Peptide-derived Method to Transport Genes and Proteins Across Cellular and Organellar Barriers in Plants
Summary
Existing methods for the modification of plants have limited applicability. The novel peptide-derived technology proposed here promises both simplicity and efficiency in the introduction of exogenous protein or genes into the desired intracellular compartments of intact plants.
Abstract
The capacity to introduce exogenous proteins and express (or down-regulate) specific genes in plants provides a powerful tool for fundamental research as well as new applications in the field of plant biotechnology. Viable methods that currently exist for protein or gene transfer into plant cells, namely Agrobacterium and microprojectile bombardment, have disadvantages of low transformation frequency, limited host range, or a high cost of equipment and microcarriers. The following protocol outlines a simple and versatile method, which employs rationally-designed peptides as delivery agents for a variety of nucleic acid- and protein-based cargoes into plants. Peptides are selected as tools for development of the system due to their biodegradability, reduced size, diverse and tunable properties as well as the ability to gain intracellular/organellar access. The preparation, characterization and application of optimized formulations for each type of the wide range of delivered cargoes (plasmid DNA, double-stranded DNA or RNA, and protein) are described. Critical steps within the protocol, possible modifications and existing limitations of the method are also discussed.
Introduction
Plant genetic engineering is conventionally used for transferring beneficial traits to plants. In recent years, this technology has been applied to convert plants into bio-factories for the production of pharmaceutically important and commercially valuable proteins, many of which cannot be chemically synthesized and are very costly to produce using animal or microbial systems1. By the introduction of new genes, the plant’s own metabolism can be manipulated for the production of various biopharmaceuticals like antibodies, metabolic enzymes, hormones, antigens or vaccine2.
Established gene transfer technologies for plants are the Agrobacterium-mediated delivery3, bombardment with DNA-coated microprojectiles (biolistics)4, and electroporation5 or polyethylene glycol6 treatment of protoplasts. Techniques requiring protoplasts are generally avoided because they are time-consuming, cumbersome and yield inconsistent results7. As a result, virtually all plant modification work at present utilizes either Agrobacterium or microprojectiles for gene transfer. The Agrobacterium method is more extensively used but not applicable to many economically important plant species. Meanwhile, microprojectile bombardment is more versatile due to a broad range of susceptible plants but requires specialized equipment and often causes severe tissue damage. Furthermore, these methods involve either a random (biolistics) or complex (Agrobacterium) delivery mechanism and have variable transformation rates8. Hence, a novel plant transformation technology that is simple yet effective, and applicable to different plant types is required.
Currently, plants are subjected to genetic modification primarily by the delivery of exogenous DNA encoding a desired trait, rather than by direct delivery of a target protein. The higher stability of DNA over proteins is a prime advantage; nevertheless, potential problems associated with DNA delivery include the random insertion of exogenous DNA into the plant genome and unintended transmission of antibiotic resistance genes to pathogenic bacteria via horizontal gene transfer9. For genome-editing purposes, the ability to edit plant genomes without introducing foreign DNA into cells may circumvent regulatory concerns related to genetically modified plants. Thus, an alternative DNA-free strategy for the modification of plants by direct delivery of protein will be able to cater to these needs.
Here we introduce a peptide-based system, originally developed for human gene therapy10-14, for the targeted delivery of exogenous genes or proteins in intact plants. Peptides are able to protect DNA from nuclease degradation and can mediate gene transfer across cell as well as organellar membranes15-17. They also have diverse and tunable properties besides being non-cytotoxic18-20. More importantly, with the use of peptides, genes can be precisely targeted to intracellular organelles such as the mitochondria21 or plastids (chloroplasts)22 for expression-a task not achievable by biolistic or Agrobacterium-mediated transformation. Depending on the cargo type, this new plant modification technology can be exploited to deliver proteins23 and either express (plasmid21,24 or double-stranded DNA25) or down-regulate (double-stranded RNA26) specific genes within the plant, throughout its cytosolic space24-26 or within a specific organellar compartment21. The designed carrier peptides consist of a cationic domain in the form of either polylysine (K8) or polylysine-co-histidine (KH)9 for binding and/or condensation of negatively-charged cargoes, which is conjugated to cell penetrating (BP100 peptide) or mitochondria transit (Cytcox peptide) sequences.
Protocol
1. Preparation of Peptide-Based Formulations
- Prepare stock solutions of each peptide as follows: (KH)9-BP100 (1 mg/ml or 800 nM), Cytcox-(KH)9 (1 mg/ml), BP100 (1 mg/ml) and (BP100)2K8 (70 µM). Weigh the required amount of each peptide in a 1.5 ml microcentrifuge tube and add autoclaved ultrapure water to dissolve the peptide. Mix well by repeated pipetting until a clear solution is obtained.
- Amplify and purify the plasmid DNA (pDNA), double-stranded DNA (dsDNA) and double-stranded RNA (dsRNA) according to standard molecular methodologies. In 1.5 ml microcentrifuge tubes, make stock solutions with a concentration of 1 mg/ml (pDNA and dsDNA) or 400 nM (dsRNA).
- Prepare the protein stock solution with a concentration of 7 µM, by dissolving 1 mg of protein (e.g., alcohol dehydrogenase, ADH) powder in 1 ml of sodium carbonate solution (0.1 M, pH 9). Label the protein with fluorescent probes such as rhodamine B (RhB) according to standard protocols to enable microscopic visualization of the protein delivered into cells.
- Combine the respective components in a 1.5 ml microcentrifuge tube.
- For peptide-pDNA formulations targeting the cytoplasm, add 6.4 µl of (KH)9-BP100 (1 mg/ml) to 20 µl of pDNA (1 mg/ml) and mix well by pipetting. Allow the mixture to stabilize for 15 min at 25 °C. Add 773.6 µl of autoclaved ultrapure water to dilute the solution to a final volume of 800 µl. Use the pDNA construct designed for nuclear expression (P35S-RLuc-TNOS, Table 1).
- For peptide-pDNA formulations targeting the mitochondria, add 6.6 µl of Cytcox-(KH)9 (1 mg/ml) to 20 µl of pDNA (1 mg/ml) and mix well by pipetting. Allow the mixture to stabilize for 15 min at 25 °C and then add 2.4 µl of BP100 (1 mg/ml). Allow the mixture to stabilize for 15 min at 25 °C for another 15 min. Add 771 µl of autoclaved ultrapure water to dilute the solution to a final volume of 800 µl. Use the pDNA construct designed for mitochondrial expression (pDONR-cox2:rluc, Table 1).
- For peptide-dsDNA formulations, add 5.1 µl of (KH)9-BP100 (1 mg/ml) to 8 µl of dsDNA (1 mg/ml) and mix well by pipetting. Allow the mixture to stabilize for 15 min at 25 °C. Add 786.9 µl of autoclaved ultrapure water to dilute the solution to a final volume of 800 µl.
- For peptide-dsRNA formulations, add 50 µl of (KH)9-BP100 (800 nM) to 50 µl of dsRNA (400 nM) and mix well by pipetting. Allow the mixture to stabilize for 15 min at 25 °C. Add 700 µl of RNase-free water to dilute the solution to a final volume of 800 µl.
- For peptide-protein formulations, add 16 µl of (BP100)2K8 (70 µM) to 16 µl of ADH (7 µM) and mix well by pipetting. Allow the mixture to stabilize for 15 min at 25 °C. Add 768 µl of autoclaved ultrapure water to dilute the solution to a final volume of 800 µl.
- Allow the formulations to stabilize for 15 min at 25 °C.
2. Characterization of Peptide-Based Formulations
- Transfer each solution (800 µl) into a cuvette for dynamic light scattering (DLS) analysis. Determine the hydrodynamic diameter and polydispersity index of formed complexes with a zeta nanosizer, using a 633 nm He-Ne laser at 25 °C with a backscatter detection angle of 173°.
- Following size measurements, transfer each solution (800 µl) into a folded capillary cell for zeta potential measurements at default parameters of dielectric constant, refractive index and viscosity of water at 25 °C.
- Observe a small volume of complex solution, used for DLS analysis, by atomic force microscopy (AFM). Deposit 10 µl of complex solution onto the freshly exposed cleaved surface of a mica sheet and leave the mica to air dry overnight in a covered plastic petri dish.
- Acquire an image of the complexes in air at 25 °C using a silicon cantilever with a spring constant of 1.3 N/m in tapping mode27,28.
3. Infiltration of Plant Leaves
- Use 3 week old soil-grown plants (Arabidopsis thaliana24, Nicotiana benthamiana24 or poplar26). Transfect at least 3 leaves to serve as triplicate for the quantification of gene expression or protein delivery.
- Load a 1 ml needleless plastic syringe with 100 µl of complex solution for the transfection of one leaf. Position the tip of the syringe on the abaxial surface of the leaf.
- Press the syringe tip against the leaf slightly and depress the syringe plunger slowly while exerting a counter-pressure with the index finger of a latex-gloved hand from the opposite side. Successful infiltration can be observed as the spreading of a water-soaked area in the leaf. Label the infiltrated leaves for ease of identification.
- Incubate the transfected leaf for optimized durations following infiltration with peptide-pDNA (12 hr), peptide-dsDNA (12 hr), peptide-dsRNA (9 – 48 hr) and peptide-protein (6 hr) formulations under the following conditions: 16 hr light/8 hr dark at 22 °C for A. thaliana and poplar, or 24 hr constant light at 29 °C for N. benthamiana.
4. Evaluation of Transfection Efficiency
- Excise the whole transfected leaf of smaller plants (e.g., A. thaliana) or a 1 cm2 section of the infiltrated region for larger plants (e.g., N. benthamiana).
- For transfection experiments using Renilla luciferase (Rluc) reporter vector, determine the transfection efficiency quantitatively using an Rluc Assay Kit.
- Place each excised leaf or leaf section in a 1.5 ml microcentrifuge tube. Add 100 µl of 1× Rluc Assay Lysis Buffer per tube. In the same manner, prepare lysates of non-transfected control leaves (triplicate).
- Grind the leaf using a homogenization pestle and incubate the resultant lysate at 25 °C for 6 – 10 hr.
- Centrifuge the lysate at 12,470 × g in a microcentrifuge for 1 min. Transfer 20 µl of the cleared lysate to a well in a 96-well microplate, and use the remaining volume for quantification of total protein concentration using a BCA Protein Assay Kit according to the manufacturer's protocol.
- Add 100 µl of 1× Rluc Assay Substrate (diluted using the Rluc Assay Buffer) into the well and mix by slow pipetting. Place the microplate in a multimode microplate reader and initiate measurement.
- Subtract the background luminescence (mean of the non-transfected triplicate) from each experimental sample's luminescence. Calculate the ratio of photoluminescence (relative light units, RLU) to the amount of protein (mg).
- For transfection experiments using green fluorescent protein (GFP) reporter vector or fluorescently labeled protein (e.g., ADH-RhB), observe the fluorescence using a confocal laser scanning microscope.
- Cut the edges of a whole leaf (to aid the removal of air spaces), while a sectioned leaf can be used as is. Remove the plunger from a 10 ml syringe and place each excised leaf or leaf section in the syringe.
- Replace the plunger and push it gently to the bottom of the syringe without crushing the leaf. Draw water into the syringe until it is approximately half filled.
- Point the syringe upwards and push in the plunger to remove air from the syringe through the tip. Cover the tip of the syringe and pull the plunger down slowly to expel air from the leaf. Repeat this process several times until the leaf appears translucent.
- Cover the surface of a microscope slide with adhesive tape. Cut a square area on the tape large enough to accommodate the leaf sample using a blade, and peel the square piece of tape off with forceps to create a specimen chamber. The tape will serve as a spacer between the slide and coverslip.
- Place the leaf in the chamber with the abaxial surface facing upward and fill the remaining chamber area with water. Seal the leaf within the chamber with a glass coverslip and secure the edges of the coverslip with adhesive tape.
- Examine the leaf sample using the confocal laser scanning microscope under a 40X objective or a 63X water immersion objective. GFP or RhB fluorescence can be visualized at excitation wavelengths of 488 nm or 555 nm, respectively.
Representative Results
An array of nucleic acid and protein cargoes were successfully introduced into various plants using the designed peptides as delivery vectors. Electrostatic interactions between cationic peptides and negatively-charged cargoes resulted in the formation of transfection complexes that can be directly infiltrated into plant leaves using a needleless syringe (Figure 1). Optimized formulations (empirically determined in these studies21,23-26) for the transfection of plant cells are listed in Table 1, where each type of the wide range of delivered cargoes (pDNA, dsDNA, dsRNA and protein) is represented. The mean diameters of all peptide-based formulations were in the approximate range of 150 – 300 nm. Based on DLS analysis, all formulations displayed relatively low size polydispersities, indicating that the formed peptide-cargo aggregates have a uniform size distribution. The morphologies of complexes between peptide and pDNA (Figure 2A) or protein (Figure 2B), on mica, were imaged by AFM. Homogeneous globular complexes were observed for both peptide-pDNA and peptide-protein combinations, in agreement with the data from DLS measurements. In terms of the zeta potential of complexes (Table 1), pDNA- and dsDNA-derived complexes had net negative surface charges while dsRNA-based complexes had a near-neutral surface charge. Peptide-protein complexes, on the other hand, were positively charged.
The efficiencies of peptide-pDNA and peptide-dsDNA formulations in mediating the transfection of A. thaliana or N. benthamiana as model plant systems were evaluated quantitatively as well as qualitatively. The RLuc gene expression assay was employed for quantification of gene expression levels (Table 1), therefore, for this experiment pDNA or dsDNA encoding the RLuc gene must be used for complexation with the respective carrier peptides. Using the (KH)9-BP100/pDNA formulation, nuclear-targeted delivery and expression of pDNA can be achieved, following an incubation period of 12 hr, with an estimated RLU/mg value of approximately 1 × 105. For mitochondrial-targeted delivery and expression of pDNA, a combination of peptides, Cytcox-(KH)9 and BP100, is required for complex formation. With the same optimized incubation period of 12 hr, however, a much lower level of transfection (approximately 1 × 103 RLU/mg) was attained. Meanwhile, similar incubation period (12 hr) and gene expression level (approximately 1 × 103 RLU/mg) was required/recorded for dsDNA-based complexes, also formulated using the (KH)9-BP100 peptide. Qualitative assessments of gene expression were carried out by direct microscopic observation of leaves treated with complexes prepared using pDNA or dsDNA encoding the GFP reporter gene. In cells transfected with non-targeted peptide-pDNA complexes, diffuse green fluorescence corresponding with GFP expression was clearly observed and found to localize in the cytosol (Figure 3A). Distinct differences in the localization pattern of GFP fluorescence were evident in cells infiltrated with mitochondrial-targeted peptide-pDNA complexes. Here, punctate green fluorescence that colocalize with the mitochondrial stain was visible, confirming the specificity of gene expression exclusively in the mitochondrial compartment of cells (Figure 3B).
In the case of peptide-protein formulations, conjugation of the protein cargo (ADH) to a fluorophore (RhB) will enable visualization of the delivered protein in the intracellular compartment. Within a short incubation period of 6 hr, ADH-RhB protein (blue) was found to be distributed throughout the cytosol and vacuole of infiltrated cells (Figure 3C). Meanwhile, rapid and efficient down-regulation of gene expression could be accomplished in various plants using peptide-dsRNA formulations. In the first experiment, A. thaliana leaf was infiltrated with peptide-dsRNA complexes to silence the chalcone synthase gene (CHS) responsible for anthocyanin (red pigment) biosynthesis under drought conditions. The difference in appearance of A. thaliana leaves under normal (Figure 3D, a) and drought conditions (Figure 3D, b) provided an easy means to evaluate CHS silencing using the optimized peptide-dsRNA formulation (arrow 1 indicates the infiltrated region). In the second experiment, peptide-dsRNA complexes were infiltrated into the leaves of transgenic A. thaliana expressing yellow fluorescent protein (YFP). An apparent reduction in YFP expression could be seen in the epidermal cells 9 hr post-transfection (Figure 3E, F). The effectiveness of the formulation in down-regulating gene expression in a different plant system (poplar, 12 hr post-transfection) was also verified (Figure 3G, H).
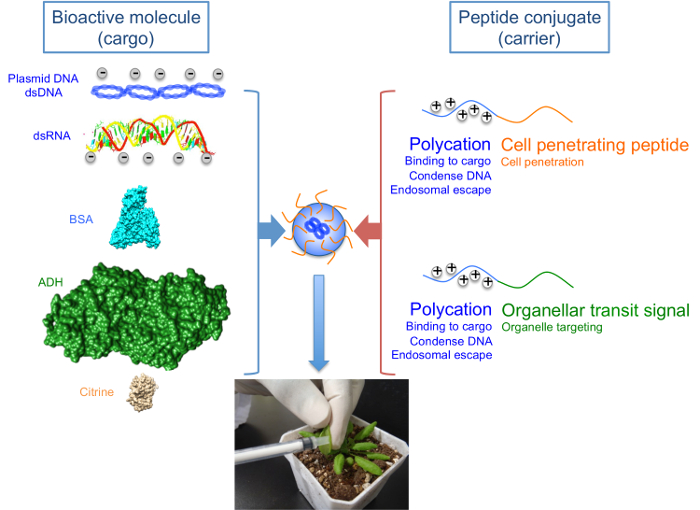
Figure 1: Peptide-based Formulations for the Delivery of Nucleic Acid and Protein Cargoes into Living Plants.
The designed carrier peptides consist of polycations conjugated to cell penetrating or organellar transit sequences. Polycations enable binding and/or condensation of negatively-charged cargoes as well as escape from the endosomal compartment following internalization into cells. Delivery of cargoes into cells and subsequently to specific organelles is mediated by cell penetrating sequences and organellar transit sequences, respectively. Various cargoes that could be successfully delivered into the plant include nucleic acids such as pDNA, dsDNA and dsRNA, as well as model proteins like bovine serum albumin (BSA), alcohol dehydrogenase (ADH) and citrine (a variant of yellow fluorescent protein). Bioactive molecules are able to form transfection complexes with peptide conjugates via electrostatic interactions, which are introduced into plant leaves by syringe infiltration. Please click here to view a larger version of this figure.
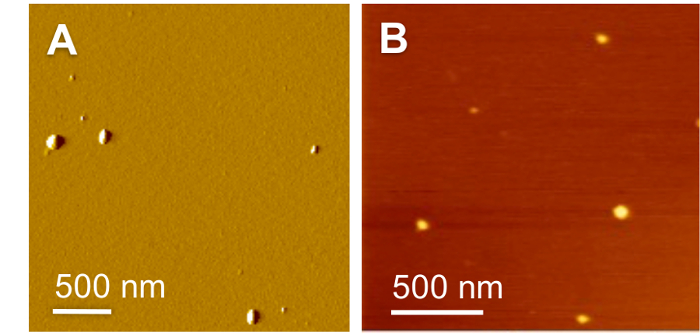
Figure 2: Morphologies of the Peptide-based Formulations.
(A) AFM amplitude image of (KH)9-BP100/pDNA formulation at N/P 0.5. (B) AFM height image of (BP100)2K8/ADH formulation at molar ratio 10. Reproduced with permission from published sources23,24. Please click here to view a larger version of this figure.
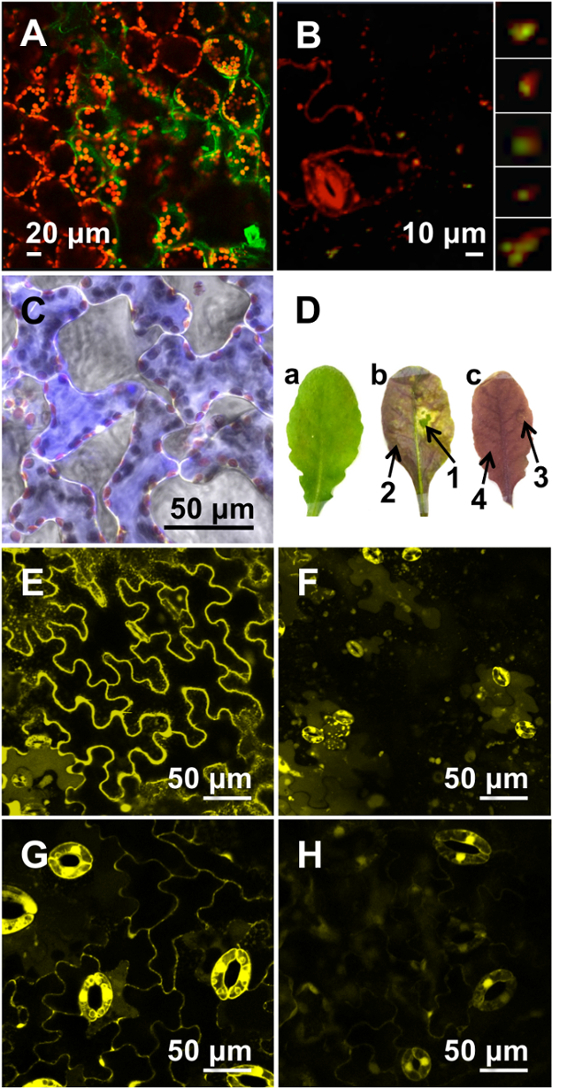
Figure 3: Microscopic Evaluation of Transfection Efficiencies using Optimized Peptide-based Formulations.
(A) Cytosolic GFP expression (green), clearly distinguished from chloroplast autofluorescence (red), was observed in the spongy mesophyll cells of A. thaliana leaves infiltrated with (KH)9-BP100/pDNA formulation (N/P 0.5; 12 hr). (B) GFP expression (green) was detected in the mitochondria (red) in the epidermal cells of A. thaliana leaf infiltrated with Cytcox-(KH)9/BP100/pDNA formulation (N/P 0.5 for each peptide; 12 hr). Enlarged images of mitochondria with GFP expression are shown in the extreme right panel. (C) Delivery of ADH-RhB (blue) into the spongy mesophyll cells of A. thaliana leaves mediated by (BP100)2K8 at a peptide/protein molar ratio of 10, visualized 6 hr post-infiltration. (D) A. thaliana leaf before (a) and after (b) treatment with (KH)9-BP100/dsRNA formulation (molar ratio 2; 48 hr), which resulted in the suppression of anthocyanin biosynthesis pathway. A similar formulation containing GFP5 dsRNA was infiltrated into the leaf as negative control (c). Arrows 1 and 3 indicate the infiltrated area while arrows 2 and 4 indicate the non-infiltrated area within the leaf. (E) A. thaliana leaves expressing yellow fluorescent protein (YFP) and (F) the diminished YFP fluorescence following infiltration with (KH)9-BP100/dsRNA formulation (molar ratio 2; 9 hr). (G) Transgenic poplar leaves expressing yellow fluorescent protein (YFP) and (H) the diminished YFP fluorescence following infiltration with (KH)9-BP100/dsRNA formulation (molar ratio 2; 12 hr). Reproduced with permission from published sources21,23,24,26. Please click here to view a larger version of this figure.
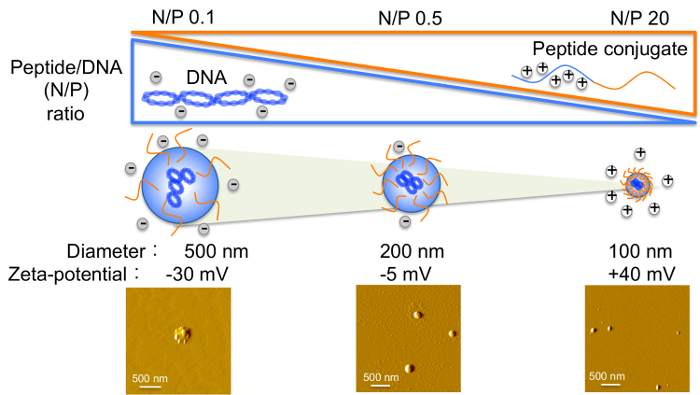
Figure 4: Variation in Peptide-to-DNA (N/P) Ratio and the Effect on Biophysical Properties of Complexes.
With increasing peptide to DNA ratio, peptide-based formulations decrease in size while their zeta-potential values transition from negative to positive. Please click here to view a larger version of this figure.
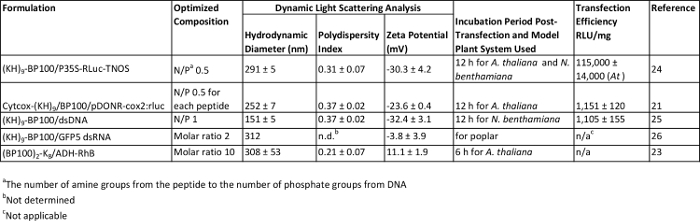
Table 1: Characterization and Evaluation of Various Peptide-based Formulations. Please click here to view a larger version of this table.
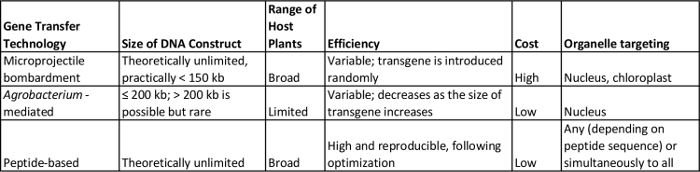
Table 2: A Comparison of Peptide-based and other Existing DNA Delivery Technologies for Intact Plants. Please click here to view a larger version of this table.
Discussion
Critical steps within the protocol that have been identified empirically are discussed. By syringe infiltration, the peptide-based formulations are introduced into the airspaces inside the plant leaf through the stomata. To ensure maximal uptake of the solution, the infiltration process should be carried out when the plants are under conditions that are conducive to stomatal opening i.e., provided with sufficient water and during the light period. With regards to the preparation of transfection complexes, for formulations that involve a combination of two peptides (for mitochondrial-targeted DNA delivery), the sequence for addition of each component is crucial and should not be inverted.
There are many plausible modifications to the procedure. Another option for the infiltration of plant cells is the use of vacuum infiltration, which is able to introduce the complex solution into whole plants and/or partial tissues including apical meristems. For this alternative procedure, seedlings are submerged in the transfection solution, and based on the pressure generated by the vacuum, the peptide-cargo complexes are forced through the stomata and into the plant cells (Yoshizumi, T., unpublished data).
Transient gene expression, based on studies using animal cells, has been shown to increase with higher DNA concentrations29-31. It is important to note that the ratio of peptide to DNA affects the biophysical properties (size, surface charge) of complexes (Figure 4), which influences transfection efficiency; hence, the optimal ratio needs to be maintained even with increased DNA concentration.
One of the main advantages in using peptides as a gene/protein delivery agent is that its sequence is amenable to tuning to fulfill a desired function. For example, the mitochondrial-targeting domain of the carrier peptide described in this study can be replaced with chloroplastic or peroxisomal-targeting sequences for localization to these organelles. While chloroplast transformation is possible in several plants using biolistics32-34, neither the Agrobacterium nor the biolistic method could introduce genes into the mitochondria or other organelles besides the nucleus (Table 2).
There are no rigid host-range limitations for peptide-based transfection, unlike the Agrobacterium-based method (Table 2). So far, formulations for the transfection of A. thaliana, N. benthamiana and poplar have been optimized (Table 1), but the method can also be applied for Nicotiana tabacum, tomato cultivar Micro-Tom, rice (based on preliminary experiments), and other mono- and dicotyledonous plants.
As yet, there is no known limitation in transgene size when peptides are used as transfection vectors. In contrast, large DNA molecules have been found to reduce the transformation efficiency of Agrobacterium-mediated methods35,36, and an upper size limit for transgenes of approximately 200 kb has been reported37-39. Using biolistics, on the other hand, large DNA fragments may be sheared during preparation or delivery into plants38. Although no upper limit has been determined for biolistic transformation so far, physical constrains have been found to restrict the size of DNA that can be transferred to much less than 150 kb40. The major benefits of using the peptide-based system for gene transfer compared to existing approaches employing either Agrobacterium or microprojectile bombardment, discussed above, are summarized in Table 2.
A few limitations exist for this method as it stands. Firstly, targeted delivery of pDNA to specific organelles, such as the mitochondria, has been proven possible by a simple combination of DNA-binding, cell-penetrating and organelle-transit sequences although the efficiency was low. Transgene expression could be detected, by confocal microscopy, only in a small population of mitochondria within cells. Hence, further modifications are necessary to: (i) enhance the translocation of more complexes across the cell/organellar membrane, and (ii) improve the dissociation and transfer of pDNA from the carrier peptide into the target organelle for expression. Secondly, using this DNA delivery system, the transient expression of exogenous reporter genes was successfully achieved in the cytosolic and mitochondrial compartment of cells. Stable incorporation and expression of the introduced genes in the plant nuclear/organellar genome have not been established yet, however, due to the absence of suitable selection strategies.
While acknowledging that there are areas for improvement or further development, the peptide-derived strategy described here remains a simple and versatile technique that has paved the way for the delivery of various cargoes into diverse plant types.
Declarações
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge funding from Japan Science and Technology Agency Exploratory Research for Advanced Technology (JST-ERATO), the New Energy and Industrial Technology Development Organization (NEDO), and the Cross-ministerial Strategic Innovation Promotion Program (SIP), Japan.
Materials
| (KH)9-BP100 peptide | Custom-synthesized by RIKEN Brain Science Institute | N/A | Sequence: KHKHKHKHKHKHKHKH- KHKKLFKKILKYL |
| (BP100)2-K8 peptide | Custom-synthesized by RIKEN Brain Science Institute | N/A | Sequence: KKLFKKILKYLKKLFKKIL- KYLKKKKKKKK |
| BP100 peptide | Custom-synthesized by RIKEN Brain Science Institute | N/A | Sequence: KKLFKKILKYL |
| Cytcox-(KH)9 peptide | Custom-synthesized by RIKEN Brain Science Institute | N/A | Sequence: MLSLRQSIRFFKKHKHKH- KHKHKHKHKHKH |
| P35S-GFP(S65T)-TNOS and P35S-RLuc-TNOS | N/A | N/A | Encodes green fluorescent protein and Renilla luciferase genes, respectively, under the control of CaMV 35S constitutive promoter (Ref: Lakshmanan et al. Biomacromolecules 2013, 14, 10) |
| pDONR-cox2:gfp and pDONR-cox2:rluc | N/A | N/A | Encodes green fluorescent protein and Renilla luciferase genes, respectively, under the control of cox2 mitochondrial-specific promoter (Ref: Chuah et al. Sci Rep 2015, 5, 7751) |
| dsDNA (PCR-amplified from pBI221-P35S-Rluc-TNOS) | N/A | N/A | Encodes green fluorescent protein and Renilla luciferase genes, respectively, under the control of CaMV 35S constitutive promoter (Ref: Lakshmanan et al. Plant Biotechnol 2015, 32, 39) |
| GFP5 siRNA | N/A | N/A | For RNA interference-mediated silencing of green fluorescent protein (Ref: Numata et al. Plant Biotechnol J 2014, 12, 1027) |
| CHS siRNA | N/A | N/A | For RNA interference-mediated silencing of chalcone synthase (Ref: Numata et al. Plant Biotechnol J 2014, 12, 1027) |
| 1 mL and 10 mL Plastic Syringes | TERUMO Corporation | SS-01T, SS-10ESZ | |
| 1.5 mL Microcentrifuge Tube | AS ONE Corporation | 151212 | |
| 96-Well Flat-Bottom Plate | Asahi Glass Co., Ltd. | 3860-096 | |
| Adhesive Tape | Sekisui Chemical Co., Ltd. | No. 835 | |
| Alcohol Dehydrogenase | Sigma-Aldrich Co., LLC. | A-7011 | |
| Atomic Force Microscope | Seiko Instruments Inc. | SPI3800, SPA 300HV | |
| Atomic Force Microscope | Hitachi High-Tech Science Corporation | AFM5300E | |
| BCA Protein Assay Kit | Thermo Fisher Scientific Inc. | 23227 | |
| Cantilever | Hitachi High-Tech Science Corporation | K-A102001593 | |
| Confocal Laser Scanning Microscope | Carl Zeiss | LSM700 | |
| Cork Borer | Sigma-Aldrich Co., LLC. | Z165220 | For excision of leaves into 1-cm diameter disks |
| Coverslip | Matsunami Glass Ind., Ltd. | C02261 | |
| Cuvette | Sarstedt | 759116 | |
| Folded Capillary Cell | Malvern Instruments, Ltd. | DTS1070 | |
| Forceps | Shimizu Akira Inc. | Stainless pincet 150 | |
| Homogenization Pestle | Ieda Trading Corp. | 9993 | |
| Mica | Nisshin EM Co., Ltd. | LC23Z | |
| Microcentrifuge | Beckman Coulter | BKA46472 | |
| Microplate Reader | Molecular Devices Corporation | Spectra MAX M3 | |
| Microscope Slide | Matsunami Glass Ind., Ltd. | S011120 | |
| Pipette | Eppendorf Research® plus | 3120000909 | |
| Pipette Tips | AS ONE Corporation | 2-5138-01, 2-5138-02, 2-5138-03 | |
| Plastic Petri Dish | AS ONE Corporation | 1-7484-01 | |
| Renilla Luciferase Assay Kit | Promega corporation | E2810 | |
| Rhodamine B Isothiocyanate | Sigma-Aldrich Co., LLC. | 283924 | |
| RNase-Free Water | Qiagen | 129112 | |
| Sodium Carbonate | Wako Pure Chemical Industries, Ltd. | 199-01585 | |
| Surgical Blade and Handle | FEATHER Safety Razor Co., Ltd. | Stainless steel No. 14 (blade), No. 3L (handle) | |
| Syringe Tip Cap | Musashi Engineering Inc. | NC-3E | |
| Weighing Balance | Sartorius | CPA225D | |
| Zeta Potentiometer | Malvern Instruments, Ltd. | Zetasizer Nano-ZS |
Referências
- Altman, A., Hasegawa, P. M. . Plant biotechnology and agriculture: Prospects for the 21st century. , (2011).
- Horn, M. E., Woodard, S. L., Howard, J. A. Plant molecular farming: systems and products. Plant Cell Rep. 22, 711-720 (2004).
- Gelvin, S. B. Agrobacterium-mediated plant transformation: the biology behind the "gene-jockeying" tool. Microbiol. Mol. Biol. Rev. 67, 16-37 (2003).
- Klein, T. M., Wolf, E. D., Wu, R., Sanford, J. C. High-velocity microprojectiles for delivering nucleic acids into living cells. Nature. 327, 70-73 (1987).
- Fromm, M., Taylor, L. P., Walbot, V. Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc. Natl. Acad. Sci. U. S. A. 82, 5824-5828 (1985).
- Krens, F. A., Molendijk, L., Wullems, G. J., Schilperoort, R. A. In vitro transformation of plant protoplasts with Ti-plasmid DNA. Nature. 296, 72-74 (1982).
- Birch, R. G. Plant transformation: problems and strategies for practical application. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 297-326 (1997).
- Newell, C. A. Plant transformation technology. Developments and applications. Mol. Biotechnol. 16, 53-65 (2000).
- Nielsen, K. M., Bones, A. M., Smalla, K., van Elsas, J. D. Horizontal gene transfer from transgenic plants to terrestrial bacteria–a rare event?. FEMS Microbiol. Rev. 22, 79-103 (1998).
- Chuah, J. A., Kaplan, D., Numata, K., Cai, W. Chapter 25, Engineering peptide-based carriers for drug and gene delivery. Engineering in Translational Medicine. , 667-689 (2014).
- Nitta, S., Numata, K. Biopolymer-based nanoparticles for drug/gene delivery and tissue engineering. Int. J. Mol. Sci. 14, 1629-1654 (2013).
- Numata, K. Poly(amino acid)s/polypeptides as potential functional and structural materials. Polym. J. 47, 537-545 (2015).
- Numata, K., Kaplan, D. L. Silk-based delivery systems of bioactive molecules. Adv. Drug Deliv. Rev. 62, 1497-1508 (2010).
- Numata, K., Subramanian, B., Currie, H. A., Kaplan, D. L. Bioengineered silk protein-based gene delivery systems. Biomaterials. 30, 5775-5784 (2009).
- Chugh, A., Eudes, F., Shim, Y. S. Cell-penetrating peptides: nanocarrier for macromolecule delivery in living cells. IUBMB Life. 62, 183-193 (2010).
- Eggenberger, K., Mink, C., Wadhwani, P., Ulrich, A. S., Nick, P. Using the peptide Bp100 as a cell-penetrating tool for the chemical engineering of actin filaments within living plant cells. ChemBioChem. 12, 132-137 (2011).
- Numata, K., Kaplan, D. L. Silk-based gene carriers with cell membrane destabilizing peptides. Biomacromolecules. 11, 3189-3195 (2010).
- Numata, K., Hamasaki, J., Subramanian, B., Kaplan, D. L. Gene delivery mediated by recombinant silk proteins containing cationic and cell binding motifs. J. Control. Release. 146, 136-143 (2010).
- Numata, K., Mieszawska-Czajkowska, A. J., Kvenvold, L. A., Kaplan, D. L. Silk-based nanocomplexes with tumor-homing peptides for tumor-specific gene delivery. Macromol. Biosci. 12, 75-82 (2012).
- Numata, K., Reagan, M. R., Goldstein, R. H., Rosenblatt, M., Kaplan, D. L. Spider silk-based gene carriers for tumor cell-specific delivery. Bioconjugate Chem. 22, 1605-1610 (2011).
- Chuah, J. A., Yoshizumi, T., Kodama, Y., Numata, K. Gene introduction into the mitochondria of Arabidopsis thaliana via peptide-based carriers. Sci. Rep. 5, 7751 (2015).
- Numata, K., Yoshizumi, T., Kodama, Y. Plant transformation method. Patent. , (2015).
- Ng, K. K., et al. Intracellular delivery of proteins in intact plants via fusion peptides. PLoS ONE. 11, e0154081 (2016).
- Lakshmanan, M., Kodama, Y., Yoshizumi, T., Sudesh, K., Numata, K. Rapid and efficient gene delivery into plant cells using designed peptide carriers. Biomacromolecules. 14, 10-16 (2012).
- Lakshmanan, M., Yoshizumi, T., Sudesh, K., Kodama, Y., Numata, K. Double-stranded DNA introduction into intact plants using peptide-DNA complexes. Plant Biotechnol. 32, 39-45 (2015).
- Numata, K., Ohtani, M., Yoshizumi, T., Demura, T., Kodama, Y. Local gene silencing in plants via synthetic dsRNA and carrier peptide. Plant Biotechnol. J. 12, 1027-1034 (2014).
- Numata, K., et al. Enzymatic degradation processes of poly[(R)-3-hydroxybutyric acid] and poly[(R)-3-hydroxybutyric acid-co-(R)-3-hydroxyvaleric acid] single crystals revealed by atomic force microscopy: effects of molecular weight and second-monomer composition on erosion rates. Biomacromolecules. 6, 2008-2016 (2005).
- Numata, K., et al. Adsorption of biopolyester depolymerase on silicon wafer and poly[(R)-3-hydroxybutyric acid] single crystal revealed by real-time AFM. Macromol. Biosci. 6, 41-50 (2006).
- Kawai, S., Nishizawa, M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol. Cell. Biol. 4, 1172-1174 (1984).
- Liu, F., Song, Y., Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6, 1258-1266 (1999).
- Luo, D., Saltzman, W. M. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat. Biotechnol. 18, 893-895 (2000).
- Boynton, J. E., et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 240, 1534-1538 (1988).
- Sikdar, R. S., Serino, G., Chaudhuri, S., Maliga, P. Plastid transformation. Arabidopsis thaliana Plant Cell Rep. 18, 20-24 (1998).
- Svab, Z., Hajdukiewicz, P., Maliga, P. Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. U. S. A. 87, 8526-8530 (1990).
- Liu, Y. G., et al. Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. U. S. A. 96, 6535-6540 (1999).
- Park, H. S., Lee, B. M., Salas, G. M., Srivatanakul, M., Smith, H. R. Shorter T-DNA or additional virulence genes improve Agrobactrium-mediated transformation. Theor. Appl. Genet. 101, 1015-1020 (2000).
- Frary, A., Hamilton, C. M. Efficiency and stability of high molecular weight DNA transformation: an analysis in tomato. Transgenic Res. 10, 121-132 (2001).
- Que, Q., et al. Maize transformation technology development for commercial event generation. Front. Plant Sci. 5, 379 (2014).
- Miranda, A., Janssen, G., Hodges, L., Peralta, E. G., Ream, W. Agrobacterium tumefaciens transfers extremely long T-DNAs by a unidirectional mechanism. J. Bacteriol. 174, 2288-2297 (1992).
- Loeb, T. A., Spring, L. M., Steck, T. R., Reynolds, T. L., Bajaj, Y. P. S. Transgenic wheat (Triticum spp.). Transgenic Crops I. , 14-36 (2000).

