish.
- For cells on coverslips
7. Confocal microscopy
- Define the tracks by specifying the fluorophores used.
- Choose the scanning mode and speed.
- Adjust the laser power, gain, and offset values for each fluorophore by comparing them with respective negative controls: for virus, mock-infected cells; for cellular proteins, samples stained with isotype control antibodies from an appropriate host.
- To acquire a 3D image, activate z-stack mode, and set the top and bottom limits. Set the step size/number.
NOTE: For more details on coronavirus imaging, see19.
Representative Results
The immuno-RNA-FISH protocol described in this manuscript was carried out using two cellular systems: a Vero cell line and a 3D HAE culture. The major steps for both cellular models are shown in Table 2. The RNA-FISH protocol for visualization of SARS-CoV-2 in HAE cultures includes steps that are typical for tissue samples, i.e., permeabilization with 100% MeOH and rehydration through a graded series of MeOH-PBS and 0.1% Tween solutions. Immunofluorescence was performed after RNA-FISH was complete. Zstack images were acquired and processed.
Figure 1 shows immuno RNA FISH in mock-inoculated control Vero cells or cells infected with SARS-CoV-2. Figure 2 shows immuno RNA FISH in mock-inoculated control HAE cultures or cultures infected with SARS-CoV-2. Figure 3 shows optimization of the permeabilization protocol in Vero cells: 70% ethanol overnight at -20 °C or 0.1% Tween-20 in PBS for 5 min at RT. Permeabilization with detergent results in a clear, specific signal for SARS-CoV-2 subgenomic RNA, whereas using ethanol results in a blurry unfocused image.
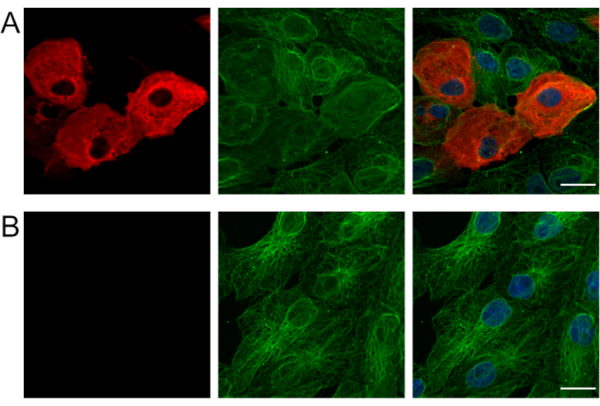
Figure 1: Immuno-RNA-FISH to detect SARS-CoV-2 RNA and β-tubulin in Vero cells. Localization of SARS-CoV-2 subgenomic RNA in (A) infected and (B) mock-inoculated Vero cells. Viral RNA was visualized by FISH (red). β-tubulin is stained with antibodies against mouse β5tubulin (1:100, overnight incubation at 4 °C) and with Alexa fluorophore 488-conjugated secondary antibodies (1:400, 1 h incubation at RT).Nuclei were stained with DAPI (blue). Each image is a single confocal plane. Scale bar = 20 µm. Abbreviations: SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; FISH = fluorescence in situ hybridization; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
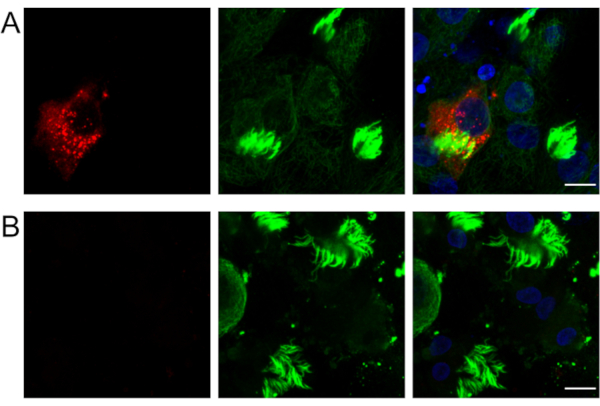
Figure 2: Human airway epithelial cells infected with SARS-CoV-2. Localization of SARS-CoV-2 subgenomic RNA in (A) infected and (B) mock-inoculated HAE cultures. Viral RNA was visualized by FISH (red). Ciliated cells are visualized using antibodies against mouse β5-tubulin (1:100, overnight incubation at 4 °C) and with Alexa fluorophore 488-conjugated secondary antibodies (1:400, 1 h incubation at RT). Nuclei were stained with DAPI (blue). Each image represents a max projection reconstructed from confocal image stacks (thickness = 3 µm). Scale bar = 10 µm. Abbreviations: SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; FISH = fluorescence in situ hybridization; HAE = human airway epithelium; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
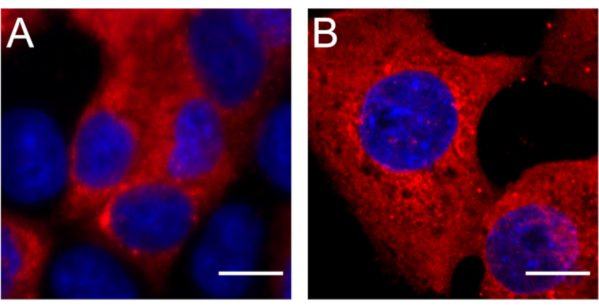
Figure 3: Optimization of permeabilization conditions for Vero cells. Permeabilization of Vero cells with (A) 70% ethanol and (B) with 0.1% Tween-20 in PBS. Permeabilization with detergent results in a clear specific signal for SARS-CoV-2 subgenomic RNA, whereas ethanol results in a blurry image. Viral RNA is shown in red. Nuclei were stained with DAPI (blue). Each image represents a max projection reconstructed from confocal image stacks (thickness = 3 µm). Scale bar = 10 µm. Abbreviations: SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; PBS = phosphate-buffered saline; DAPI = 4′,6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
Supplemental Figure 1: SARS-CoV-2 N gene sequence (5'-3') Please click here to download this file.
| Buffer | Volume of methanol [mL] | Volume of PBST [mL] |
| 75% MeOH/25% PBST | 75 | 25 |
| 50% MeOH/50% PBST | 50 | 50 |
| 25% MeOH/75% PBST | 25 | 75 |
| 100% PBST | 0 | 100 |
| Total | 100 mL | |
Table 1: Preparation of gradient methanol/PBST solutions for rehydration. To rehydrate human airway epithelium samples after overnight incubation in absolute methanol (MeOH), a slow exchange of the environment is necessary. To do this, slow exchange must occur by incubating with buffers in which the proportions of MeOH and PBST (0.1% Tween-20 in 1x phosphate-buffered saline) change gradually. Reagent volumes sufficient to prepare 100 mL of each solution, enough to perform several experiments, are listed.
| Module | Step | Vero cells | HAE cultures |
| RNA Fluorescence in situ hybridization (RNA FISH) | Fixation | (3.7% PFA) 10-40 min at room temperature or overnight at room temperature | |
| Permeabilization | (PBST: 0.1% Tween-20 in 1x PBS) 10 min at room temperature | (0.1% Tween-20 in 1x PBS) 2 × 5 min at room temperature | |
| (100% MeOH) overnight at -20 °C | |||
| Rehydration | (Graded methanol (MeOH)/PBST) 5 x 5 min, 50% 5x SSCT/PBST wash 5 min, 5x SSCT wash 5 min on ice | ||
| Detection (pre-hybridization) | (Probe hybridization buffer) 30 min at 37 °C, 200-300 µL | (Probe hybridization buffer) 5 min on ice, then 30 min at 37 °C, 100 µL | |
| Detection | (Probe solution) 12-18 h at 37 °C, 30 – 50 µL | (Probe solution) 12-18 h at 37 °C, 100 µL | |
| Probe washings | (Probe wash buffer) 4 x 5 min | (Probe wash buffer) 4 x 15 min | |
| (5 × SSCT) 2 x 5 min | |||
| Amplification (pre-amplification) | (Amplification buffer) 30 min at room temperature, 200-300 µL | (Amplification buffer) 30 min at room temperature, 100 µL | |
| Amplification | (Amplifiers solution) 12-18 h at room temperature in dark place, 30-50 µL | (Amplifiers solution) 12-18 h at room temperature in dark place, 100 µL | |
| Amplifiers washing | (5x SSCT) 5 x 5 min | (5x SSCT) 2 x 5 min, 2 x 15 min, 1 x 5 min | |
| ImmunoFluorescence (IF) | Blocking | (1% BSA in PBST) 30 min at 37 °C | |
| Primary antibody incubation | (Antibody solution of apropriate concentration in blocking solution) 2 h at room temperature / overnight at 4 °C, 30-50 µL | (Antibody solution of apropriate concentration in blocking solution) 2 h at room temperature / overnight at 4 °C, 100 µL | |
| Primary antibody washing | (PBST) 3 x 5 min at room temperature | ||
| Secondary antibody incubation | (Antibody solution of apropriate concentration in blocking solution) 1 h at 37 °C, 30-50 µL | (Antibody solution of appropriate concentration in blocking solution) 1 h at 37 °C, 100 µL | |
| Secondary antibody washing | (PBST) 3 x 5 min at room temperature | ||
| Nuclear staining | (DAPI solution) 10 min at room temperature, then 2 x 5 min with 1x PBS | ||
Table 2: Workflow of the Immuno-RNA-FISH protocol in cell lines and HAE cultures. Immuno-RNA-FISH is feasible in both cellular models, but requires different approaches. The main steps are shown, along with the buffers used (in parentheses), followed by the duration and temperature of incubation. In several steps, critical differences in the volume of incubation reagent per sample are given to simplify the calculations. If the volume is not specified, it is selected arbitrarily so that it completely covers the sample (usually 200 µL) with agitation. Abbreviations: FISH = fluorescence in situ hybridization; HAE = human airway epithelium; PFA = paraformaldehyde; DAPI = 4′,6-diamidino-2-phenylindole; BSA = bovine serum albumin; PBS = phosphate-buffered saline; MeOH = methanol.
Discussion
Immuno-RNA-FISH is a reliable method for double-staining of RNA and cellular proteins. Here, a modified immuno-RNA-FISH protocol has been described that allows detection of SARS-CoV-2 RNA and cellular proteins in cell lines and HAE cultures. This protocol can be adapted for use in different cell models including cell monolayers or specific tissues. The method relies on the concept of an HCR initiated by appropriate probe localization. Next, the use of split-initiator probes to begin amplification of the signal by fluorescently labeled amplifiers results in minimal-to-no background fluorescence when observed using a confocal microscope. Amplifiers can be labeled with different fluorescent dyes and are compatible with different probes designed to recognize various targets; therefore, they may be used simultaneously. The procedures described in this protocol are simple, but time-consuming (3-4 days). Nevertheless, the results are characterized by a low noise-to-signal ratio, unlike other protocols that use directly labeled fluorescent probes.
Vero cells and HAE cultures were used here. Different protocols are required for cells on a coverslip and cells in tissue culture. Most of the differences are encountered when handling the cells (whether on coverslip or a membrane) and the amounts of material used. General RNA-FISH protocols require permeabilization using ethanol or methanol solutions as well as an overnight incubation at -20 °C. Importantly, using detergent for permeabilization is more beneficial for immunofluorescence, shortens the procedure by 1 day, and allows more efficient planning of the experiment. The primary approach was to follow general protocols involving permeabilization with alcohol or detergent to see if any undesirable effects were noticeable. Importantly, overnight permeabilization of Vero cells with 70% ethanol solution resulted in an unspecific, blurred signal; by contrast, permeabilization with Tween20 allowed clear and specific visualization of SARS-CoV-2 RNA and shortened the protocol by 1 day (Figure 3).
The same approach was tested on HAE cultures after overnight incubation with absolute methanol at -20 °C (according to general RNA-FISH protocols for tissue samples) and 0.1% Tween20 for 5 min at RT. Incubation with Tween20 resulted in a non-specific signal, which disqualifies this reagent (data not shown). Overnight incubation with methanol led to a highly specific signal with no artifacts. Importantly, detachment of the Transwell membrane was observed because methanol dissolved the glue. This problem was handled by detaching the membrane and proceeding with the coverslip protocol. Classical RNA-FISH procedures use proteinase K to improve sensitivity as this removes proteins and clears RNA-protein complexes, making cells penetrable by chemicals and dyes. The present protocol omitted this step as proteinase K prevents protein staining. No differences were observed in the sensitivity of RNA-FISH when proteinase K was absent (data not shown).
Performing immunofluorescence assays following RNA-FISH did not affect the RNA signal and resulted in successful combination of both methods. Therefore, the complete protocol represents a convenient way of visualizing RNA and its interactions with proteins at the single-cell level. Of note, fixation of cells (required for immuno-RNA-FISH) does not allow time-lapse experiments to examine dynamic events at the single-cell level. Visualization of SARS-CoV-2 RNA allows analysis of SARS-CoV-2 replication within a cell and, when coupled with immunofluorescence, allows the study of intracellular SARS-CoV-2 RNA/host protein interactions including interplay with the epigenome. Finally, this protocol has a wide variety of applications including the detection of SARS-CoV-2 and other emerging viruses at single-cell resolution. Thanks to sensitive and specific HCR technology, it can also be used as a diagnostic tool.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Ministry of Science and Higher Education for research on SARS-CoV-2, and by grants from the National Science Center (grants UMO2017/27/B/NZ6/02488 to K.P. and UMO-2018/30/E/NZ1/00874 to A.K.-P.).
Materials
| Equipment | |||
| Confocal Microscope LSM 880 | ZEISS | ||
| Grant Bio, Mini Rocker- Shaker | Fisher Scientific | 12965501 | |
| Incubator Galaxy170R | New Brunswick | CO170R-230-1000 | |
| Thermomixer Comfort | Eppendorf | 5355 000 011 | |
| Materials | |||
| 15 mm x 15 mm NO. 1 coverslips | LabSolute | 7695022 | |
| 1.5 mL tubes | FL-MEDICAL | 5.350.023.053 | |
| 12-well plate | TTP | 92412 | |
| Conical centrifuge tube | Sarstedt | 5.332.547.254 | |
| parafilm | Sigma | P7793-1EA | |
| serological pipets | VWR Collection | 612-5523P, 612-5827P | |
| slide glass | PTH CHEMLAND | 04-296.202.03 | |
| Transwell ThinCerts | Grainer bio-one | 665641 | |
| Reagents | |||
| Alexa fluorophore 488-conjugated secondary antibodies | Invitrogen | ||
| β5-tubulin | Santa Cruz Biotechnology | sc-134234 | |
| DAPI | Thermo Scientific | D1306 | |
| Disodium phosphate | Sigma | S51136-500G | |
| EGTA | BioShop | EGT101.25 | |
| HCR Amplification Buffer | Molecular Instruments, Inc. | BAM01522 | Buffer can be also prepared doi:10.1242/dev.165753: Supplementary information |
| HCR amplifier B1-h1 Alexa Fluor 647 | Molecular Instruments, Inc. | S013922 | |
| HCR amplifier B1-h2 Alexa Fluor 647 | Molecular Instruments, Inc. | S012522 | |
| HCR Probe Hybridization Buffer | Molecular Instruments, Inc. | BPH03821 | Buffer can be also prepared doi:10.1242/dev.165753: Supplementary information |
| HCR probe set for SARS-CoV-2 Ncapsid | Molecular Instruments, Inc. | PRE134 | |
| HCR Probe Wash Buffer | Molecular Instruments, Inc. | BPW01522 | Buffer can be also prepared doi:10.1242/dev.165753: Supplementary information |
| HEPES | BioShop | HEP001.100 | |
| Magnesium sulfate heptahydrate | Sigma | 63138-250G | |
| Methanol | Sigma | 32213-1L-M | |
| Monopotassium phosphate | Sigma | P5655-100G | |
| Paraformaldehyde | Sigma | P6148-1KG | |
| PIPES | BioShop | PIP666.100 | |
| Potassium Chloride | Sigma | P5405-250G | |
| Prolong Diamond Antifade Mounting Medium | Invitrogen | P36970 | |
| Sodium Chloride | BioShop | SOD001.5 | |
| Trisodium Citrate 2-hydrate | POCH | 6132-04-3 | |
| Tween-20 | BioShop | TWN580.500 | |
| Software | |||
| Fluorescence Spectraviewer | Modeling spectral parameters | ||
| ImageJ Fiji | Acquiring and processing z-stack images |
Referências
- Choi, H. M. T., et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 145, (2018).
- Milewska, A., et al. Replication of severe acute respiratory syndrome coronavirus 2 in human respiratory epithelium. Journal of Virology. 94, (2020).
- Banach, B. S., et al. Human airway epithelial cell culture to identify new respiratory viruses: coronavirus NL63 as a model. Journal of Virological Methods. 156 (1-2), 19-26 (2009).
- Milewska, A., et al. Entry of human coronavirus NL63 into the cell. Journal of Virology. 92, (2018).
- El Baba, R., Herbein, G. Management of epigenomic networks entailed in coronavirus infections and COVID-19. Clinical Epigenetics. 12, 118 (2020).
- Pinto, B. G. G., et al. ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. Journal of Infectious Diseases. 222 (4), 556-563 (2020).
- Verdecchia, P., Cavallini, C., Spanevello, A., Angeli, F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. European Journal of Internal Medicine. 76, 14-20 (2020).
- Gordon, D. E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 583, 459-468 (2020).
- Ferron, F., Decroly, E., Selisko, B., Canard, B. The viral RNA capping machinery as a target for antiviral drugs. Antiviral Research. 96 (1), 21-31 (2012).
- Mehta, S., Jeffrey, K. L. Beyond receptors and signaling: epigenetic factors in the regulation of innate immunity. Immunology and Cell Biology. 93 (3), 233-244 (2015).
- Poppe, M., et al. The NF-kappaB-dependent and -independent transcriptome and chromatin landscapes of human coronavirus 229E-infected cells. PLoS Pathogens. 13, 1006286 (2017).
- Schafer, A., Baric, R. S. Epigenetic landscape during coronavirus infection. Pathogens. 6 (1), 8 (2017).
- Carletti, T., et al. Viral priming of cell intrinsic innate antiviral signaling by the unfolded protein response. Nature Communications. 10, 3889 (2019).
- Heil, F., et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 303 (5663), 1526-1529 (2004).
- Sze, A., et al. Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host and Microbe. 14 (4), 422-434 (2013).
- Kikkert, M. Innate immune evasion by human respiratory RNA viruses. Journal of Innate Immunity. 12, 4-20 (2020).
- Kennedy, E. M., et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host and Microbe. 22 (6), 830 (2017).
- Fulcher, M. L., Gabriel, S., Burns, K. A., Yankaskas, J. R., Randell, S. H. Well-differentiated human airway epithelial cell cultures. Human Cell Culture Protocols in Methods in Molecular Medicine. 107, 183-206 (2005).
- Milewska, A., Owczarek, K., Szczepanski, A., Pyrc, K. Visualizing coronavirus entry into cells. Methods in Molecular Biology. 2203, 241-261 (2020).

