激光捕获微分块RNA-测序空间和时间组织特异性基因表达分析
Summary
这里介绍的是植物组织激光捕获微切除(LCM)的协议。LCM 是一种以无污染方式隔离组织区域的微观技术。该过程包括组织固定、石蜡嵌入、分切、LCM和RNA提取。RNA用于下游组织特异性,时间解析的转录组分析。
Abstract
复杂多细胞生物体的发育由具有不同转录特征的不同细胞类型控制。为了确定控制发育过程的转录调控网络,有必要测量这些单个细胞类型的空间和时间基因表达特征。因此,深入了解基因表达的时空控制对于了解生物发育过程如何被调节至关重要。在这里,我们描述了一种激光捕获微分裂(LCM)方法,在发芽过程中从三个大麦胚胎器官中分离出少量细胞,然后进行转录分析。该方法包括组织固定、组织处理、石蜡嵌入、分切、LCM和RNA提取,然后是实时PCR或RNA-seq。这种方法使我们能够从不同数量的细胞(十到数百个)获得种子器官转录组的空间和时间特征,提供比典型的大宗组织分析更大的组织特异性。根据这些数据,我们能够定义和比较转录调控网络,以及预测单个组织的候选调控转录因子。该方法应适用于其他植物组织,优化程度极小。
Introduction
植物的发育和生长涉及存在于复杂细胞环境中的不同细胞内的转录调节网络的协调作用。为了了解这些调控网络的活动,我们需要了解不同细胞类型中不同发育阶段的空间和时间基因表达。然而,由于隔离和分析少量细胞的技术挑战,基因表达分析更常见于整个器官或散装组织样本中。我们在此描述的方法允许通过将 LCM 与 RNA-seq 耦合获得空间和时间组织特异性转录组分析。
LCM是20年前由埃默特-巴克及其同事1开发的。该技术使研究人员能够使用直接的微观可视化和利用窄束激光1将单个细胞或细胞群从环境中精确分离。此后,该方法已广泛应用于癌症生物学和病理学,2、3。2许多植物研究小组还将LCM用于不同的植物种类和不同的组织类型4、5、6、7、8、9、10、11。5,6,7,8,9,10,114最近,几篇论文还利用LCM对欧迪科特和单体种子研究胚胎、内分体和其他种子结构,在种子发育和发芽过程中,10、12、13。,1310,大多数其他常用的单细胞分离方法,如微移液、细胞分选、磁分离和微流体平台,都依赖于酶消化或机械均质来分离细胞。这可能干扰基因表达,引入技术人工制品,混淆数据解释14,15。14,这些方法还要求以前对每种细胞类型的标记基因的了解,以将分离的细胞与其空间位置和真实细胞类型联系起来。另一组技术依赖于基于亲和力的亚细胞结构隔离,而不是整个细胞,例如,INTACT(细胞类型中标记的核分离)和 TRAP(翻译核糖体亲和力纯化)16,17,17。然而,在没有成熟转化协议的植物物种中,核或核糖体的相关性标记和纯化在技术上具有挑战性。LCM利用快速组织固定,以保持转录水平和传统的组织学识别,直接可视化细胞在其正常组织/器官范围内,这允许离散细胞在短时间内分离18,19。,19
此处提出的协议是将特定细胞或细胞类型从谷类种子的组织部分分离的优化方法,可应用于大多数可组织学识别的细胞。LCM提供一种无接触细胞分离方法,大大减少污染,提高回收RNA的完整性。此外,该方法还说明了 LCM 在从少量生物材料开始的大规模基因组广泛研究中的力量。我们还描述了RNA的线性扩增,用于生成足够的输入材料,用于下游转录/转录组分析。
此 LCM RNA-seq 协议中,空间和时间组织特异性转录组有十个主要步骤,包括组织样本固定、脱水、石蜡渗透、嵌入、分切、LCM、RNA 提取、RNA扩增、RNA定量和 qRT-PCR 和/或RNA-seq(图 1)。
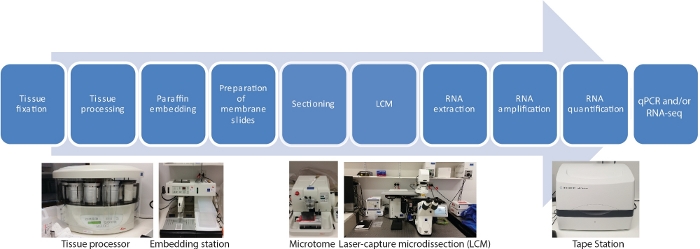
图1:LCM的流程图,后跟RNA-seq或qRT-PCR。LCM 是一种空间精确且无接触的技术,利用微显可视化下的激光束从固定组织部分采集细胞。这个过程从固定组织样品开始,然后使用乙醇和二甲苯的梯度系列进行脱水,最后以石蜡渗透完成。通过使用组织处理器,该过程可以完全自动化。一旦组织与石蜡渗透,它就被嵌入一个模子中,使用嵌入站与熔融石蜡一起嵌入。使用设置为所需厚度的微切进行。在从捕获的细胞中提取RNA之前,立即准备幻灯片并进行LCM。RNA提取后,直接进行两轮RNA扩增,然后进行qRT-PCR和/或RNA-seq。 请单击此处查看此图的较大版本。
Protocol
Representative Results
Discussion
许多组织特异性基因表达研究受到样品手工解剖的有限,这种解剖耗时,劳动密集型,具有很高的污染风险,只能利用人体操作者足够灵巧的样品来收获。LCM 是一种精确且无接触的技术,用于在微观可视化下使用机械操作的激光束从固定组织部分采集细胞。
良好的样品制备对于 LCM 至关重要。这个过程依赖于样品的正确固定和嵌入,以保持组织形态和RNA完整性之间的良好平?…
Declarações
The authors have nothing to disclose.
Acknowledgements
这项工作得到了澳大利亚植物能源生物学研究中心(CE140100008)对JW的支持。M.G.L 得到了拉特罗贝大学启动助学金的支持。我们感谢拉特罗贝基因组学平台在高通量测序和数据分析方面的支持。我们感谢马修·塔克副教授就在我们的实验室中建立LCM提供专家建议。
Materials
| Acetic acid 100 % ACS/R. | AnalaR NORMAPUR (BioStrategies) | VWRC20104.323 | |
| AdhesiveCap 200 opaque | Zeiss | 415190-9181-000 | |
| Clear base moulds 8 X 10 | Leica | 3803015 | |
| Diethyl pyrocarbonate | Sigma-Aldrich | 40718-25ML | |
| High Sensitivity RNA ScreenTape | Agilent | 5067-5579 | |
| Lowprofile disp.blades DB80LS | Leica | 14035843489 | |
| MembraneSlide 1.0 PEN | Zeiss | 415190-9041-000 | |
| MessageAmp II aRNA Amplification Kit | Ambion (ThermoFisher) | AMB17515 | |
| On-Column DNase I Digestion Set | Sigma-Aldrich | DNASE70 | |
| Ovation RNA-Seq System V2 | NuGen (Integrated Science) | 7102-08 | |
| Paraffin (Surgipath Paraplast) | Leica | 39601006 | |
| PicoPure RNA Isolation Kit | ABI (ThermoFisher) | KIT0214 | |
| RNaseZap RNase Decontamination Solution | Ambion (ThermoFisher) | AM9780 | |
| Xylene | AnalaR NORMAPUR (BioStrategies) | VWRC28975.360 | |
| Leica Benchtop Tissue Processor | Leica Biosystems | TP1020 | |
| Leica Heated Paraffin Embedding Module | Leica Biosystems | EG1150H | |
| Leica Cold Plate | Leica Biosystems | EG1150C | |
| Safemate Class 2 Biological Safety Cabinets | LAF Technologies Pty Ltd | Safemate 1.5 | |
| Leica Fully Automated Rotary Microtome | Leica Biosystems | RM2265 | with PALMRobo v 4.6 software |
| Zeiss PALM MicroBeam LCM system | Zeiss miscroscopy | ||
| TapeStation | Agilent | TapeStation 2200 |
Referências
- Emmert-Buck, M. R., et al. Laser capture microdissection. Science. 274 (5289), 998-1001 (1996).
- Alevizos, I., et al. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene. 20 (43), 6196-6204 (2001).
- Cong, P., et al. In situ localization of follicular lymphoma: description and analysis by laser capture microdissection. Blood, The Journal of the American Society of Hematology. 99 (9), 3376-3382 (2002).
- Blokhina, O., et al. Laser capture microdissection protocol for xylem tissues of woody plants. Frontiers in Plant Science. 7, 1965 (2017).
- Casson, S., Spencer, M., Walker, K., Lindsey, K. Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. The Plant Journal. 42 (1), 111-123 (2005).
- Chen, Z., et al. LCM-seq reveals the crucial role of LsSOC1 in heat-promoted bolting of lettuce (Lactuca sativa L.). The Plant Journal. 95 (3), 516-528 (2018).
- Jiao, Y., et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nature Genetics. 41 (2), 258-263 (2009).
- Kivivirta, K., et al. A protocol for laser microdissection (LMD) followed by transcriptome analysis of plant reproductive tissue in phylogenetically distant angiosperms. Plant Methods. 15 (1), 1-11 (2019).
- Li, P., et al. The developmental dynamics of the maize leaf transcriptome. Nature Genetics. 42 (12), 1060-1067 (2010).
- Liew, L. C., et al. Temporal tissue-specific regulation of transcriptomes during barley (Hordeum vulgare) seed germination. The Plant Journal. 101 (3), 700-715 (2020).
- Matas, A. J., et al. Tissue-and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. The Plant Cell. 23 (11), 3893-3910 (2011).
- Sakai, K., et al. Combining laser-assisted microdissection (LAM) and RNA-seq allows to perform a comprehensive transcriptomic analysis of epidermal cells of Arabidopsis embryo. Plant Methods. 14 (1), 10 (2018).
- Zhan, J., et al. RNA Sequencing of Laser-Capture Microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. The Plant Cell. 27 (3), 513-531 (2015).
- Hwang, B., Lee, J. H., Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Experimental & Molecular Medicine. 50 (8), 1-14 (2018).
- Zeb, Q., Wang, C., Shafiq, S., Liu, L. . Single-Cell Omics. , 101-135 (2019).
- Deal, R. B., Henikoff, S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nature Protocols. 6 (1), 56 (2011).
- Heiman, M., Kulicke, R., Fenster, R. J., Greengard, P., Heintz, N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nature Protocols. 9 (6), 1282 (2014).
- Bevilacqua, C., Ducos, B. Laser microdissection: A powerful tool for genomics at cell level. Molecular Aspects of Medicine. 59, 5-27 (2018).
- Nelson, T., Tausta, S. L., Gandotra, N., Liu, T. Laser microdissection of plant tissue: what you see is what you get. Annual Reviews in Plant Biology. 57, 181-201 (2006).
- Day, R. C., Grossniklaus, U., Macknight, R. C. Be more specific! Laser-assisted microdissection of plant cells. Trends in Plant Science. 10 (8), 397-406 (2005).
- Takahashi, H., et al. A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. Journal of Plant Research. 123 (6), 807-813 (2010).
- Schroeder, A., et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology. 7 (1), 3 (2006).
- Ferreira, E. N., et al. Linear mRNA amplification approach for RNAseq from limited amount of RNA. Gene. 564 (2), 220-227 (2015).
- Schneider, J., et al. Systematic analysis of T7 RNA polymerase based in vitro linear RNA amplification for use in microarray experiments. BMC Genomics. 5 (1), 29 (2004).
- Shanker, S., et al. Evaluation of commercially available RNA amplification kits for RNA sequencing using very low input amounts of total RNA. Journal of Biomolecular Techniques. 26 (1), 4 (2015).
- Bhattacherjee, V., et al. Laser capture microdissection of fluorescently labeled embryonic cranial neural crest cells. Genesis. 39 (1), 58-64 (2004).
- Clément-Ziza, M., Munnich, A., Lyonnet, S., Jaubert, F., Besmond, C. Stabilization of RNA during laser capture microdissection by performing experiments under argon atmosphere or using ethanol as a solvent in staining solutions. RNA. 14 (12), 2698-2704 (2008).
- Blokhina, O., et al. Parenchymal Cells Contribute to Lignification of Tracheids in Developing Xylem of Norway Spruce. Plant Physiology. 181 (4), 1552-1572 (2019).
- Schad, M., Lipton, M. S., Giavalisco, P., Smith, R. D., Kehr, J. Evaluation of two-dimensional electrophoresis and liquid chromatography-tandem mass spectrometry for tissue-specific protein profiling of laser-microdissected plant samples. Electrophoresis. 26 (14), 2729-2738 (2005).
- Schad, M., Mungur, R., Fiehn, O., Kehr, J. Metabolic profiling of laser microdissected vascular bundles of Arabidopsis thaliana. Plant Methods. 1 (1), 2 (2005).
- Latrasse, D., et al. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics & Chromatin. 10 (1), 22 (2017).
- Turco, G. M., et al. DNA methylation and gene expression regulation associated with vascularization in Sorghum bicolor. The New Phytologist. 214 (3), 1213-1229 (2017).
- Gomez, S. K., Harrison, M. J. Laser microdissection and its application to analyze gene expression in arbuscular mycorrhizal symbiosis. Pest Management Science: Formerly Pesticide Science. 65 (5), 504-511 (2009).
- Roux, B., et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. The Plant Journal. 77 (6), 817-837 (2014).
- Tang, W., Coughlan, S., Crane, E., Beatty, M., Duvick, J. The application of laser microdissection to in planta gene expression profiling of the maize anthracnose stalk rot fungus Colletotrichum graminicola. Molecular Plant-Microbe Interactions. 19 (11), 1240-1250 (2006).

