A Nanobar-Supported Lipid Bilayer System for the Study of Membrane Curvature Sensing Proteins in vitro
Summary
Here, a nanobar-supported lipid bilayer system is developed to provide a synthetic membrane with a defined curvature that enables the characterization of proteins with curvature sensing ability in vitro.
Abstract
Membrane curvature plays important roles in various essential processes of cells, such as cell migration, cell division, and vesicle trafficking. It is not only passively generated by cellular activities, but also actively regulates protein interactions and is involved in many intracellular signaling. Thus, it is of great value to examine the role of membrane curvature in regulating the distribution and dynamics of proteins and lipids. Recently, many techniques have been developed to study the relationship between the curved membrane and protein in vitro. Compared to traditional techniques, the newly developed nanobar-supported lipid bilayer (SLB) offers both high-throughput and better accuracy in membrane curvature generation by forming a continuous lipid bilayer on patterned arrays of nanobars with a pre-defined membrane curvature and local flat control. Both the lipid fluidity and protein sensitivity to curved membranes can be quantitatively characterized using fluorescence microscopy imaging. Here, a detailed procedure on how to form a SLB on fabricated glass surfaces containing nanobar arrays and the characterization of curvature-sensitive proteins on such SLB are introduced. In addition, protocols for nanochip reusing and image processing are covered. Beyond the nanobar-SLB, this protocol is readily applicable to all types of nanostructured glass chips for curvature sensing studies.
Introduction
Membrane curvature is a critical physical parameter of a cell that occurs in a variety of cellular processes such as morphogenesis, cell division, and cell migration1. It is widely recognized now that membrane curvature is beyond a simple result of cellular events; instead, it has emerged as an effective regulator of protein interactions and intracellular signaling. For example, several proteins involved in clathrin-mediated endocytosis were found to preferentially bind to the curved membrane, resulting in the formation of a hotspot for endocytosis2. There are many different causes of membrane deformation such as membrane pulling by the cytoskeletal forces, the presence of lipid asymmetry with different sized head groups, the existence of transmembrane proteins with conical shape, the accumulation of membrane-shaping proteins like BAR-domain proteins (named after Bin, amphiphysin, and Rvs proteins), and the insertion of amphipathic helices domain into the membrane1. Interestingly, these proteins and lipids not only deform the membrane but can also sense the membrane curvature and exhibit preferential accumulation1. Therefore, it is crucial to study whether and how membranes with different curvatures alter the distribution and dynamics of proteins and lipids attached to them and the potential impacts on the related intracellular processes.
Many techniques have been developed to analyze the interaction between curved membrane and proteins in both live cell and in vitro systems. The live cell system provides a real cell environment with rich lipid diversity and dynamic protein signaling regulation2,3,4,5,6,7. However, such a sophisticated system is difficult to study due to the uncertainties and fluctuations in the intracellular environment. Hence, the in vitro assays using an artificial membrane composed of known lipid species and purified proteins have become powerful reconstitution systems to characterize the relationship between proteins and curved membranes. Traditionally, liposomes of different diameters are generated by extrusion to detect curvature-sensitive proteins via either a co-sedimentation assay using centrifugal force or a co-flotation assay with a density gradient to avoid protein aggregation8,9. However, the curvature of the extruded liposomes is limited by the available pore size of the membrane filter used in the extruder10. Single liposome curvature (SLiC) assay has been proven to overcome this limitation, in which liposomes with different diameters are fluorescence-labeled and immobilized onto the surface so that the curvature can be marked by the fluorescent intensity11. However, strong variability in lipid composition has been observed in small vesicles, which affects the accuracy of the curvature measurement12. Tether-pulling experiments provide a more accurate measurement of the curvature on the transient tether pulled from giant unilamellar vesicles (GUVs) using an optical tweezer, where the curvature can be well controlled by the membrane tension generated13,14. This method is suitable to study either positive- or negative-curvature sensing proteins, but is constrained by the throughput of tube generation10. Supported membrane tubes (SMrT) assay affords simultaneous generation of multiple membrane tubes that are extruded from the same lipid reservoir by microfluidic flows. Nevertheless, the membrane curvature varies intrinsically along the nanotube, which compromises the accuracy of fluorescence-intensity-based curvature measurement15,16. In comparison, using small unilamellar vesicles (SUVs, diameter <100 nm17) to form a supported lipid bilayer (SLB) on surfaces containing designed topographies generated a single bilayer membrane with curvatures predetermined by nanofabrication or nanomaterials in high accuracy18,19,20.
Here, we present a protocol for the formation of the SLB on fabricated nanochip surfaces with nanobar arrays and how it can be used to probe the curvature sensitivity of proteins in vitro. As shown in Figure 1, there are six essential components of the assay: A) Cleaning and assembly of the chip with a microfluidic chamber; B) Preparation of SUVs with defined lipid composition; C) Formation of the SLB on a nanochip and binding with curvature sensitive proteins; D) Imaging and characterization of the SLB and curvature sensitive proteins under fluorescence microscopy; E) Cleaning the chip for reuse; F) Image processing for quantitative analysis of protein curvature sensing ability. The detailed protocol is described step-by-step below.
Protocol
1. Cleaning of nanochip
- Place the nanochip in a 10 mL beaker with the patterned side facing up.
NOTE: This quartz nanochip has been fabricated via electron beam lithography as described before21. The geometry and arrangement of the nanostructure on the chip can be custom designed. The sizes of the gradient nanobars used here are 2000 nm in length, 600 nm in height, and 100 to 1000 nm in width (100 nm step-set). - Carefully add 1 mL of 98% sulfuric acid to the beaker, and ensure the acid fully covers the front and backside of the chip.
NOTE: 98% sulfuric acid is extremely corrosive and can cause rapid tissue destruction and serious chemical burns upon contact with the skin or eyes. Use it in the fume hood with proper protection. - Slowly rotate the beaker and add 200 µL of 30% hydrogen peroxide drop by drop until the whole beaker becomes hot. Ensure that sulfuric acid and hydrogen peroxide are well mixed to form piranha solution for the removal of organic molecules from the nanochip17. There are alternative techniques for generating the SLB on clean hydrophilic surfaces, such as UV light and ozone exposure as described earlier23.
NOTE: The reaction is extremely exothermic and can cause the solution to boil, so add hydrogen peroxide to the sulfuric acid drop by drop and keep rotating. - Place the beaker in a secondary glass container and keep the nanochip immersed in the piranha solution overnight to clean the impurities thoroughly.
NOTE: Place the beaker in the fume hood without any cover in case the reaction can generate gas. - Take the beaker out and carefully pipette the piranha solution into an acid waste container.
- Load 5 mL of deionized water into the beaker to dilute the residual acid and discard it into the acid waste. Repeat this step five times and use 5 M NaOH to neutralize the acid waste.
- Grab the chip with tweezers and wash with a continuous stream of deionized water to remove residual acid thoroughly. Blow-dry the chip with 99.9% nitrogen gas for SLB formation22.
2. Generation of small unilamellar vesicles (SUVs)
- Add 100 µL of chloroform into an amber vial.
NOTE: Chloroform is a highly volatile, colorless liquid, and it can be toxic if inhaled or swallowed. Use it in the fume hood with the mask and gloves on. - Dissolve 500 µg of the lipid mixture in chloroform. The composition of the lipid mixture used here is 89.5 mol% of egg phosphatidylcholines (PC), 10 mol% of brain phosphatidylserine (PS), and 0.5 mol% of Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (Texas Red DHPE).
- Blow-dry the lipid mixture in the vial with 99.9% nitrogen gas until the chloroform is volatilized and the lipid mixture is dry.
NOTE: This step should be performed in the fume hood. Avoid liquid spatter when blow-drying. - Place the lipid mixture present in the amber vial without the lid in the aluminum foil-covered vacuum desiccator. Then vacuum dry the sample with a pump for 3 h to completely volatilize the chloroform.
- Add 250 µL of phosphate-buffered saline (PBS) buffer to the vial. Vortex the solution until it is homogeneous.
NOTE: PBS buffer consists of 150 mM NaCl, without calcium, magnesium, and phenol red; the pH is adjusted to 7.2 for making the PC and PS lipid bilayer. - Sonicate the lipid mixture for 30 min at a frequency of 50 kHz in a bath sonicator. Then transfer the lipid mixture into a 1.5 mL centrifuge tube and seal with parafilm.
- Freeze the lipid mixture in liquid nitrogen for 20 s and then thaw at 42 °C for 2 min in a water bath. Repeat the freeze-thaw cycles 30 times. After that, the lipid mixture looks like a clear liquid.
CAUTION: Liquid nitrogen has a boiling point of about −195.8 ˚C. It can cause frostbite or cryogenic burns. Use it in the unconfined space with heat insulation gloves on. - Rinse two glass gas-tight syringes and the connector of the mini-extruder with ethanol, chloroform, and methanol respectively. Repeat the rinsing sequence five times to pre-clean the apparatus. Leave them in the fume hood until the organic solvent is completely volatilized.
- Assemble the mini-extruder apparatus and pass 500 µL of PBS buffer through the connector to pre-wet the apparatus, then discard the buffer from another syringe. This step removes impurities and facilitates extrusion.
- Assemble the mini-extruder apparatus with a 100 nm pore-sized polycarbonate filter membrane.
NOTE: The pore size of the filter membrane determines the diameter of liposomes. - Take 500 µL of PBS buffer with one of the syringes and gently push it through the filter to fill the second empty syringe at the other end. Repeat the extrusion back and forth three times to check the apparatus leakage and discard the buffer from the second syringe.
- Pass the lipid mixture through the mini extruder to replace the PBS buffer. Then extrude back and forth 20 times to form SUVs. The multilamellar character of the vesicles may be retained with insufficient number of extrusion times, which will affect SLB formation23.
- Collect the SUVs from the second syringe to reduce the contamination with larger particles and transfer it into a 1.5 mL centrifuge tube.
NOTE: Remove the syringe straight out of the connector in case the syringe breaks off. - Wrap the tube with aluminum foil to prevent bleaching of fluorescent-labeled lipids and store them at 4 ˚C. Usually, the SUVs can be stored for up to 7 days.
3. Formation of the SLB on a nanochip
- Take out the cleaned nanochip from deionized water carefully with a pair of tweezer and blow-dry with 99.9% nitrogen gas.
- Perform surface cleaning of the nanochip with air plasma treatment for 1 h.
- Assemble the nanochip in a polydimethylsiloxane (PDMS) chamber, which consists of two pieces of PDMS-a middle PDMS and a top PDMS (Figure 1A). The middle PDMS is thin (~0.5 mm) with a large oval-shaped opening in the center to ensure sufficient exposure to the nanobar pattern. The top PDMS is thick (~4.0 mm) with two small holes within the large oval opening as an inlet and an outlet for liquid handling.
- Place the chip on a clean surface with the pattern facing up.
- Gently cover the middle PDMS with the chip, and make sure that the whole pattern is exposed to the central area of the large oval-shaped opening in the middle PDMS.
- Cover the top PDMS with the middle PDMS and keep its two small holes within the region of the large oval hole of the middle PDMS. Then the PDMS chamber is assembled.
NOTE: PDMS is the most widely used silicon-based organic polymer. It is biocompatible, transparent, as well as deformable to be designed as a customized shape for chip assembly and imaging under the microscope. More importantly, it can be covalently stuck to another PDMS layer or glass tightly after plasma treatment, making it suitable as the chamber to prepare the SLB. This step ensures that each layer of PDMS is tightly fitted to avoid gaps and leakage.
- Load the SUVs into the PDMS chamber from one of the two small holes in the top PDMS with a pipette and incubate for 15 min at room temperature to form the SLB.
- Gently pipette the PBS buffer into the PDMS chamber from one side of the small hole and remove the waste with a cotton bud from the other hole to wash away the unbound SUVs. Then acquire the SLB formed on the nanochip (the quality of the SLB is tested by fluorescence recovery after photobleaching (FRAP) as shown in step 4.4 and discussed in the representative results section).
NOTE: When pipetting the PBS buffer into the chamber, the pipette tip should be full of the buffer and in contact with the liquid surface to avoid injecting any bubbles.
4. Imaging the SLB and the curvature sensing protein binding on the nanochip
NOTE: This section will depend on the microscope system available for the experiment. Here, an overall guideline on how to perform the imaging will be described. The detailed settings can be changed between the different microscope setups.
- Set up the laser scanning confocal microscopy using a 100x (N.A.1.4) oil objective. Open the ZEN software to select the excitation laser power that can excite the fluorescence of the lipid and protein. Choose Acquisition mode > Smart Setup > Texas Red DHPE / EGFP.
- Adjust the focus with the focus knob to locate the nanobars on the chip until the nanobar edges are sharp under the lipid channel.
- Set the scanning parameters as below to obtain a control image of the lipid channel before adding protein: Frame Size = 512 px x 512 px (512 pixels x 512 pixels, a pixel size of 124 nm), Bits per Pixel = 16 (16-bit depth), Scan speed = 5, and Averaging = 4 (averaging mode of four times/line).
- Conduct the FRAP assay by bleaching the fluorescent-labeled lipid bilayer on a random single nanobar area.
- Select the Experiment Regions and Bleaching checkboxes. Draw a circular area of 5 µm diameter which can include the whole nanobar at the center (nanobar size is 2 µm) and add to the Experiment Regions. Input time-lapse imaging parameters as the following example: choose Scan speed = 9 and Averaging = 1. Choose Time Series > Duration = 100 cycles and Interval = 2 s. Input bleaching parameters as the following example: select the Start after # images checkbox and choose three images.
- Select the laser checkboxes that perform FRAP and change the power to 100.0%. Click Start Experiment for the FRAP experiment.
NOTE: FRAP is a common method to characterize the mobility of cellular molecules. If the SLB has good fluidity, the fluorescence in the bleached area will gradually recover as bleached fluorophores diffuse out and unbleached fluorophores from other areas diffuse in.
- Load the protein solution into the PDMS chamber and incubate for 5 min at room temperature to allow the binding of the protein on the SLB.
NOTE: The proteins used in Figure 2G,H to demonstrate lipid composition facilitate protein binding on nanobar-curved SLBs are 74 µM GFP and 79 µM GFP-His. These two proteins are green fluorescence proteins without or with His-tag. The proteins used for curvature sensing study in Figure 4 and Figure 5 are 16 µM F-BAR, 16 µM IDRFBP17, and 16 µM FBAR+IDRFBP17. These three proteins are the domains from FBP17, which is a typical BAR protein that is intuitively assumed as both curvature generators and sensors. Each protein volume is 20 µL. - Re-focus the nanobars and repeat step 4.3 to take images of both lipid and protein channels for protein curvature sensing detection.
- Repeat step 4.4 to conduct the FRAP assay on both lipid and protein channels and perform time-lapse imaging to characterize the mobility of curvature sensing protein.
5. Reuse of nanochip
- Grab the nanochip with tweezers and carefully detach it from the PDMS chamber in a 50 mL beaker filled with deionized water.
NOTE: This is a critical step. Prevent the tweezers from touching the nanostructure, and do not force the chip to separate to avoid cracks. - Wash the nanochip thoroughly with anhydrous ethanol to remove organic residues attached on the surface.
NOTE: Use clean tissue paper to remove water on the surface before washing with anhydrous ethanol.
CAUTION: Anhydrous ethanol is a highly volatile and slightly hazardous chemical. Avoid contact with the skin and eyes. Use it in the fume hood with the mask and gloves on. - Wash the chip thoroughly with plenty of deionized water to remove residual ethanol. Then blow-dry the chip with 99.9% nitrogen gas.
NOTE: It is important to remove all traces of ethanol on the chip before proceeding to the next step because the piranha acid can react violently with organic solvent and may cause explosions. - Place the chip in a clean 10 mL beaker with the pattern side facing up and clean the chip with piranha solution for reuse which follows the same steps as 'cleaning of nanochip'.
6. Image quantification
- Rotate the images acquired by a microscope so that the nanobar array is in a vertical position for subsequent analysis in Fiji software.
- Use a custom-written MATLAB code 'c_pillarmask_averaging' to locate the individual nanobar by a square mask (51 pixels x 51 pixels) both in the lipid channel and protein channel.
NOTE: This MATLAB code is specially designed for the nanochip. It can be obtained on GitHub via the link: https://github.com/wtzhaolab/GNB_SLB_MX. - Subtract the background signals of each image with the three ROIs (5 pixels x 5 pixels) at the corners of the image by the meshgrid function in MATLAB code 'BarGra_avg'. This operation aims to correct potential uneven background noises caused by the microscope setup or chip leveling on the microscope stage.
- Generate the average images of the same-sized nanobars using MATLAB code 'BarGra_avg', where each point is the mean of the values at that point in all the individual nanobar square masks. Generate 3D surface plots of the average images in Fiji software to show the average signal distribution around the nanobars intuitively. Choose Analyze > Surface Plot > Input 100 for Polygon Multiplier, and select Shade, Draw Axis, and Smooth checkboxes.
- Segment each nanobar into three areas, which include two nanobar-end areas and one nanobar-center area to extract their fluorescence intensities respectively by MATLAB code 'avg_nanobar_quantification'. The sizes of the nanobar ROIs are adjusted according to the dimension of the nanobars using the 'position' file.
- Divide the protein intensities by lipid intensities extracted from the MATLAB code 'avg_nanobar_quantification' at the bar-end area to get the nanobar-end density which excludes the surface area effect.
- Plot the nanobar-end density with different concentrations in GraphPad Prism to get the binding curve. Open the Analyze menu. Choose Nonlinear regression (curve fit) > Binding – Saturation > Specific binding with Hill slope function to fit the curve with the Hill equation and then calculate the KD and Hill coefficient value.
- The lipid and protein intensities are normalized to their corresponding intensities at the 600 nm nanobars center acquired in the same image using the MATLAB code 'avg_nanobar_quantification'.
- Use the brightest nine pixels of normalized protein intensities at the nanobar-end area divided by the bar-center area to quantify the protein curvature sensing with same-sized nanobars, the value is named as "end-to-center ratio". Use the ratio of normalized protein intensity to normalize lipid intensity to quantify the protein curvature sensing range with different-sized nanobars, the value is named as "Normalized Nanobar-End Density". These values are calculated by the MATLAB code 'avg_nanobar_quantification'.
- Load the FRAP stack image in Fiji software. Choose the FRAP area and generate an ROI. Choose the More > Multi measure function to measure the intensity of the FRAP area for each time. Plot the intensity in GraphPad Prism to generate the FRAP recovery curve.
Representative Results
Nanobar design is recommended for probing positive curvature sensing proteins, which contains a half circle at each end with curvature defined by the nanobar width and one flat/zero curvature control locally at the center (Figure 2A,B). Successful formation of the SLB on nanobars results in evenly distributed lipid marker signals across the entire nanobar surface as shown in Figure 2C. Signals from multiple nanobars can be combined by averaging the individual nanobar images (Figure 2D) so that random variations between different nanobars can be minimized. An earlier electron microscopy study showed that cell plasma membrane formed conformal coating around the entire nanostructures24. Therefore, the synthetic lipid bilayer is believed to attach tightly on the nanostructure surface as well, which gives diffraction-limited lines on the 200 nm nanobar as observed in Figure 4E. In addition, the membrane fluidity on the nanobar-curved SLB can also be characterized by FRAP assay, where the fluorescent signal on a single nanobar can be bleached individually for membrane fluidity characterization (Figure 2E,F).
The formation of the SLB on nanobars requires the generation of SUVs similarly to the SLB formation on flat surfaces as reported earlier17. The lipid composition of SUVs determines the surface chemistry of the nanobar-curved SLB that can be altered for membrane labeling and different protein binding mechanisms. For example, Texas Red DHPE can be added in ~0.5 mol% in SUVs so that the enrichment of the membrane surface area on nanobars can be easily imaged by fluorescence microscopy (Figure 2C). Besides visualization, lipid composition can also be changed to facilitate protein binding on nanobar-curved SLBs. For example, to facilitate the anchorage of His-tagged proteins, nickel-nitrilotriacetic acid (Ni-NTA) can be brought to the SLB on the nanobars by incorporating 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl] (nickel salt) (18:1 DGS-NTA(Ni)) into SUVs (~1 mol%)25. Without this functional group on lipids, green fluorescence protein (GFP) cannot bind to the SLB with negatively charged PS, resulting in a darker bar-shaped hole at each nanobar location due to volumetric depletion of GFP signals in the solution (Figure 2G). In comparison, by adding 1 mol% 18:1 DGS-NTA(Ni) in the SLB, the His-tag labeled GFP can strongly anchor on the membrane to clearly follow the SLB shape curved by nanobars (Figure 2H). The lipid diffusion did not show a detectable difference upon protein binding as probed by FRAP assay (Supplementary Figure 1). It is worth noting that the FRAP measurement is based only on the intensity of the 0.5 mol% Texas Red DHPE in the SLB, which may not represent the 1 mol% DGS-NTA(Ni) used for the binding of the His-tag protein. In addition, charged lipids such as PS can enhance protein binding through electrostatic interaction26, while signaling lipids such as phosphatidylinositol (4,5)-bisphosphate (PIP2) can also be integrated in SUVs to facilitate specific protein binding (e.g., FCHo2, for which PIP2 is found essential for its membrane curvature sensing27).
The quality of SLB formation in terms of surface uniformity and membrane fluidity is critical for probing the protein curvature sensing ability on nanobars. There are three typical issues that may compromise SLB uniformity and fluidity. One is the binding of excess SUVs on the SLB due to inefficient washing that generates puncta both on the nanobars and on the flat membrane between the nanobars (Figure 3A,B). It significantly affects the quantification of protein binding on curved or flat surfaces on nanobars for curvature sensing assessment. Another issue is the unexpected introduction of air bubbles inside the PDMS chamber during the washing step, which may destroy the SLB along the trajectory of the air-liquid interface and leave scratch-like features easily identifiable with extremely low lipid signals on the nanobar surface (Figure 3C,D). Moreover, improperly cleaned nanobar substrates leave randomly distributed chemical residues on the surface that prevent lipid adsorption for SLB formation. It leads to the generation of membrane defects that may or may not be above the optical resolution for microscopy visualization (Figure 3E). However, membrane fluidity will be sensitively affected by membrane defect formation28 so that FRAP assay can be used to verify the surface cleanness (Figure 3F,G).
To demonstrate the characterization of curvature sensing protein using nanobar array, the typical F-BAR domain is compared with a recently identified intrinsically disordered region in FBP17 (IDRFBP17). F-BAR is fluorescently labeled with Alexa Fluor 488 tetrafluorophenyl ester dyes while IDRFBP17 is labeled with Alexa Fluor 647 C2 Maleimide. As shown in Figure 4A, both protein domains show increased accumulation on SLB-coated individual nanobars with 300 nm width. Here, the SLB contains 10% PS to electrostatically enhance the protein binding. The protein's curvature sensing ability can be identified by the preferential enrichment at the curved nanobar ends with higher fluorescent intensity than the signals at the flat nanobar centers, which is more obviously shown after image averaging from more than 100 nanobars (Figure 4B). The curvature sensing ability can be quantitatively reflected by calculating the end-to-center ratio on each nanobar (i.e., the fluorescence intensity at the nanobar end versus the nanobar center). As shown in Figure 4C, both proteins have a higher end-to-center ratio than lipids, which is around 1. When comparing the IDRFBP17 and F-BAR, it can be seen that the higher end-to-center ratio of IDRFBP17 (1.385 ± 0.011, mean ± SEM) indicates stronger curvature sensing than F-BAR (1.144 ± 0.004, mean ± SEM).
To examine the range of curvatures that can be sensed by the proteins, the SLB is generated on nanobar arrays with gradient widths from 200 nm to 600 nm (Figure 4D) where lipids showed homogeneous coating on different-sized nanobars (Figure 4E). Three proteins (IDRFBP17, F-BAR, and F-BAR+IDRFBP17) are incubated on the gradient nanobar array, and all of them showed preferential accumulation at the nanobar-end curved membrane sites when the curvature decreased below 400 nm in diameter (Figure 4E,F). Among these three protein domains, IDRFBP17 gives the highest nanobar-end density, indicating its strongest curvature sensing ability, while F-BAR shows the lowest value (Figure 4F). Furthermore, the binding curve of the curvature sensing protein can also be plotted by gradually increasing the protein concentration (Figure 4G), which shows that IDRFBP17 has a strong cooperative curvature sensing ability. When fitting its binding curves to the Hill equation, the KD is found in the µM range (0.6 ± 0.1 µM) and the Hill coefficient (H) is between 2 and 3, suggesting an ultrasensitive binding of IDRFBP17 to nanobar-induced membrane curvatures. The sharper the curvature, the higher the binding capacity at equilibrium.
The dynamic membrane diffusion and membrane association of curvature sensing proteins around the nanobar-shaped SLB sites can also be studied by FRAP. Compared to the fast recovery of lipid signals on nanobars (Figure 5A,E), the F-BAR could not recover within 2 min (Figure 5B,E), suggesting its significantly decreased membrane mobility and association dynamic at the curved membrane sites. Surprisingly, different from the behavior of F-BAR, IDRFBP17 signal showed obvious recovery within the same time frame (Figure 5C,E), indicating the dynamic nature of IDRFBP17 accumulated at the curved membranes. However, after washing away the unbound IDRFBP17 in the solution, the same recovery in the IDRFBP17 channel couldn't be observed as before (Figure 5D,E). It indicates that the recovery is dominated by the exchange with the solution-containing IDRFBP17 molecules and less by lateral diffusion of the membrane-bound molecules.
Overall, these results demonstrate that this nanobar-SLB system is a powerful tool to characterize membrane curvature-sensing proteins in vitro.
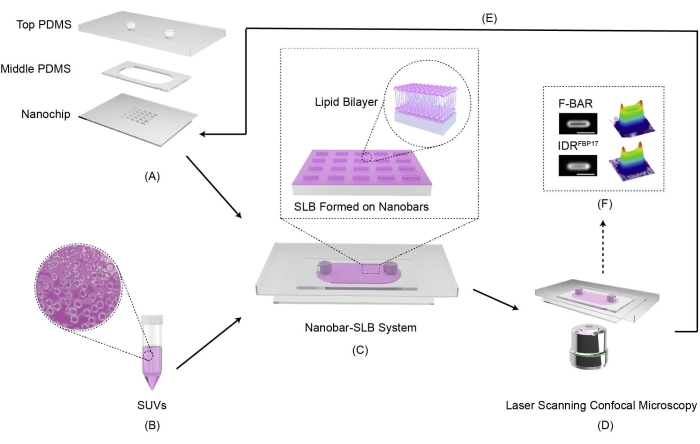
Figure 1: Illustration of the nanobar-SLB system. (A) The PDMS chamber consists of the top and middle layers with the cleaned nanochip assembled on it. (B) The Texas Red DHPE labeled SUVs with zoomed-in detail. (C) The intact model of the nanobar-SLB system and the zoom-in area illustrates that the SLB forms on nanobars with a uniform single layer of the lipid bilayer. (D) Imaging and characterization process under laser scanning confocal microscopy. (E) Cleaning the nanochip for further reuse. (F) Imaging processing for protein curvature sensing quantification. Please click here to view a larger version of this figure.
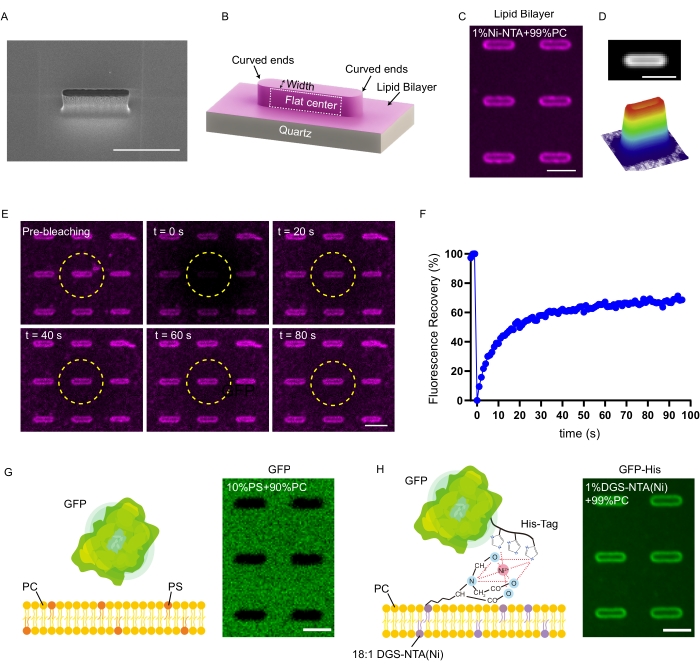
Figure 2: The formation of the curved SLB on nanobars with protein binding. (A) SEM image of an individual nanobar. (B) Schematic depiction of the nanobar-SLB system. (C) Fluorescent images of the SLB labeled with Texas Red DHPE on the nanochip. (D) Average image of SLB distribution on nanobars (top) and a 3D surface plot of the average image (bottom). This figure has been modified from Su et al.22 and reproduced with permission. (E) Time-lapse imaging of the SLB FRAP on the nanobar. The bleaching region is indicated by a yellow dashed circle. (F) Normalized nanobar intensity plot by FRAP measurement. (G) Schematic depiction of GFP cannot bind to the SLB with 10 mol% negative charged PS (left) and a corresponding representative image on the right. (H) Schematic depiction of His-tag labeled GFP binded to the SLB with 1 mol% DGS-NTA(Ni) (left) and a corresponding representative image on the right. Scale bars: 2 µm. Please click here to view a larger version of this figure.
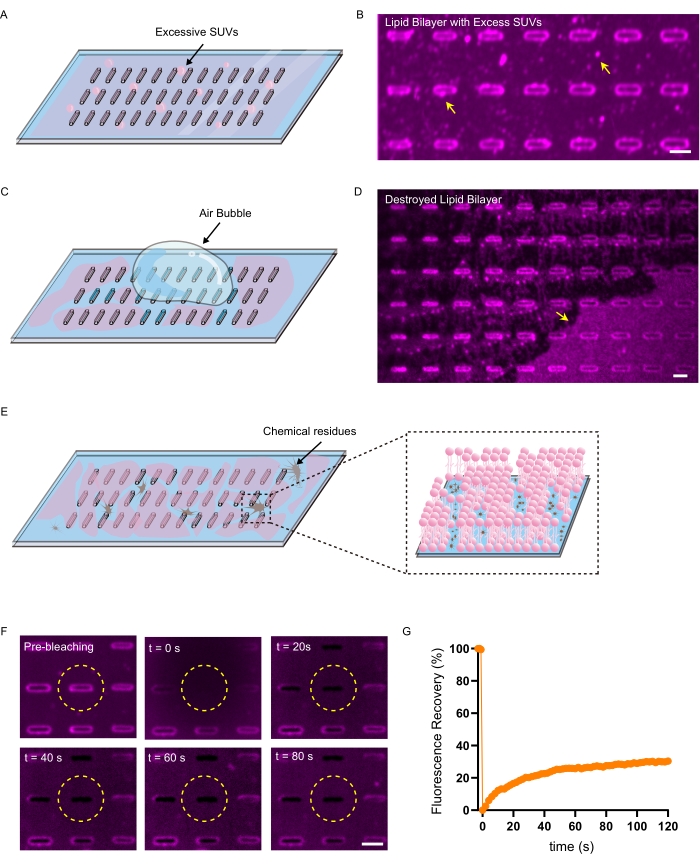
Figure 3: Typical issues that compromise the SLB quality. (A) Schematic of SLB preparation without efficient washing. (B) Representative image of the SLB bind with excess SUVs. The yellow arrows show the puncta which are excess SUVs on the SLB surface. (C) Schematic of SLB preparation with air bubbles injected. (D) Representative image of SLB destroyed by the trajectory of the air-liquid interface. The yellow arrow shows the incomplete SLB on the substrate. (E) Schematic of SLB preparation with uncleaned nanobar substrates. The zoom-in area shows a discontinuous lipid bilayer due to the surface residues prevent lipid adsorption. (F) Time-lapse imaging of SLB FRAP analysis on uncleaned nanobar substrates. The bleaching region is indicated by a yellow dashed circle. (G) Normalized nanobar intensity plot by FRAP measurement. Scale bars: 2 µm. Please click here to view a larger version of this figure.
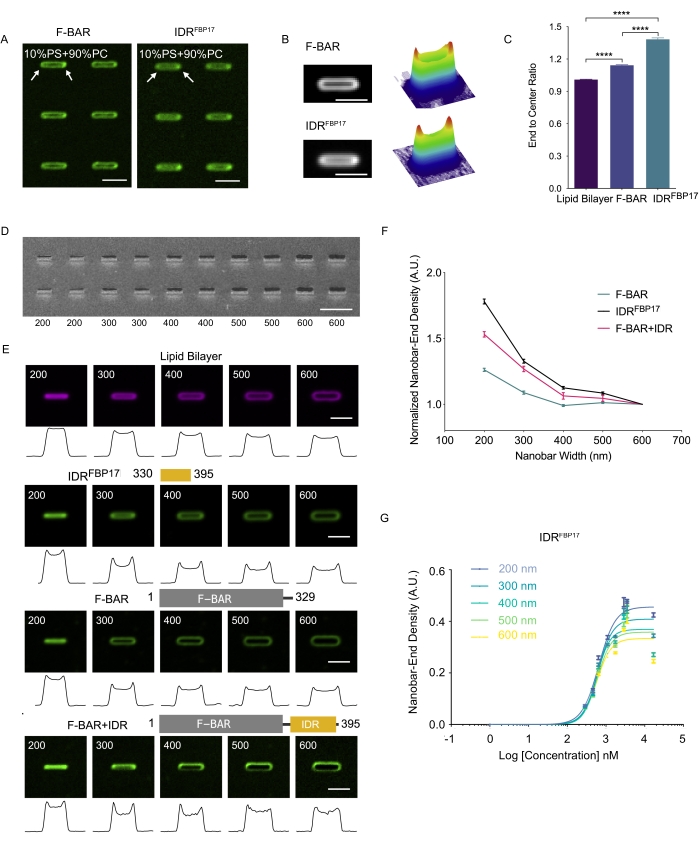
Figure 4: Protein curvature sensing characterization by the nanobar-SLB system. (A) Fluorescent images of F-BAR (left) and IDRFBP17 (right) accumulate on SLB-coated nanobars (10% PS and 90% PC). Scale bar: 2 µm. (B) Average images of F-BAR (left top) and IDRFBP17 (left bottom) accumulate on nanobars and corresponding 3D surface plots (right top and right bottom, respectively). Scale bar: 2 µm. (C) End-to-center ratio of the lipid bilayer, F-BAR and IDRFBP17 fluorescence intensity around the 300 nm width nanobar. A Welch's t-test was performed for statistical analysis. ****p < 0.0001. (n = 3) (D) SEM image of gradient nanobar arrays with widths from 200 nm to 600 nm. Scale bar: 5 µm. (E) Annotation of protein construct (top), average images of lipid bilayer, IDRFBP17, F-BAR, and F-BAR+IDRFBP17 distribution on gradient nanobars with width ranging from 200 nm to 600 nm (middle) and intensity profiles along the nanobars of each average image (bottom). Scale bar: 2 µm. (F) IDRFBP17, F-BAR, and F-BAR+IDRFBP17 normalized nanobar-end density quantified with different widths of nanobars respectively. Each data is shown as the mean ± SEM (n ≥ 60). (G) The binding curve of IDRFBP17 fitted with the Hill equation. Each data is shown as the mean ± SEM (n = 3). This figure has been modified from Su et al.22 and reproduced with permission. Please click here to view a larger version of this figure.
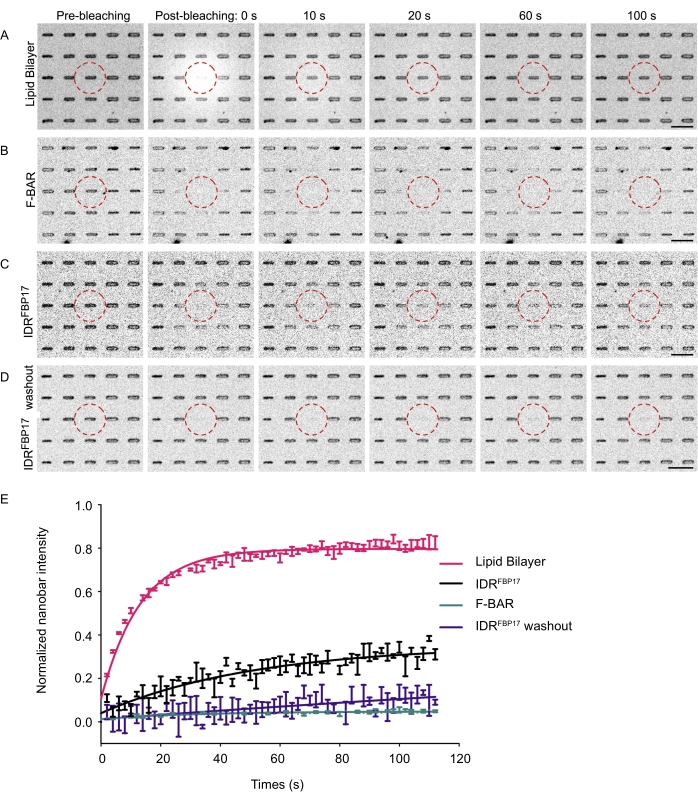
Figure 5: The membrane-binding dynamics of curvature sensing proteins around the nanobar. (A–D) Time-lapse imaging of the lipid bilayer (A), F-BAR (B), IDRFBP17 (C), and IDRFBP17 washed out (D) FRAP analysis on the nanobar. The bleaching region is indicated by a red dashed circle. Scale bar: 5 µm. (E) Normalized nanobar intensity plot by FRAP measurement. This figure has been modified from Su et al.22 and reproduced with permission. Please click here to view a larger version of this figure.
Supplementary Figure 1: Membrane diffusion before and after adding protein. (A) Time-lapse imaging of FRAP test on the POPC lipid bilayer with 1 mol% 18:1 DGS-NTA(Ni) and 0.5 mol% Texas Red DHPE around the nanobar before (top) and after (bottom) adding GFP-His protein. The bleaching region is indicated by a yellow dashed circle. (B) Normalized nanobar intensity plot by FRAP measurement. Scale bars: 2 µm. Please click here to download this File.
Discussion
The nanobar-SLB system described here offers a unique combination of the advantages in several existing in vitro assays. It efficiently reveals the preferential binding of proteins to highly curved membranes as the liposome floatation or sedimentation assay but requires much fewer samples and offers more accurately defined curvature on individual nanobars8,29. It also offers a wide range of precisely controlled curvature for simultaneous comparison as the SLiC assay, with less concern about lipid composition change in different curvatures and undetectable morphological or curvature changes with sizes below the diffraction limit of light, as the SLB is mobile and continuously covers all the sizes of nanobars tightly30. Nanobar-SLB also allows observation of dynamic behaviors of protein on the curved membrane as the tether-pulling assays, but not limited by the throughput of tube generation and experience in optical trapping manipulation13. Although the SMrT assay generates membrane tubes in high throughput, the membrane curvature varies along the nanotube which makes it harder to study the curvature sensitivity of proteins16. More interestingly, compared to other SLB-based systems on the patterned substrate18,19,20,27, the nanobar-SLB provides a flat bar center next to the defined curvature at the bar end, which effectively serves as local control of zero curvature to minimize the impact of surface density variations on quantitative analysis. The ability to customize curvature combinations using high-resolution electron-beam lithography can generate not only local zero curvature controls, but also positive and negative curvatures as demonstrated earlier in live cell studies2.
There are potential limitations worth noting when using this method. Firstly, nanofabrication on the chip is a relatively complex procedure, with cleanroom facilities and specialized equipment needed (in particular, an E-beam writer). This high cost led us to reuse the chip; this results in a longer time spent cleaning the chip surface and assembling the chip with PDMS compared with one-time use. The availability of the nanochip is another type of limitation. Improper operation when using the chip such as facing the surface towards the bench may cause nanostructure attrition, especially for the high-aspect-ratio structures. Moreover, the lipid bilayer formed on the substrate is a continuous membrane; the tension can easily propagate everywhere. For studies that require manipulation of the membrane tension, techniques such as tether pulled from GUVs via the attached micropipette system provide more convenient solutions10.
To obtain optimal results, the following steps are also critical. (i) Vesicle size is critical for SLB formation31,32. Previous literature shows a higher propensity of fusing for smaller sized vesicles (< 90 nm) compared to larger size using a quartz crystal microbalance with dissipation (QCM-D) test32. Thus, compared with LUVs of 300 nm, the diameters of ~50 nm will be easier for SLB formation. Considering this issue, at least 20 times in both freeze-thaw and extrusion steps are needed to form homogeneous SUVs for SLB formation. FRAP test is also necessary in every experiment to validate the membrane mobility. (ii) During SUV preparation, all the equipment should be kept clean to avoid the contamination of dust-like small particles. Especially for the mini-extruder, the needles are easily clogged in a dirty environment. (iii) To prepare SUVs with good quality, the chloroform in the lipid mixture needs to be completely removed to avoid destroying lipid vesicles. (iv) Plasma cleaning of the chip needs to be conducted freshly before SLB formation to ensure a highly hydrophilic surface for efficient SUV breakage to form a continuous bilayer. Long-time exposure in the environment after plasma treatment may cause non-specific binding of dust particles or organic residues to the chip surface, compromising the hydrophilicity of the surface. (v) To assemble the PDMS chamber for SLB formation and protein incubation, the cleanness and flatness of each PDMS piece are important to ensure a good seal of the fluidic channel. Any leakage of the chamber may cause the SLB to dry out or change the protein concentration. (vi) Imaging focus adjustment is critical to ensure consistent intensity measurement, since the height of the nanobar (600 nm) is comparable to the focal depth of the laser scanning confocal microscopy in the z axis33. The focus should be re-adjusted each time the array moves horizontally to maintain the sharpness of the nanobar edges in the images.
Above all, the nanobar-SLB system introduced here provides a powerful method to study the membrane curvature sensing proteins. Various parameters affecting the protein interaction with the curved membrane can be studied quantitatively across a range of curvatures, such as curvature-sensitive domains (e.g., BAR domain and amphiphilic helix) of the protein8,30, lipid composition (e.g., PS and PIP2) of the membrane27,34, pH, ionic strength and temperature of the environment35,36, as well as the protein-protein interaction for complex formation (e.g., CHMP2B and CHMP2A might contribute differently to the ESCRT-III-catalyzed membrane remodeling processes)37,38. Dynamic studies can also be conducted using the nanobar-SLB system to probe membrane binding kinetic11, membrane protein diffusion or assembly39, as well as enzymatic reactions40 or phase separation behavious13,41,42. In addition, the artificial lipid bilayer is generally considered as symmetric with each lipid monolayer containing same lipid composition, which is quite different from the live cell membrane43. Inducing an asymmetry bilayer to better mimic the plasma membrane can be obtained by changing the material of the surface, buffer condition, lipid composition, or temperature44,45,46,47. It is of great value to verify whether an asymmetric bilayer can be formed on the nanobar-SLB system to study protein behavior on the curved membrane, which deserves further studies.
Declarações
The authors have nothing to disclose.
Acknowledgements
We thank Nanyang NanoFabrication Centre (N2FC) and the Centre for Disruptive Photonic Technologies (CDPT) at Nanyang Technological University (NTU) for supporting nanostructure fabrication and SEM imaging, the Protein Production Platform (PPP) at the School of Biological Sciences NTU for protein purification, and the School of Chemical and Biomedical Engineering NTU for the confocal microscope. This work is funded by the Singapore Ministry of Education (MOE) (W. Zhao, RG112/20, RG95/21, and MOE-T2EP30220-0009), the Institute for Digital Molecular Analytics and Science (IDMxS) supported by MOE funding under the Research Centres of Excellence scheme (W. Zhao), the Human Frontier Science Program Foundation (W. Zhao, RGY0088/2021), the NTU Start-up Grant (W. Zhao), School of Chemical and Biomedical Engineering NTU for the research scholarship (X. Miao), and China Scholarship Council for the research scholarship (J. Wu).
Materials
| Anhydrous Ethanol | Sigma-Aldrich | 100983 | |
| Aluminum foil | Diamond | RN0879999FU | |
| Amber Vial | Sigma-Aldrich | 27115-U | |
| Brain PS: L-α-phosphatidylserine (Brain, Porcine) (sodium salt) | Avanti Polar Lipids, Inc. | 840032 | |
| 10 mL Beaker | Schott-Duran | SCOT211060804 | |
| 50 mL Beaker | Schott-Duran | SCOT211061706 | |
| 1000 mL Beaker | Schott-Duran | SCOT211065408 | The second container |
| Chloroform | Sigma-Aldrich | V800117 | |
| Cotton buds | Watsons | ||
| 18:1 DGS-NTA(Ni): 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) | Avanti Polar Lipids, Inc. | 790404 | |
| Egg PC: L-α-phosphatidylcholine (Egg, Chicken) | Avanti Polar Lipids, Inc. | 840051 | |
| F-BAR | Protein Production Plaftorm, School of Biological Sciences, Nanyang Technological University, Singapore | Proteins and peptide | |
| F-BAR+IDR | Protein Production Plaftorm, School of Biological Sciences, Nanyang Technological University, Singapore | Proteins and peptide | |
| GFP | Protein Production Plaftorm, School of Biological Sciences, Nanyang Technological University, Singapore | Proteins and peptide | |
| GFP-His | Protein Production Plaftorm, School of Biological Sciences, Nanyang Technological University, Singapore | Proteins and peptide | |
| GraphPad Prism | GraphPad | V9.0.0 | |
| Hydrogen Peroxide, 30% (Certified ACS) | Thermo Scientific | H325-500 | |
| IDR from human FBP17 | Sangon Biotech (Shanghai) Co., Ltd. | ||
| ImageJ | National Institutes of Health | 1.50d | |
| Laser Scanning Confocal Microscopy | Zeiss | LSM 800 with Airyscan | 100x (N.A.1.4) oil objective. |
| Methanol | Fisher scientific | 10010240 | |
| Mini-extuder | Avanti Polar Lipids, Inc. | 610000-1EA | |
| 1.5 mL Microtubes | Greiner | 616201 | |
| MATLAB | Mathworks | R2018b | |
| Nuclepore Hydrophilic Membrane,0.1 μm | Whatman | 800309 | |
| Phosphate Bufferen Saline (PBS) | Life Technologies Holdings Pte Ltd. | 70013 | |
| Polydimethylsiloxane (PDMS) Base | Dow Corning Corporation | SYLGARD 184 | |
| Polydimethylsiloxane (PDMS) Crosslinker | Dow Corning Corporation | SYLGARD 184 | |
| Plasma Cleaner | HARRICK PLASMA | PDC-002-HP | |
| Quartz Nanochip | Donghai County Alfa Quartz Products CO., LTD | ||
| Sodium Hydroxide | Sigma-Aldrich | 795429 | |
| Sulfuric acid | Sigma-Aldrich | 258105 | |
| Texas Red DHPE: Texas Red 1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine, Triethylammonium Salt | Life Technologies Holdings Pte Ltd. | T1395MP | |
| Tweezer | Gooi | PDC-002-HP | |
| Ultrasonic Cleaners | Elma | D-78224 | |
| Voterx | Scientific Industries | G560E | |
| Vacuum Desiccator | NUCERITE | 5312 | |
| Water Bath | Julabo | TW8 |
Referências
- McMahon, H. T., Gallop, J. L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438 (7068), 590-596 (2005).
- Zhao, W. et al. Nanoscale manipulation of membrane curvature for probing endocytosis in live cells. Nature Nanotechnology. 12 (8), 750-756 (2017).
- Galic, M. et al. External push and internal pull forces recruit curvature-sensing N-BAR domain proteins to the plasma membrane. Nature Cell Biology. 14 (8), 874-881 (2012).
- Rosholm, K. R. et al. Membrane curvature regulates ligand-specific membrane sorting of GPCRs in living cells. Nature Chemical Biology. 13 (7), 724-729 (2017).
- Lou, H. Y. et al. Membrane curvature underlies actin reorganization in response to nanoscale surface topography. Proceedings of the National Academy of Sciences. 116 (46), 23143-23151 (2019).
- Cail, R. C., Shirazinejad, C. R., Drubin, D. G. Induced nanoscale membrane curvature bypasses the essential endocytic function of clathrin. Journal of Cell Biology. 221 (7), e202109013 (2022).
- Mu, H. et al. Patterning of oncogenic ras clustering in live cells using vertically aligned nanostructure arrays. Nano Letter. 22 (3), 1007-1016 (2022).
- Peter, B. J. et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 303 (5657), 495-499 (2004).
- Bigay, J., Casella, J. F., Drin, G., Mesmin, B., Antonny, B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. The EMBO Journal. 24 (13), 2244-2253 (2005).
- Ebrahimkutty, M. P., Galic, M. Receptor-free signaling at curved cellular membranes. Bioessays. 41 (10), e1900068 (2019).
- Bhatia, V. K. et al. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. The EMBO Journal. 28 (21), 3303-3314 (2009).
- Larsen, J., Hatzakis, N. S., Stamou, D. Observation of inhomogeneity in the lipid composition of individual nanoscale liposomes. Journal of the American Chemical Society. 133 (28), 10685-10687 (2011).
- Prevost, C. et al. IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nature Communications. 6, 8529 (2015).
- Simunovic, M. et al. How curvature-generating proteins build scaffolds on membrane nanotubes. Proceedings of the National Academy of Sciences. 113 (40), 11226-11231 (2016).
- Holkar, S. S., Kamerkar, S. C., Pucadyil, T. J. Spatial control of epsin-induced clathrin assembly by membrane curvature. Journal of Biological Chemistry. 290 (23), 14267-14276 (2015).
- Dar, S., Kamerkar, S. C., Pucadyil, T. J. Use of the supported membrane tube assay system for real-time analysis of membrane fission reactions. Nature Protocols. 12 (2), 390-400 (2017).
- Nair, P. M., Salaita, K., Petit, R. S., Groves, J. T. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nature Protocols. 6 (4), 523-539 (2011).
- Lee, I. H., Kai, H., Carlson, L. A., Groves, J. T., Hurley, J. H. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proceedings of the National Academy of Sciences. 112 (52), 15892-15897 (2015).
- Beber, A. et al. Membrane reshaping by micrometric curvature sensitive septin filaments. Nature Communications. 10 (1), 420 (2019).
- Bridges, A. A., Jentzsch, M. S., Oakes, P. W., Occhipinti, P., Gladfelter, A. S. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. Journal of Cell Biology. 213 (1), 23-32 (2016).
- Li, X. et al. A nanostructure platform for live-cell manipulation of membrane curvature. Nature Protocols. 14 (6), 1772-1802 (2019).
- Su, M. et al. Comparative study of curvature sensing mediated by F-BAR and an intrinsically disordered region of FBP17. iScience. 23 (11), 101712 (2020).
- Mayer, L. D., Hope, M. J., Cullis, P. R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochimica et Biophysica Acta. 858 (1), 161-168 (1986).
- Santoro, F. et al. Revealing the cell-material interface with nanometer resolution by focused ion beam/scanning electron microscopy. ACS Nano. 11 (8), 8320-8328 (2017).
- Platt, V. et al. Influence of multivalent nitrilotriacetic acid lipid-ligand affinity on the circulation half-life in mice of a liposome-attached His6-protein. Bioconjugate Chemistry. 21 (5), 892-902 (2010).
- Williams, D., Vicogne, J., Zaitseva, I., McLaughlin, S., Pessin, J. E. Evidence that electrostatic interactions between vesicle-associated membrane protein 2 and acidic phospholipids may modulate the fusion of transport vesicles with the plasma membrane. Molecular Biology of the Cell. 20 (23), 4910-4919 (2009).
- El Alaoui, F. et al. Structural organization and dynamics of FCHo2 docking on membranes. Elife. 11, e73156 (2022).
- Seu, K. J. et al. Effect of surface treatment on diffusion and domain formation in supported lipid bilayers. Biophysical Journal. 92 (7), 2445-2450 (2007).
- Hung, Y. F. et al. Amino terminal region of dengue virus NS4A cytosolic domain binds to highly curved liposomes. Viruses. 7 (7), 4119-4130 (2015).
- Hatzakis, N. S. et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nature Chemical Biology. 5 (11), 835-841 (2009).
- Johnson, J. M., Ha, T., Chu, S., Boxer, S. G. Early steps of supported bilayer formation probed by single vesicle fluorescence assays. Biophysical Journal. 83 (6), 3371-3379 (2002).
- Jing, Y., Trefna, H., Persson, M., Kasemo, B., Svedhem, S. Formation of supported lipid bilayers on silica: relation to lipid phase transition temperature and liposome size. Soft Matter. 10 (1), 187-195 (2014).
- Cole, R. W., Jinadasa, T., Brown, C. M. Measuring and interpreting point spread functions to determine confocal microscope resolution and ensure quality control. Nature Protocols. 6 (12), 1929-1941 (2011).
- Itoh, T. et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Developmental Cell. 9 (6), 791-804 (2005).
- Florentsen, C. D. et al. Annexin A4 trimers are recruited by high membrane curvatures in giant plasma membrane vesicles. Soft Matter. 17 (2), 308-318 (2021).
- Sarkar, Y., Majumder, R., Das, S., Ray, A., Parui, P. P. Detection of curvature-radius-dependent interfacial pH/polarity for amphiphilic self-assemblies: positive versus negative curvature. Langmuir. 34 (21), 6271-6284 (2018).
- Raiborg, C., Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 458 (7237), 445-452 (2009).
- Alqabandi, M. et al. The ESCRT-III isoforms CHMP2A and CHMP2B display different effects on membranes upon polymerization. BMC Biology. 19 (1), 66 (2021).
- Leitenberger, S. M., Reister-Gottfried, E., Seifert, U. Curvature coupling dependence of membrane protein diffusion coefficients. Langmuir. 24 (4), 1254-1261 (2008).
- Bozelli, J. C., Jr. et al. Membrane curvature allosterically regulates the phosphatidylinositol cycle, controlling its rate and acyl-chain composition of its lipid intermediates. Journal of Biological Chemistry. 293 (46), 17780-17791 (2018).
- Parthasarathy, R., Yu, C. H., Groves, J. T. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir. 22 (11), 5095-5099 (2006).
- Yuan, F. et al. Membrane bending by protein phase separation. Proceedings of the National Academy of Sciences. 118 (11), e2017435118 (2021).
- London, E. Membrane structure-function insights from asymmetric lipid vesicles. Accounts of Chemical Research. 52 (8), 2382-2391 (2019).
- Rossetti, F. F., Textor, M., Reviakine, I. Asymmetric distribution of phosphatidyl serine in supported phospholipid bilayers on titanium dioxide. Langmuir. 22 (8), 3467-3473 (2006).
- Richter, R. P., Maury, N., Brisson, A. R. On the effect of the solid support on the interleaflet distribution of lipids in supported lipid bilayers. Langmuir. 21 (1), 299-304 (2005).
- Wacklin, H. P., Thomas, R. K. Spontaneous formation of asymmetric lipid bilayers by adsorption of vesicles. Langmuir. 23 (14), 7644-7651 (2007).
- Lin, W. C., Blanchette, C. D., Ratto, T. V., Longo, M. L. Lipid asymmetry in DLPC/DSPC-supported lipid bilayers: a combined AFM and fluorescence microscopy study. Biophysical Journal. 90 (1), 228-237 (2006).

