Multiomics Analysis of TMEM200A as a Pan-Cancer Biomarker
Summary
Here, a protocol is presented in which multiple bioinformatic tools are combined to study the biological functions of TMEM200A in cancer. In addition, we also experimentally validate the bioinformatics predictions.
Abstract
The transmembrane protein, TMEM200A, is known to be associated with human cancers and immune infiltration. Here, we assessed the function of TMEM200A in common cancers by multiomics analysis and used in vitro cell cultures of gastric cells to verify the results. The expression of TMEM200A in several human cancer types was assessed using the RNA-seq data from the UCSC Xena database. Bioinformatic analysis revealed a potential role of TMEM200A as a diagnostic and prognostic biomarker.
Cultures of normal gastric and cancer cell lines were grown and TMEM200A was knocked down. The expression levels of TMEM200A were measured by using quantitative real-time polymerase chain reaction and western blotting. In vitro loss-of-function studies were then used to determine the roles of TMEM200A in the malignant behavior and tumor formation of gastric cancer (GC) cells. Western blots were used to assess the effect of the knockdown on epithelial-mesenchymal transition (EMT) and PI3K/AKT signaling pathway in GC. Bioinformatic analysis showed that TMEM200A was expressed at high levels in GC.
The proliferation of GC cells was inhibited by TMEM200A knockdown, which also decreased vimentin, N-cadherin, and Snai proteins, and inhibited AKT phosphorylation. The PI3K/AKT signaling pathway also appeared to be involved in TMEM200A-mediated regulation of GC development. The results presented here suggest that TMEM200A regulates the tumor microenvironment by affecting the EMT. TMEM200A may also affect EMT through PI3K/AKT signaling, thus influencing the tumor microenvironment. Therefore, in pan-cancers, especially GC, TMEM200A may be a potential biomarker and oncogene.
Introduction
Cancer has emerged as a persistent public health issue endangering human health globally1due to its high morbidity and mortality rates worldwide, posing a heavy financial and medical burden to society2. Significant advancements in cancer therapy have been achieved in recent years thanks to the discovery of cancer markers3, and researchers have developed novel diagnostic methods and new drugs to treat cancer. However, some patients with cancer still have poor prognoses because of factors such as medication resistance, side effects of drugs, and chemical sensitivity4. Therefore, there is an urgent need for identifying new biomarkers for screening and treating early-stage cancers5.
Membrane proteins are proteins that can bind and integrate into cells and organelle membranes6. These can be grouped into three categories depending on the strength of binding to the membrane and their location: lipid-anchored proteins, integral proteins, and peripheral membrane proteins7,8. A transmembrane (TMEM) protein is an integral membrane protein that consists of at least one transmembrane segment9, which passes either completely or partially through the biological membrane.
Although the mechanisms of action of proteins belonging to the TMEM family are not well understood, these proteins are known to be involved in several types of cancers10. Several TMEM proteins are associated with migratory, proliferative, and invasive phenotypes, and their expression is often associated with a patient’s prognosis11. Therefore, TMEM family members have become the target of research. A comprehensive review of existing reports on TMEM revealed that they are mostly associated with inter- and intracellular signaling12, immune-related diseases, and tumorigenesis10. Many TMEMs also possess important physiological functions, for example, ion channels in the plasma membrane, activation of signal transduction pathways, as well as the mediation of cell chemotaxis, adhesion, apoptosis, and autophagy10. Therefore, we hypothesized that TMEM proteins may be important prognostic markers in the detection and treatment of tumors.
TMEM200A expression is significantly elevated in gastric cancer (GC). Higher expression of TMEM200A13, which has eight exons and a full length of 77.536 kb on chromosome 6q23.1, has been linked to a poor prognosis for overall survival (OS) in cases of GC. Yet the changes in its expression have rarely been reported in oncology studies. This article compares and analyzes the usefulness of TMEM200A as a therapeutic target and tumor diagnostic marker in various cancer studies using different publicly available datasets. We assessed the effectiveness of TMEM200A as a pan-cancer diagnostic and prognostic biomarker as well as its expression levels in various human cancer types using RNA-seq data from the UCSC Xena and TCGA databases, as well as by real-time quantitative polymerase chain reaction (qRT-PCR) and western blotting.
The effect of TMEM200A expression levels on mutation rates, regulatory processes, tumor diagnosis and prognosis, immune infiltration, and immunotherapy was further investigated using a mix of computational tools and dataset websites. CBioPortal and the Catalog of Somatic Mutations in Cancer Cells (COSMIC) databases were used to examine mutations in TMEM200A. Sangerbox and TISIDB websites were utilized to understand how TMEM200A influences immune infiltration. The Tumor Immune Single Cell Center (TISCH) online tool and CancerSEA database were used to investigate the function of TMEM200A. Finally, to assess the impact of TMEM200A on the malignant behavior and tumor development function of GC cells, a loss-of-function experiment was conducted in an in vitro assay. Additionally, western blotting was performed to assess how TMEM200A knockdown affected the PI3K/AKT signaling pathway and the epithelial-mesenchymal transition (EMT) in GC.
Protocol
1. The Cancer Genome Atlas (TCGA) database
NOTE: The Cancer Genome Atlas (TCGA) database contains the sequencing data of genes in different tumor tissues14. RNA-seq data in TCGA for the study of TMEM200A transcripts per part per million (TPM) formats were extracted from the UCSC Xena website15 (https://xenabrowser. net/datapages/) and log2 transformed for comparing the expressions between samples.
- Go to the UCSC Xena website interface.
- Click on the Launch Xena tab.
- Click on the DATA SETS tab at the top of the screen.
- From the 129 cohorts and 1,571 datasets on this page, click on the tab for a total of 33 cancers such as GDC TCGA Acute Myeloid Leukemia (LAML), GDC TCGA Adrenocortical Cancer (ACC), GDC TCGA Bile Duct Cancer (CHOL), and more.
- From the gene expression RNAseq menu, select and click on the HTSeq – FPKM GDC Hub tab.
NOTE: HTSeq – FPKM GDC Hub represents data that have been corrected by consent. - From the download menu, click on its corresponding link.
NOTE: By the above method, RNA-Seq data were downloaded for 33 different cancer types. - Unpack the zip archive of RNA-seq data for 33 different cancer types in a single file in the desktop folder of the computer.
- Unify the unzipped txt files in the same folder and place the Perl scripts in this folder.
- Open the Perl software, copy and paste the path of the folder into the Perl software where the mouse cursor is located, press the Enter key, enter the nome of the Perl code and the name of the gene (TMEM200A), and then press the Enter key to get a new txt file(singleGeneExp.txt).
NOTE: This new txt file (singleGeneExp.txt) contains gene expression of TMEM200A in 33 cancers in different patients. - Open the R code (diffR) and copy and paste the path where the singleGeneExp.txt file is located to the line where setwd is located in the R code.
- Open the R software, run the modified R code (diffR), and draw the picture through the "ggpubr" package.
2. The TIMER2.0 database
- Go to the TIMER2.0 (http://timer.cistrome.org) database 16 website interface.
- Click the Cancer Exploration tab.
- Enter the gene ID (TMEM200A) in the search box and click the Enviar tab to get the differential expression images of TMEM200A in different types of tumors.
3. Human Protein Atlas (HPA)
- Go to the Human Protein Atlas (HPA) (www.proteinatlas.org) 17web interface.
- Type ID (TMEM200A) in the search box and click the Buscar button. Select TISSUE sub-atlas.
- Hover over the page and scroll down to find the Protein Expression Overview tab.
NOTE: The Protein Expression Overview section shows the protein expression levels of TMEM200A in 45 organs in the human body.
4. The HumanMethylation450 Illumina Infinium DNA methylation platform array
NOTE: The HumanMethylation450 Illumina Infinium DNA methylation platform array was used to collect data on methylation. We could assess the TMEM200A DNA methylation levels using the SMART (http://www.bioinfo-zs.com/smartapp/) database18.
- Go to the SMART website interface.
- Enter the gene ID (TMEM200A) in the search box to obtain the distribution of TMEM200A in chromosomes and the methylation level of TMEM200A in different types of malignancies.
- Scroll down the page to the Click to check CpG-aggregated methylation menu and click the Plot button. At the bottom of the page, find and click on the Download Figure button. Download the figure.
5. The UALCAN database
- Go to the UALCAN website interface.
NOTE: Box plots of the TMEM200A promoter methylation levels in various malignancies were generated through the UALCAN database (http://ualcan.path.uab.edu/analysisprot.html)19 - At the top of the page, click on the TCGA button.
- Enter TMEM200A in the Gene ID input box on the screen. In the TCGA dataset menu, scroll down and select the type of cancer to be queried.
- After clicking the Explore button, click on its subgroup analysis Methylation in the Links for analysis menu to get the methylation results of this tumor.
NOTE: By this method, we obtained methylation results for 22 tumors in the UALCAN database.
6. The Catalog of Somatic Mutations in Cancer Cells (COSMIC) database
- Go to the Catalog of Somatic Mutations in Cancer Cells (COSMIC) website interface.
NOTE: Data on coding mutations, noncoding mutant genomic rearrangements, and fusion genes in the human genome are available from the Catalog of Somatic Mutations in Cancer Cells (COSMIC) database20 (https://cancer.sanger.ac.uk/cosmic/), which is also used to determine the frequency of various TMEM200A mutations in various cancers. - Enter the gene ID (TMEM200A) in the search box and click the SEARCH button to go to a new screen.
- In the Genes menu, locate and click on the link where the gene ID name of the TMEM200A appears.
- Find the Mutation distribut grouping column on the new page to get the mutation status of TMEM200A in the tumor.
7. CBioPortal
- Go to the CBioPortal website interface.
NOTE: Cancer genomics data may be found, downloaded, analyzed, and visualized using the cBioPortal (www.cioportal.org) database. Somatic mutations, DNA copy number changes (CNAs), mRNA and microRNA (miRNA) expression, DNA methylation, protein abundance, and phosphoprotein abundance are only a few of the genomic data types that are integrated by cBioPortal21. With cBioPortal, a broad range of studies may be carried out; however, the major emphasis is on different analyses relating to mutations and their visualization. - Select the Pan-cancer analysis of whole genomes dataset from the PanCancer Studies subsection of the Select Studies for Visualization & Analysis section. Then, click the Query By Gene button.
- Enter the target gene ID (TMEM200A) in the query box of Enter Genes. Click the Submit Query button.
- Click on the Cancer Type Detailed button at the top of the page to get the mutations of TMEM200A in various types of cancers.
8. Sangerbox 3.0 tool
- Go to the Sangerbox 3.0 tool interface.
NOTE: The correlation of TMEM200A expression with immune infiltrating cells, immunosuppressive agents, immunostimulatory factors, and MHC molecules (major histocompatibility complex) in all cancers was analyzed by CIBERSORT immune infiltration using the pan-cancer immune infiltration data in Sangerbox 3.0 tool22 (http://vip.sangerbox.com/). - From the toolbar on the left side of the page, select and click on the Immunocellular Analysis (CIBERSORT) tab from the Pan-Cancer Analysis menu.
- Enter the target gene ID (TMEM200A) in the query box of Enter Genes. Click the Enviar button.
9. TISIDB database
- Go to the TISIDB website interface.
NOTE: The TISIDB database23 (http://cis.hku.hk/TISIDB/) was employed to examine the correlation of TMEM200A expression with immune infiltrating cells, immunosuppressive agents, immunostimulatory factors, and MHC molecules in various cancers. - Enter the target Gene Symbol (TMEM200A) in the query box of Enter Genes. Click the Enviar button.
- Click on the Lymphocytes, Immunomodulators, Chemokines, and Subtype sections at the top of the page. Download the relevant images at the bottom of the page.
10. The TIDE database
- Go to the TIDE website interface.
NOTE: The TIDE database (http://tide.dfci.harvard.edu/)was used to evaluate the potential of TMEM200A as a biomarker for predicting response to tumor immune checkpoint blockade therapy. - Enter the email address on the initial screen to register the account and log in.
- Find the Biomarker Evaluation subsection at the top of the page and click on it.
- Enter the target gene ID (TMEM200A) in the query box for Enter Genes at the bottom of the page. Then, click the Enviar button.
- In the page, drag the mouse to slide the page down to find the results of the analysis.
11. The CancerSEA database
- Go to the CancerSEA website interface.
NOTE: Using single-cell sequencing data, the relationships between TMEM200A expression and the functional state of different tumor cells were investigated. This was done using the CancerSEA database24 (http://biocc.hrbmu.edu.cn/CancerSEA/). - At the top of the page, find and click on the Buscar tab.
- Enter the target gene ID (TMEM200A) in the query box of Enter Genes. Click the Enviar button.
12. The tumor immune single-cell center (TISCH) network tool
- Go to the tumor immune single-cell center (TISCH) network tool website interface.
NOTE: The correlation between the expression levels of TMEM200A and cancer cells was also shown by using the tumor immune single-cell center (TISCH) network tool25. - Enter the target gene ID (TMEM200A) in the query box of Enter Genes. Click the Explore button.
- In the Cancer type menu on this page, select and click on the All Cancers dataset. Then, scroll down the page and click the Buscar button.
13. GeneMANIA
- Go to the GeneMANIA website interface.
NOTE: The correlation between TMEM200A and its functionally relevant genes was predicted using the integration strategy for the gene multi-association network Website GeneMANIA (www.genemania.org)26 for constructing gene-gene interaction (GGI) networks. - In the search box at the top left of the page, enter the gene ID (TMEM200A) and click Search Tools.
NOTE: The GeneMANIA web tool was used to find 20 genes associated with TMEM200A.
14. Functional enrichment analysis
- Save the symbol IDs of the genes to be analyzed for enrichment in txt file format.
- Open the R code (symbolidR) and copy and paste the path where the above txt file is located to the line where setwd is located in the R code.
- Open the R software and run the modified R code (symbolidR). Convert the symbol ids of these genes to entrezIDs via the "org.Hs.eg.db" package. Get the id.txt file.
- Open the R code (GOR and KEGGR) and copy and paste the path where the id.txt file is located to the line where setwd is located in the R code.
- Open the R software and run the modified R code (GOR and KEGGR). To follow this protocol, analyze these genes for GO and KEGG enrichment by the packages "clusterProfiler," "org.Hs.eg.db," and "enrichplot" and plot the enrichment results using the package "gglot2".
15. Analysis of the differences in gene activity
NOTE: TMEM200A scoring in each cancer sample was calculated using ssGSEA (single-sample gene set enrichment analysis)27, and the differential analysis of TMEM200A gene activity in cancerous and healthy tissues was performed for various cancers.
- Based on the RNA-seq data of 33 tumor types downloaded in step 1.7, unzip all downloaded files and place them in a unified folder.
- Place the merge.txt file in the above folder.
NOTE: The data in merge.txt file from BioWolf (www.biowolf.cn) contains gene expression for all patients in all tumors in The Cancer Genome Atlas (TCGA) database. - Open the R code (ssGSEAR) and copy and paste the path where the above folder is located to the line where setwd is located in the R code.
- Take the line where geneName is in the R code (ssGSEAR) and enter the gene ID (TMEM200A).
- Open the R software and run the modified R code(ssGSEAR). Get the scoring file for gene activity: scores.txt.
NOTE: With the ssGSEA algorithm, we first construct a gene set file based on the correlation coefficients between genes according to the data provided in the merge.txt file and identify the genes with a higher correlation with the target gene (TMEM200A) from the file. Then, the genes with higher correlation are unified as the active genes of TMEM200A, and finally, we get the scoring of active genes in each sample. - Open the R code (scoreDiffR) and copy and paste the path where the scores.txt file is located to the line where setwd is located in the R code.
- Open the R software, run the modified R code(scoreDiffR), and draw the picture through the "plyr," "reshape2," and "ggpubr" packages.
16. Clinicopathological correlation and survival prognosis analysis
- Go to the UCSC Xena website interface.
- Click the Launch Xena tab.
- Click on the DATA SETS tab at the top of the screen.
- Hover over the page and scroll down to find the TCGA Pan-Cancer (PANCAN) tab.
- From the 129 cohorts and 1,571 datasets on this page, click the TCGA Pan-Cancer (PANCAN) tab.
- From the phenotype menu, select and click on the Curated clinical data tab.
- From the download menu, click on its corresponding link.
NOTE: Download clinical data for 33 cancers from the TCGA database at the above link. - Unzip the downloaded clinical data file and open the unzipped file in xls file format.
- Organize and retain the needed clinical data (stage, age, pathological stage, and patient status) in the file, delete the remaining unneeded clinical data, and save the entire file as a txt format file after renaming it clinical.
- Based on the singleGeneExp.txt obtained in step 1.9, put this file in the same folder as the organized clinical.txt file.
- Unzip the downloaded clinical data and place the singleGeneExp.txt and clinical.txt files in the same folder as the unzipped clinical files.
- Open the R code (clinicalDiffR) and copy and paste the path where the above folder is located to the line where setwd is located in the R code.
- Open the R software, run the modified R code (clinicalDiffR), and draw the picture using the "ggpubr" package.
- Go to the UCSC Xena website interface.
- Click the Launch Xena tab.
- Click on the DATA SETS tab at the top of the screen.
- From the 129 cohorts and 1,571 datasets on this page, click on the tab for a total of 33 cancers such as GDC TCGA Acute Myeloid Leukemia (LAML), GDC TCGA Adrenocortical Cancer (ACC), GDC TCGA Bile Duct Cancer (CHOL), and more.
- From the phenotype menu, select and click on the survival data tab.
- From the download menu, click on its corresponding link.
- Unpack the zip archive of survival data for 33 different cancer types in a single file in the desktop folder of the computer.
- Put the unzipped survival data in the same folder as the singleGeneExp.txt file obtained in step 1.9.
- Open the R code (preOSR) and copy and paste the path where the above folder is located to the line where setwd is located in the R code.
- Open the R software and run the modified R code (preOSR). Calculate through the "limma" package to get a data file about the survival of the TMEM200A in pan-cancer: expTime.txt.
- Open the R code (OSR) and copy and paste the path where the expTime.txt file is located to the line where setwd is located in the R code.
- Open the R software and run the modified R code (OSR). Perform a KM analysis using the "survival" and "survminer" packages based on the data in the expTime.txt file and plot the survival curves for TMEM200A in pan-cancer.
17. Univariate and multivariate Cox regression analyses with forest plot construction
- Open the R code (COXR) and copy and paste the path where we got the expTime.txt file from 16.23 is located to the line where setwd is located in the R code.
- Open the R software and run the modified R code (COXR). Based on the data in the expTime.txt file, perform univariate COX regression analyses using the "survival," "survminer," and "forestplot" packages to plot a Univariate -forest plot of TMEM200A in different cancers.
- Put the expTime.txt file from step 16.23 and clinical.txt from step 16.10 in the same folder.
- Open the R code (multicoxR) and copy and paste the path where the folder containing the expTime.txt file from step 16.23 and clinical.txt file from step 16.10 is located to the line where setwd is located in the R code.
- Open the R software and run the modified R code (multicoxR). Perform a multivariate COX regression analysis using the "survival" and "survminer" packages to plot a multivariate-forest plot of TMEM200A in different cancers based on the data in the expTime.txt and clinical.txt files.
18. Prognostic model of gastric cancer based on TMEM200A expression and clinical features
- Put the clinical.txt file from step 16.10 and the expTime.txt file from step 16.23 in the same folder.
- Open the R code (NomoR) and copy and paste the path where the above txt file is located to the line where setwd is located in the R code.
- Open the R software and run the modified R code (NomoR). Use the software packages "survival," "regplot," and "rms" for TMEM200A expression data in GC and clinical data to construct a prognostic model to predict patient survival.
19. Cell culture and siRNA transfection of TMEM200A
NOTE: Human STAD HGC-27 cells, SGC-7901 cells, and human gastric mucosal epithelial GES-1 cells were obtained commercially (see the Table of Materials), revived, inoculated in Roswell Park Memorial Institute (RPMI) 1640 complete medium (containing 10% neonatal fetal bovine serum and 1% penicillin mixture), and cultured at 37 °C in a 5% CO2 cell culture incubator. The culture medium was changed every 2-3 days. The cells in good growth condition were inoculated in six-well plates according to (2-3) × 105 cells per well.
- First, take three microcentrifuge tubes and add 8 µL of transfection reagent (see the Table of Materials) and 200 µL of basal medium to each tube. Then, add 4 µg of SiRNA1, SiRNA2, and SiRNA3 to each tube respectively.
NOTE: For TMEM200A knockdown, siRNA was designed and purchased (see the Table of Materials). - Mix the mixture in the three microcentrifuge tubes thoroughly and add to each of the three wells of the 6-well plate. Gently shake the 6-well plate to distribute the mixture evenly. Label the three wells where the mixture was added as SiRNA-1, SiRNA-2, and SiRNA-3.
- When the cells have been incubated for 4-6 h, replace half of the medium in the three sub-wells in the 6-well plate.
NOTE: When replacing half of the complete medium, aspirate half of the original complete medium and replenish with half of the fresh complete medium. - Twenty-four to 48 h later, use TMEM200A siRNA-transfected cells as the experimental group and untransfected cells as the negative control group. Perform quantitative real-time polymerase chain reaction (qRT-PCR, see section 20) to assess the effect of transfection on the cells.
20. Quantitative real-time polymerase chain reaction
- Discard the cell culture medium in each subgroup and gently wash the cells twice with 1 mL of phosphate-buffered saline (PBS).
- Isolate total RNA using an RNA isolation kit (see the Table of Materials).
- Estimate the RNA concentration and synthesize cDNA with 1 µg of RNA using a cDNA Synthesis Kit, following the manufacturer's instructions.
- Perform quantitative real-time PCR (qRT-PCR) on 10-fold dilutions of cDNA in 10 µL of reaction mix, including human-specific forward and reverse primers for GAPDH (for standardization), TMEM200A (see the Table of Materials), and qPCR Master Mix, using the Real-Time PCR Assay System.
NOTE: Perform each experiment in triplicate. - Use the following qPCR reaction conditions: initialization, 95 °C for 3 min; denaturation, 95 °C for 10 s; annealing, 60 °C for 30 s; and extension, 80 °C for 10 s; repeat the denaturation, annealing, and extension 40x. Determine the relative expression of each gene using the 2-ΔΔCt technique28.
- Repeat the experiments at least three times to obtain biological triplicates.
21. Western blot detection of relevant protein expression
- Discard the cell culture medium in each subgroup and gently wash the cells with 2 x 1 mL of PBS.
- Cool the 6-well plates containing the cells on ice and add 150 µL of precooled radio immune precipitation assay (RIPA) lysis buffer (150 mM NaCl, 0.1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl pH 8.0, and freshly added protease inhibitor cocktail). Allow the lysis to proceed on ice for 10 min.
- Utilizing a plastic cell scraper, remove adhering cells from the dish and delicately transfer the cell solution into a microcentrifuge tube that has been precooled.
- Centrifuge the cell lysate for 10 min at 1.5 × 104 g at 4 °C. Transfer the supernatant to a new 1.5 mL microcentrifuge tube.
- Utilize a BCA protein assay Kit to determine the protein content according to the manufacturer's instructions.
- Load 30 g of protein from each sample on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and run at 80 V for 0.5 h followed by 1.5 h at 120 V.
- Transfer the proteins from the gel to a 45 µm polyvinylidene difluoride (PVDF) membrane at a voltage of 300 mA for 1-1.5 h.
- After placing the PVDF membrane on a shaker and shaking it for 3 x 5 min, add Tris-buffered saline with Tween 20 (TBST, 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Tween 20) to the appropriate container with the protein side (gel side) facing up.
- Place the membrane in the blocking buffer (see the Table of Materials) and incubate it for 0.5 h at room temperature.
- Wash the membrane with TBST for 3 x 10 min.
- Incubate the membrane with primary antibodies against phospho-AKT (p-AKT; 1:1,000), total AKT 1:1,000, E-cadherin (E-ca; 1:1,000), N-cadherin (N-ca; 1:1,000), Vimentin 1:1,000, snail 1:1,000, TMEM200A 1:1,000, glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:1,000), overnight at 4 °C.
- Wash the membrane for 3 x 10 min and incubate the membrane with a rabbit or mouse secondary antibody (1:5,000) for 1 h at room temperature.
- Incubate the PVDF membrane with ECL substrate for 30 s and detect the signal using an imaging system.
22. CCK-8 assay
- Seed human STAD HGC-27 cells in good growth condition in 96-well plates.
- When the cell density reaches 60-70%, transfect the cells with TMEM200A siRNA (as in step 19.3).
- Seed the transfected cells into 96-well plates at a cell density of 5 × 10 3/well (count viable cells using a cell counter), divided into NC and TMEM200A siRNA groups (with three replicate wells in each group), and incubated at 37 °C.
- Add CCK-8 reagent after 0, 24, 48, 72, and 96 h and incubate at 37 °C for 2 h. Measure the absorbance (450 nm) using a multifunctional enzyme labeler.
Representative Results
Expression of TMEM200A in various cancers
As illustrated in Figure 1, we first analyzed the differential expression levels of TMEM200A in various cancers through different databases. TMEM200A expression was elevated in cholangiocarcinoma (CHOL), head and neck squamous cell carcinoma (HNSC), renal clear cell carcinoma (KIRC), renal papillary cell carcinoma (KIRP), hepatocellular carcinoma (LIHC), STAD, and thyroid carcinoma (THCA) compared with adjacent normal tissues solely based on TCGA data. However, TMEM200A expression decreased in bladder uroepithelial carcinoma (BLCA), cervical squamous cell carcinoma and cervical adenocarcinoma (CESC), glioblastoma multiforme (GBM), kidney chromophobe (KICH), lung squamous carcinoma (LUSC), pheochromocytoma and paraganglioma (PCPG), rectal adenocarcinoma (READ), and uterine corpus endometrial carcinoma (UCEC) (Figure 2A). In the TIMER2.0 database, TMEM200A expression increased in CHOL, HNSC, KIRC, KIRP, LIHC, and STAD. Moreover, TMEM200A expression increased in BLCA, CESC, GBM, KICH, LUSC, PCPG, READ, skin cutaneous melanoma (SKCM), and UCEC (Figure 2B).
By comparing the results of the two databases, we found that the pan-cancer expression of TMEM200A was the same in each database. We obtained the clinical follow-up data and RNA sequencing data of 33 patients with cancer from the TCGA database, calculated TMEM200A scoring in each cancer sample using ssGSEA analysis, and then calculated TMEM200A gene activity in tumor and normal tissues in many tumors. We discovered that TMEM200A was highly expressed in GBM, HNSC, kidney KIRC, and THCA (Figure 2C); hence, the gene activity of TMEM200A was ranked in each tumor tissue. It was found that the gene activity of TMEM200A was the highest in KIRC and the lowest in uveal melanoma (UVM) (Figure 2D). Subsequent HPA database assessment of TMEM200A expression levels in human tissues showed that TMEM200A expression levels were low in most normal tissues but high in the adrenal cortex, rectum, endometrium, and smooth muscle (Figure 2E).
In normal tissue cell lines, a few cell lines such as granulosa cells, bipolar cells, skeletal muscle cells, endometrial stromal cells, and T cells showed high expression of TMEM200A, whereas TMEM200A expression was low in most other cell lines (Figure 2F). We also examined immunohistochemistry (IHC) data in the HPA database to look at the expression of TMEM200A at the protein level. The data regarding staining and intensity revealed much greater and more pronounced expression levels of TMEM200A in colorectal cancer (CRC), lung cancer (LUAD), liver cancer (LIHC), breast cancer (BRCA), and prostate cancer (PRAD) tissues compared with normal tissues (Figure 2G). These findings suggested that TMEM200A was widely expressed in different cancers and affected cancer development via different mechanisms in different tumors.
Clinicopathological relationship and survival prognosis
For 33 human malignancies, we gathered information and clinical data in the TCGA pan-cancer (PanCan) cohort from the UCSC Xena database website and investigated the correlation between TMEM200A expression in PanCan and age, pathologic grade, clinicopathological stage, and patient survival status. This study revealed that, in six cancers, the expression of TMEM200A was associated with the clinicopathological stage, including BLCA, invasive breast carcinoma (BRCA), esophageal carcinoma (ESCA), KIRP, SKCM, and testicular carcinoma (TGCT) (Figure 3A). Age was correlated with six tumors, including invasive BRCA, GBM, acute myeloid leukemia (LAML), SKCM, thymic carcinoma (THYM), and endometrial cancer (UCEC) (Figure 3B). The pathological grade was associated with six tumors, including esophageal carcinoma (ESCA), HNSC, KIRC, low-grade glioma of the brain (LGG), pancreatic adenocarcinoma (PAAD), and endometrial carcinoma (UCEC) (Figure 3C). The survival status was associated with five tumors, including adrenocortical carcinoma (ACC), BLCA, KIRC, PCPG, and uveal melanoma (UVM) (Figure 3D).
We conducted studies for different cancers on OS, DSS, PFI, and DFI using one-way Cox regression analysis with a median group threshold to learn more about the correlation between TMEM200A expression and prognosis. The results of the OS analysis revealed that, among the four tumors, higher TMEM200A expression had a greater impact on OS prognosis in four tumors, including adrenocortical carcinoma (ACC) (P = 0.037), BLCA (P = 0.036), acute myeloid leukemia (LAML) (P = 0.011), and GC (STAD) (P = 0.005). In contrast, higher TMEM200A expression had a better prognosis for OS in five tumors, including KIRC (P < 0.001), KIRP (P = 0.020), low-grade glioma of the brain (LGG) (P = 0.042), PCPG (P = 0.002), and cutaneous melanoma (SKCM) (P = 0.043) (Figure 3E,F). For DSS, high TMEM200A expression had a poor prognosis in two tumor types, including adrenocortical carcinoma (ACC) (P = 0.026) and BLCA (P = 0.015). High TMEM200A expression had a better prognosis in four tumor types, including KIRC (P < 0.001), KIRP (P = 0.001), low-grade glioma of the brain (LGG) (P = 0.025), and PCPG (P = 0.007) (Figure 3G). In addition, for PFI, higher TMEM200A expression had a poor prognosis in three tumor types, including adrenocortical carcinoma (ACC) (P = 0.007), BLCA (P = 0.040), and uveal melanoma (UVM) (P = 0.020). Higher TMEM200A expression had a better prognosis in two tumor types, including KIRC (P < 0.001) and low-grade glioma (LGG) (P = 0.044) of the brain (Figure 3H). In DFI, higher expression of TMEM200A had a poorer prognosis in adrenocortical carcinoma (ACC) (P = 0.044) (Figure 3I).
Subsequently, we selected several tumors (KIRC and KIRP) for multi-COX regression analysis. The results showed that TMEM200A could individually affect the survival rate of cancer patients compared to other clinical factors (Figure 3J and K). These results showed that TMEM200A could be used as an independent bioprognostic marker in different cancers to influence the survival of patients with cancer.
DNA methylation analysis
One of the epigenetic alterations that has received the maximum attention is DNA methylation29. It has been suggested that one type of epigenetic change, DNA methylation, has a significant impact on tumor growth30. Hence, using the SMART database, we evaluated the DNA methylation levels of TMEM200A in normal and cancerous tissues. PAAD and PRAD tissues had higher levels of DNA methylation of TMEM200A than normal tissues. However, TMEM200A had lower DNA methylation levels in COAD, KIRC, KIRP, LIHC, LUSC, READ, THCA, and UCEC than in normal tissues (Figure 4A). The DNA methylation levels of TMEM200A were substantially higher in the TCGA database in KIRP, PRAD, PCPG, and THYM based on the UALCAN database (Figure 4B) but not in BLCA, BRCA, CHOL, COAD, CESC, ESCA, GBM, HNSC, KIRC, LIHC, LUAD, LUSC, PAAD, READ, TGCT, STAD, THCA, and UCEC. The methylation level of TMEM200A significantly decreased in LUSC, PAAD, READ, TGCT, STAD, THCA, and UCEC (Figure 4C). To explore the relationship between the expression level of TMEM200A and DNA methylation and clinicopathological factors even further, we again utilized the SMART database and found that the DNA methylation level of TMEM200A in BLCA, HNSC, LUAD, and STAD changed with the pathological stage (Figure 4D). Clinical factors influence the level of DNA methylation of TMEM200A in the above tumors.
Gene mutation analysis
One of the reasons for the development of tumors is the accumulation of gene mutations31. Therefore, the frequency of somatic TMEM200A mutations was analyzed by estimating the frequency of TMEM200A mutations in pan-cancer clinical samples using the cBioPortal database. TMEM200A is mutated in many cancers, such as lung cancer, uterine endometrioid carcinoma, melanoma, esophagogastric cancer, bone cancer, cervical cancer, hepatobiliary cancer, mature B-cell lymphoma, BRCA, endometrial cancer, ovarian cancer, embryonal tumor, pancreatic cancer, head and neck cancer, CRC, glioma, non-small cell lung cancer, soft tissue sarcoma, and cancer of unknown primary (Figure 5A). In addition, DNA alterations can lead to structural or amino acid changes at the protein level. Figure 5B shows the 3D structure of TMEM200A. Using the cBioPortal database, we found mutation sites in the amino acids of TMEM200A; missense mutations were the most prevalent form of mutations (Figure 5C).
We analyzed the data using Sangerbox 3.0 to verify the accuracy of the mutation sites and achieved the same outcomes as earlier. Missense mutations are the most frequent form of mutations in TMEM200A and they may occur as often as 7.8% in SKCM (Figure 5D). A missense mutation is a mutation in which a codon coding for one amino acid undergoes a base substitution and becomes a codon coding for another amino acid, resulting in a change in the amino acid type and sequence of the polypeptide chain. The result of missense mutation is usually that the polypeptide chain loses its original function. Many protein abnormalities are caused by missense mutation, which is a common type of tumor mutation. It has been found that missense mutations play a key role in the stability of proteins, and the presence of mutations can have a significant effect on the binding affinity between proteins32, altering the interactions between proteins and the surrounding correlative macromolecules33.
Thus, missense mutations most likely affect the biological function of transmembrane protein 200A in different cancers. The correlation between TMEM200A gene mutation and the clinical survival prognosis of patients with cancer was also examined. The likelihood of OS increased compared with that in the TMEM200A-mutated group, but not in the disease-free group (Figure 5E). The study of gene correlations revealed that TP53, AKAP7, TAAR5, TAAR1, MOXD1, THEMIS, VNN2,ENPP3, TMEM244, and SLC18B1 were among the genes that co-mutated with TMEM200A (Figure 5F). We were able to identify several mutation types and single-nucleotide variations (SNVs) through the COSMIC database to learn more about TMEM200A mutations in GC. The frequency of missense substitutions was the highest (38.86%) (Figure 5G), and SNV data showed that the most common SNVs in STAD were G>A (31.73%), followed by C>T (26.35%) and G>T (11.73%) (Figure 5H).
Single-cell analysis of TMEM200A in cancer
We analyzed TMEM200A expression levels in different single cells using the TISCH (Tumor Immunology Single Cell Center) website. The TISCH online tool displayed the expression of TMEM200A for 65 datasets and 28 cell types in the heat map shown in Figure 6A. The findings showed that TMEM200A was mostly expressed in CD8+ T cells and fibroblasts. The GSE72056 dataset, which includes cells from patients with metastatic cutaneous melanoma (SKCM), is noteworthy in this regard. TMEM200A was widely highly expressed in B cells, CD4+ T lymphocytes, depleted CD8+ T cells, endothelial cells, fibroblasts, and other cell types in the SKCM microenvironment (Figure 6B,C). In the GSE146771 dataset, which contains cells from patients with CRC, TMEM200A was widely highly expressed in B cells, CD4+ T lymphocytes, CD8+ T cells, depleted CD8+ T cells, endothelial cells, fibroblasts, and other cell types in the CRC microenvironment (Figure 6D,E). By using the CancerSEA database the expression levels and functional status of TMEM200A in specific tumor cells were determined (Figure 6G). TMEM200A expression was strongly correlated with the cellular functional state in a range of cancers, including glioblastoma (GBM), lung adenocarcinoma (LUAD), chronic granulocytic leukemia (CML), CRC, retinoblastoma (RB), and uveal melanoma (UM). Angiogenesis, apoptosis, differentiation, and inflammation were all positively correlated with TMEM200A expression in a majority of tumor cells, whereas DNA damage, DNA repair, invasion, and metabolism were negatively correlated with TMEM200A expression (Figure 6F).
Functional and pathway enrichment analysis of TMEM200A in various cancers
We examined the link between TMEM200A and proteins with similar functions using the GGI network to explore the function of TMEM200A in different cancers (Figure 7A). Genomic and proteomic information is analyzed by GeneMANIA using a query list of target genes. By locating genes that operate similarly to TMEM200A, the 40 proteins that directly interact with this gene were discovered. The correlation of these genes is shown by the PPI network (Figure 7B). Subsequently, GO and KEGG enrichment analyses of these 20 genes closely related to TMEM200A revealed that these adjacent genes were mostly implicated in the RNA's negative regulation of gene silencing. KEGG analysis revealed that TMEM200A was abundant in pathways, including the Wnt signaling pathway and cellular senescence (Figure 7C). GO enrichment analysis showed that, in molecular function, TMEM200A was mainly abundant in the calcium channels and negative regulation of gene silencing by RNA (Figure 7D).
Correlation analysis between TMEM200A and immune infiltration
We further investigated the correlation between TMEM200A expression and immune cell infiltration to demonstrate the correlation between TMEM200A and cancer immunity. We conducted CIBERSORT immune infiltration analysis using the Sangerbox 3.0 pan-cancer data. The results showed that pan-cancer TMEM200A expression was correlated with CD8+ T cells, CD4+ T cells, B cells, helper T cells, natural killer cells, regulatory T cells, monocytes, macrophages, dendritic cells, eosinophils, basophils, and granulocytes (Figure 8A). Subsequently, we looked at the correlation between TMEM200A expression and TIL (Tumor Infiltrating Lymphocytes) using the TISIDB database, showing a close correlation between TMEM200A expression and 28 TIL abundance in different cancer types (Figure 8B). We used the TISIDB database to assess the correlation between TMEM200A expression and immunosuppressive drugs in human malignancies to more thoroughly investigate how immunomodulators and TMEM200A expression were related. The results showed that TMEM200A expression levels were significantly correlated with immunosuppressive agents in a variety of cancers (Figure 8C). Significant correlations between TMEM200A expression and certain immunosuppressive agents were found in testicular cancer and UM (Figure 8D–F). We then performed a correlation analysis of TMEM200A expression with immunostimulatory factors and MHC molecules using the TISIDB database (Figure 8G,H). The results showed that TMEM200A expression was correlated with selected immunostimulants and MHC molecules in urothelial carcinoma of the bladder, testicular carcinoma, adrenocortical carcinoma, and UM (Figure 8I–L). The next step was to investigate the correlation between TMEM200A expression and immunological or molecular subtypes in the TCGA PanCan cohort in the TISIDB database. We found that the immunological subgroups of nine distinct cancer types expressed TMEM200A differently, including BLCA, UVM, KIRC, LIHC, LUAD, LUSC, PRAD, TGCT, and BRCA (Figure 8M). Additionally, six different cancer forms with diverse molecular subgroups, including HNSC, KIRP, LIHC, PCPG, OV, and LUSC, displayed variable expression of TMEM200A (Figure 8N).
TMEM200A and immunotherapy response analysis
The effectiveness of PD-1/PD-L1 inhibitors is highly correlated with TMB, and some patients with tumors can be somewhat predicted using TMB markers for the efficacy of immunotherapy34. Microsatellite instability (MSI) is the term used to describe the faulty DNA mismatch repair (MMR) function when microsatellite replication mistakes are not fixed and accumulate, changing the length or base composition of microsatellites35. MSI is a tumor marker of clinical significance 36. We thus assessed the impact of TMEM200A expression on immunotherapy by investigating the correlation between TMEM200A expression and TMB or MSI. The findings demonstrated a substantial positive correlation between TMEM200A expression and TMB in THYM, PRAD, OV, LAML, KIRP, and KIRC, but TMEM200A expression was negatively correlated with UCEC, PAAD, LUSC, LUAD, LIHC, LGG, HNSC, GBM, and BRCA (Figure 9A). MSI and TMEM200A expression were strongly and favorably linked in TGCT, whereas TMEM200A expression was significantly and negatively correlated with MSI in UCEC, STAD, SKCM, LUSC, LUAD, HNSC, and CHOL (Figure 9B). We also assessed the potential of TMEM200A as a biomarker to examine the effectiveness of ICB. The results showed that TMEM200A alone predicted ICB responses with an accuracy of Area Under Curve (AUC) > 0.5 in 7 out of 25 ICB subpopulations. TMEM200A showed a higher predictive value than T- and B-cell clones, both of which had a value of 5. However, the value for TMEM200A was lower than that for TIDE (AUC > 0.5 in 11 ICB subpopulations), MSI score (AUC > 0.5 in 11 ICB subpopulations), CD274 (AUC > 0.5 in 15 ICB subpopulations), CD8 (AUC > 0.5 in 17 ICB subpopulations), IFNG (AUC > 0.5 in 16 ICB subpopulations), and Merck18 (AUC > 0.5 in 17 ICB subgroups) (Figure 9C). Poor survival prognosis in the ICB_Ria 2017_PD1 Ipi_Naïve, ICB_Gide 2019_PD1, and ICB_Hugo 2016_PD1 cohorts was correlated with high TMEM200A expression. However, TMEM200A knockdown enhanced lymphocyte-mediated tumor-killing potency in the average cohort of Kearney 2018 T_PD1 and Pan 2018 OT1 (Figure 9D).
GSEA enrichment analysis of TMEM200A in different cancers
We used DEGs between TMEM200A high-expression and TMEM200A low-expression subgroups in 32 malignancies to explore TMEM200A-related cancer characteristics. We discovered a substantial correlation between primary immunodeficiency, the RIG (Retinoic acid-inducible gene)-I-like receptor signaling pathway, and TMEM200A expression in pan-cancer, especially in BLCA, COAD, KICH, GBM, LUSC, LUAD, MESO, READ, SARC, and SKCM (Figure 10). The viral cytoplasmic protein ribonucleic acid is detected by the RIG-I-like receptor signaling pathway. RIG-I-like receptors may influence the production of a variety of inflammatory cytokines in the body by activating the NF-kB signaling pathway by engaging certain intracellular junction proteins. As a result, the transmembrane protein 200A (TMEM200A) may play a crucial role in the recruitment of intracellular proteins by the RIG-I-like receptor. These findings implied a strong correlation between TMEM200A expression and immunological infiltration. Additionally, GSEA enrichment analysis showed that pan-cancer TMEM200A expression was correlated with chemokine signaling, cell adhesion, cytokine receptor connections, cell membrane sensing pathways, and autophagy control (Figure 10). Transmembrane proteins may function as adhesion molecules to affect the tumor microenvironment in addition to playing a critical role in controlling the entrance of calcium ions into the cell from calcium reservoirs 36. Therefore, the results obtained from GSEA enrichment analysis may be due to the involvement of TMEM200A in many physiological processes, including activation of signal transduction pathways, formation of plasma membrane ion channels, and regulation of cell chemotaxis, adhesion, apoptosis, and autophagy9.
We identified the top 50 STAD genes both positively and negatively correlated with TMEM200A from the LinkedOmics database, which were co-expressed in STAD, in the form of heat maps (Figure 11A,B). We evaluated the top 5 genes positively and the top 5 genes negatively associated with TMEM200A in GC (Figure 11C). Our results showed that TMEM200A expression was correlated with SPON1 (r = 0.51), CDH11 (r = 0.50), EPB41L2 (r = 0.49), LUM (r = 0.49), SAMD3 (r = 0.49), OVOL2 (r = -0.37), RASAL1 (r = -0.36), IRX5 (r = -0.35), SLC4A11 (r = -0.34), and SYTL1 (r = -0.33) co-expression. We analyzed the threshold visualization results to further understand the role of TMEM200A in STAD. The following criteria were used for screening: log2-fold change (FC) > 2.0 and adjusted P value 0.05. We found 483 DEGs, of which 385 were upregulated and 98 were downregulated, and we chose 100 of them to be visualized to create a heat map (Figure 11D). Then, for the DEGs that satisfied the screening criteria, we ran GO and KEGG enrichment analyses. The results of GO enrichment analysis showed that the enrichment of BP (Biological Process) was mainly associated with extracellular matrix and structural tissue, the enrichment of CC (Cellular Component) was mainly associated with collagen-containing extracellular matrix and basement membrane, and MF (Molecular Function) was mainly enriched in an extracellular matrix structure and glycosaminoglycan binding (Figure 11E and Figure 11G). The enrichment of KEGG analysis showed that TMEM200A was mainly enriched in the extracellular matrix receptor interactions pathway, cytochrome P450, and renin-angiotensin system (Figure 11F and Figure 11H).
TMEM200A-based prognostic model and clinical features of stomach cancer
We investigated the correlation between TMEM200A and the clinical data of patients with STAD and therefore analyzed the clinicopathological characteristics of patients in the TCGA-STAD cohort by logistic regression analysis and based on TMEM200A expression levels (Figure 12A). Age, clinical stage, and TMEM200A expression were all substantially correlated with OS in patients with STAD, according to a univariate Cox regression analysis (Figure 12B). The multivariate Cox regression analysis showed that TMEM200A expression might be a standalone predictive factor for OS in patients with STAD (HR = 1.282, 95% CI = 1.066-1.541, P = 0.008) (Figure 12C). We examined the potential of these clinicopathological characteristics to predict OS after 1, 3, and 5 years in STAD using the standard column line plot model in conjunction with several clinical parameters (Figure 12D). The calibration curves for 1-year survival probabilities were highly consistent with column line plot-predicted probabilities (Figure 12E).
TMEM200A upregulation in STAD cells and cell proliferation enhancement
By reviewing the literature related to TMEM200A, we discovered that the high expression of TMEM200A indicated a bad prognosis 13 and the GC immune microenvironment might be influenced by TMEM200A acting as an adhesion molecule37, thus promoting the invasion and metastasis of GC cells. We examined the protein levels and mRNA transcript levels of TMEM200A in STAD cell lines to confirm the findings of the aforementioned study. The GC cell line HGC-27 dramatically overexpressed TMEM200A compared with the healthy gastric mucosal epithelial cell line GES-1 at both the protein and mRNA transcript levels (Figure 13A,B), indicating that TMEM200A was overexpressed in HGC-27 cells. The following tests were conducted using HGC-27 cells as a result. Meanwhile, to prove the accuracy of the experimental results, we used qRT-PCR to compare the differential mRNA expression of TMEM200A in human GC cells SGC-7901 and gastric mucosal epithelial cells GES-1, and the results showed that TMEM200A was highly expressed in SGC-7901 cells (Figure 13B). TMEM200A was knocked down in HGC-27 cells to examine the possible involvement of TMEM200A in STAD (Figure 13C). Based on the database mining results and our correlation analysis of TMEM200A, it was found that TMEM200A was mainly involved in the extracellular matrix receptor interactions pathway, which was associated with cancer proliferation and invasion. We thus employed CCK-8 assays to ascertain how TMEM200A expression affected cell growth. The CCK-8 assays showed that TMEM200A knockdown groups (Si-RNA2 and Si-RNA3) showed a notable drop in cell viability as compared to the NC group (Figure 13D). These findings suggested that TMEM200A was upregulated in STAD, and its expression level could affect the proliferation of GC cells. Based on the results of KEGG enrichment analysis in Figure 11F, we found that TMEM200A was associated with the PI3K/AKT signaling pathway in GC, and TMEM200A, as a cell adhesion factor, may influence the mechanism of gastric carcinogenesis by affecting EMT. Therefore, we used western blotting to verify the effects of TMEM200A knockdown on EMT and the PI3K/AKT signaling pathway before and after knockdown. The results showed that P-AKT protein expression was decreased in the TMEM200A knockdown group (TMEM200A-SiRNA2) compared to the negative control group (NC), while levels of N-cadherin, Vimentin, and Snai proteins were also decreased and E-cadherin protein expression was increased (Figure 13E). All these results suggest that TMEM200A may affect EMT through the PI3K/AKT signaling pathway and thus play a role in GC.
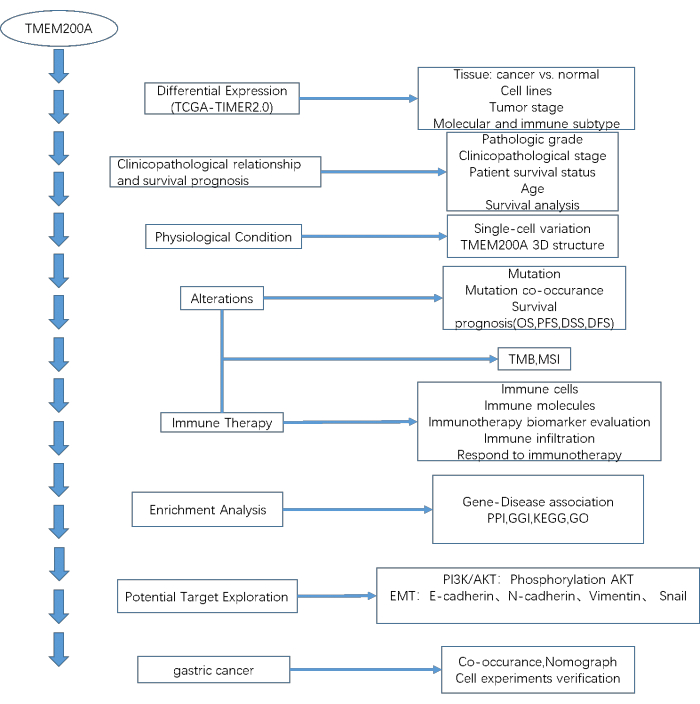
Figure 1: The workflow of the study. Abbreviations: TCGA = The Cancer Genome Atlas database; TIMER2.0 = The TIMER2.0 database; OS= Overall Survival; PFS = Progression-free Survival time; DSS = Disease-specific Survival; DFS = Disease-free Survival; TMB = Tumor Mutational Burden; MSI = Microsatellite instability; PPI = Protein-protein interaction; GGI = Gene-gene interaction; KEGG = Kyoto Encyclopedia of Genes and Genomes; GO = Gene Ontology. Please click here to view a larger version of this figure.
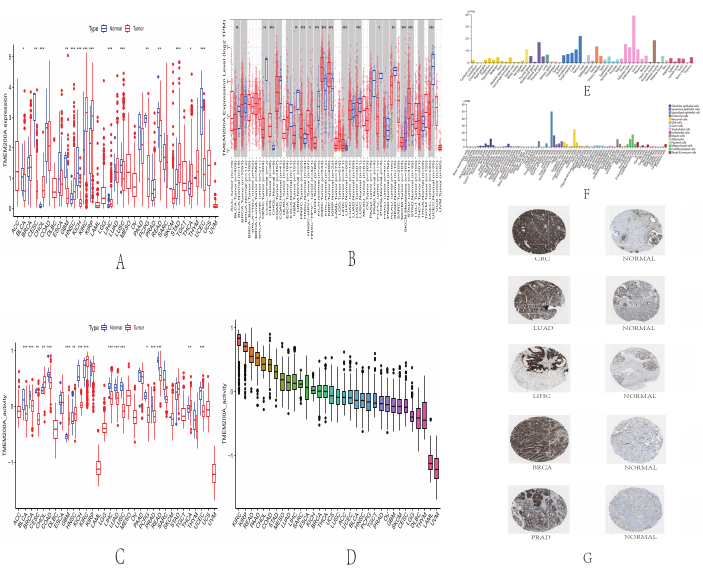
Figure 2: Expression levels of TMEM200A in different cancers. (A) Up- or downregulated expression of TMEM200A from the TCGA dataset in different human malignancies. (B) Analysis of TMEM200A expression levels in tumor and normal tissues using the TIMER2.0 database. (C) Differential analysis of TMEM200A gene activity in various human cancers from the TCGA dataset. (D) Ranking of gene activity levels of TMEM200A in various human cancers based on ssGSEA scoring. (E) TMEM200A expression levels from the HPA database in healthy tissues. (F) HPA database's TMEM200A expression levels in normal cell lines. (G) IHC outcomes for TMEM200A protein expression in cancer from the HPA database. Abbreviations: TCGA = The Cancer Genome Atlas; ssGSEA = Single Sample Gene Set Enrichment Analysis; HPA = Human Protein Atlas; IHC = Immunohistochemistry. Please click here to view a larger version of this figure.
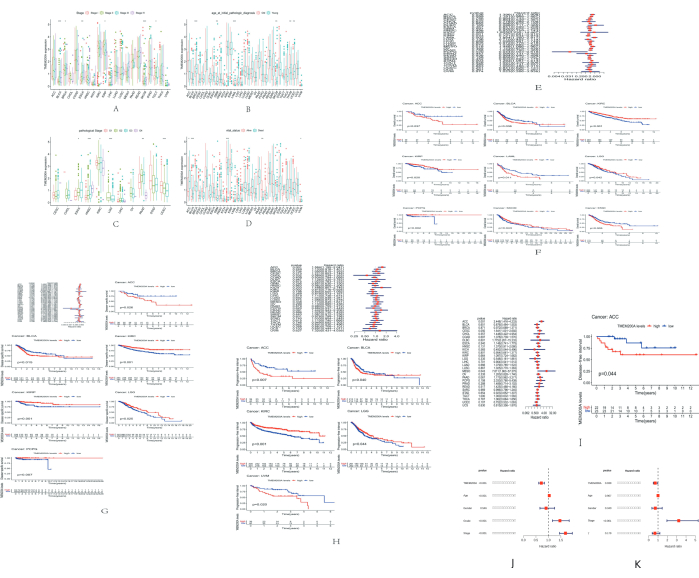
Figure 3: Correlation of TMEM200A expression with clinicopathological characteristics and prognosis of different tumors. (A) Correlation of TMEM200A expression in the TCGA pan-cancer (PanCan) cohort with clinicopathological staging in each tumor was analyzed using the UCSC Xena database. (B) Correlation of TMEM200A expression in the TCGA PANCAN cohort with patient age in each tumor. (C) Correlation of TMEM200A expression in the TCGA PANCAN cohort with tumor pathological grade in each tumor. (D) Correlation of TMEM200A expression in the TCGA PanCan cohort with the survival status of patients in each tumor. (E) Correlation analysis of TMEM200A expression with pan-cancer overall survival in various cancers using the Cox regression model. (F) Correlation between TMEM200A expression and pan-cancer OS prognosis in the TCGA PanCan cohort. (G) Correlation between TMEM200A expression and disease-specific survival. (H) Correlation between TMEM200A expression and progression-free interval prognosis in the TCGA PanCan cohort. (I) Correlation between TMEM200A expression and disease-free interval. (J) A multifactorial COX regression analysis was performed on KIRC. (K) A multifactorial COX regression analysis was performed on KIRP. Abbreviations: TCGA = The Cancer Genome Atlas; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; OS = Overall survival; DSS = Disease-specific survival; PFI = Progression-free interval; DFI = Disease-free interval. Please click here to view a larger version of this figure.
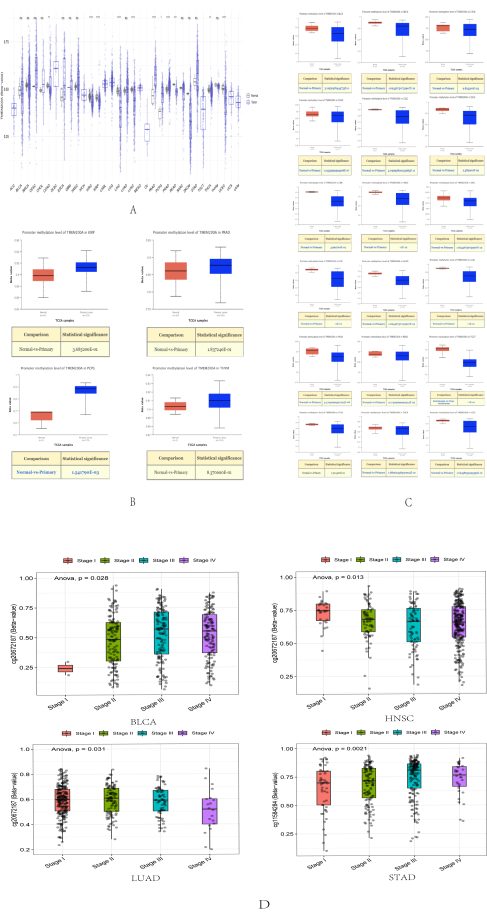
Figure 4: DNA methylation analysis of TMEM200A in different cancers. (A) Promoter methylation levels of TMEM200A were examined in several cancer types, according to the SMART database. (B) Higher promoter methylation levels of TMEM200A in different cancer types than in normal tissues were examined in accordance with the UALCAN database. (C) Using the UALCAN database, it was determined that normal tissues had lower promoter methylation levels of TMEM200A in several cancer types. (D)The DNA methylation level of TMEM200A in BLCA, HNSC, LUAD, and STAD changed with the pathological stage. Abbreviations: BLCA=Bladder Urothelial Carcinoma; HNSC= Head and Neck squamous cell carcinoma; LUAD=Lung adenocarcinoma; STAD= Stomach adenocarcinoma. Please click here to view a larger version of this figure.
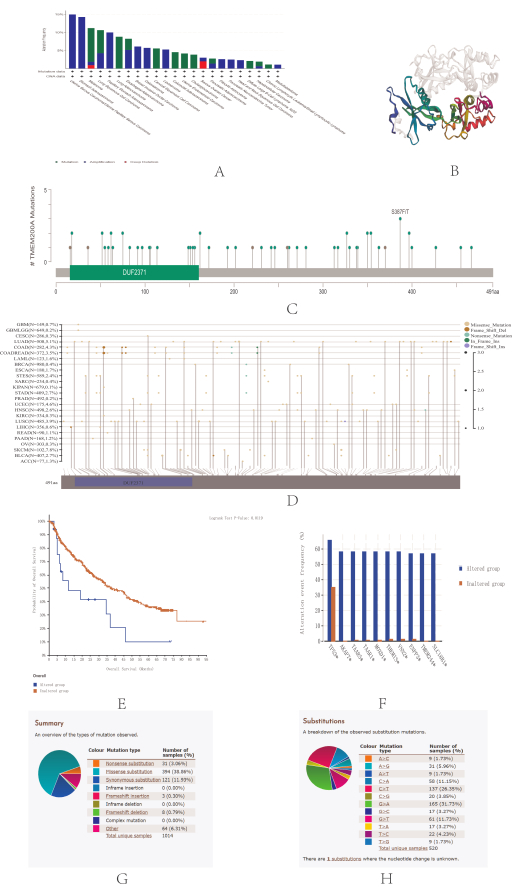
Figure 5: TMEM200A gene mutations in different cancers. (A) Analysis of TMEM200A gene mutation types in different cancers using the cBioPortal database. (B) 3D protein structure of TMEM200A. The binding area is indicated by the colored portion, whereas the other sections of TMEM200A are shown by the gray portion. (C) Isoforms and distribution of TMEM200A somatic mutations. X-axis, amino acid sites; y-axis, number of TMEM200A mutations; green dots, missense mutations; and gray dots, truncated mutations. (D) Validation of the isoforms and distribution of TMEM200A somatic mutations using data in Sangerbox 3.0. (E) For all TCGA tumors, the correlation between mutation status and patients' overall survival prediction was investigated using the cBioPortal database, with red squares indicating the TMEM200A mutation group and blue squares indicating the unmutated group. (F) Genes co-mutated with TMEM200A were analyzed using the cBioPortal tool: TP53, AKAP7, TAAR5, TAAR1, MOXD1, THEMIS, VNN2, ENPP3, TMEM244, and SLC18B1. (G) Analysis of TMEM200A mutation types in GC using the COSMIC database. (H) An analysis of TMEM200A SNV types in GC was conducted using the COSMIC database. Abbreviations: TCGA = The Cancer Genome Atlas; OS = Overall Survival; COSMIC = Catalogue of Somatic Mutations in Cancer Cells; GC = Gastric cancer. Please click here to view a larger version of this figure.
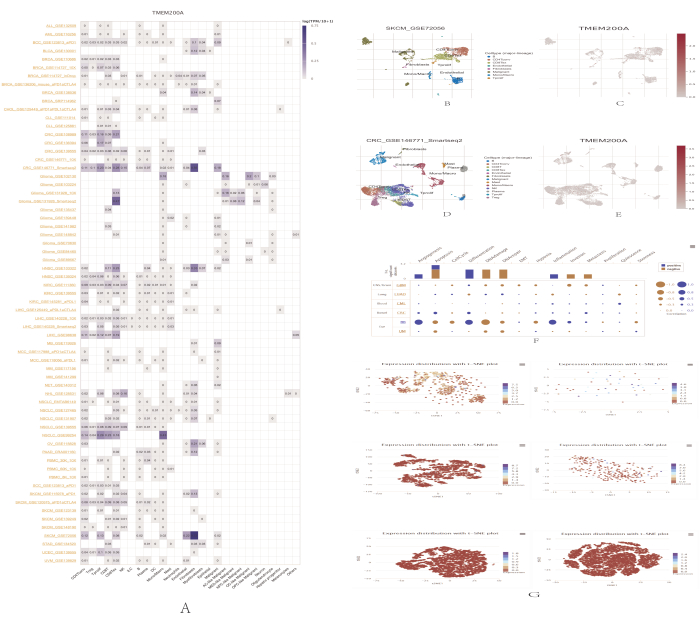
Figure 6: Single-cell correlation analysis of TMEM200A expression in different cancers. (A) Summary of the expression of TMEM200A on the TISCH website. (B) Distribution of eight distinct cell types in the metastatic cutaneous melanoma GSE72056 dataset. (C) Expression levels of TMEM200A in cutaneous metastatic melanoma cells from the GSE72056 dataset. (D) Distribution of 13 distinct cell types in the colorectal cancer dataset from GSE146771. (E) TMEM200A expression levels in colorectal cancer cells from the GSE146771 dataset. (F) Correlation of TMEM200A expression with single-cell functional status in multiple tumors was demonstrated using the CancerSEA database. (G) T-SNE maps illustrating the levels of TMEM200A expression in individual tumor cells, including GBM, LUAD, CML, CRC, RB, and UM. Abbreviations: GBM = Glioblastoma; LUAD = Lung adenocarcinoma; CML = Chronic granulocytic leukemia; CRC = Colorectal cancer; RB = Retinoblastoma; UM = Uveal melanoma. Please click here to view a larger version of this figure.
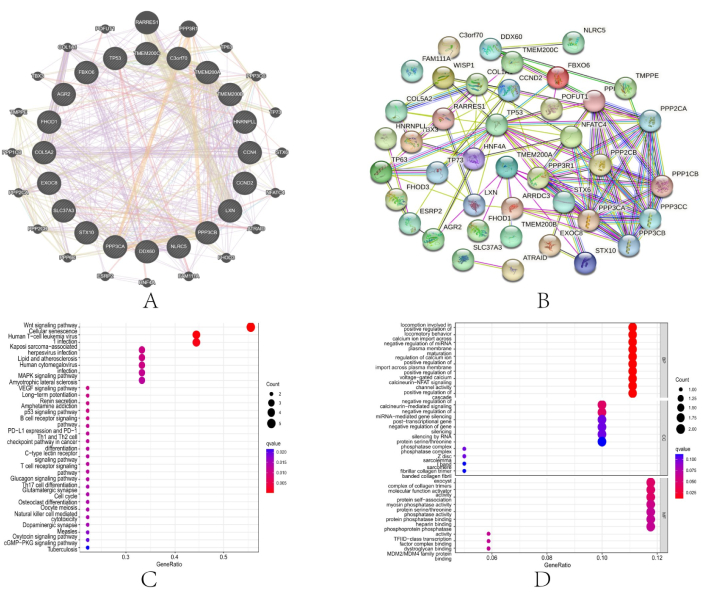
Figure 7: Co-expression network and functional enrichment analysis of TMEM200A at the gene level. (A) GGI network of TMEM200A and its co-expressed genes. (B) PPI network. (C,D) TMEM200A and the co-expressed genes were analyzed for GO and KEGG pathway enrichment. Abbreviations: PPI = Protein-protein interaction; GGI = Gene-gene interaction; KEGG = Kyoto Encyclopedia of Genes and Genomes; GO = Gene Ontology. Please click here to view a larger version of this figure.
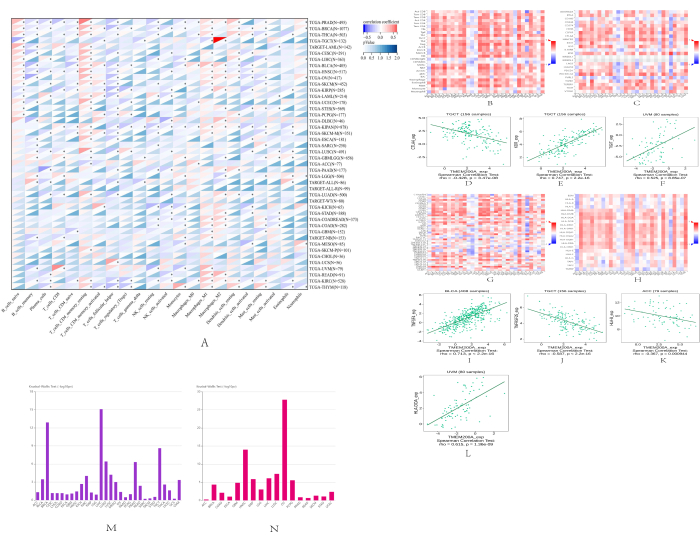
Figure 8: Correlation between the expression of TMEM200A and immune cells, immunosuppressive agents, immunostimulatory substances, and MHC molecules in various cancer types. (A) Sangerbox 3.0 heat map demonstrating the correlation between TMEM200A expression and immune infiltrating cells. (B) Heat map showing the correlation between immune infiltrating cells and TMEM200A expression in the TISIDB database. (C) Heat map showing the correlation between immunosuppressive cells and TMEM200A expression in the TISIDB database. (D–F) Correlation between TMEM200A expression and certain immunosuppressive agents in testicular cancer and uveal melanoma. (G) Heat map showing the correlation between immunostimulatory factors and TMEM200A expression in the TISIDB database. (H) Heat map of the correlation between TMEM200A expression and MHC molecules in the TISIDB database. (I–L) Correlation between TMEM200A expression and some immunostimulatory factors and MHC molecules in the urothelial carcinoma of the bladder, testicular carcinoma, adrenocortical carcinoma, and uveal melanoma. (M) Correlation between pan-cancer immune subgroups and TMEM200A expression. (N) Correlation between pan-cancer molecular subgroups and TMEM200A expression. Abbreviations: MHC = Major histocompatibility complex. Please click here to view a larger version of this figure.
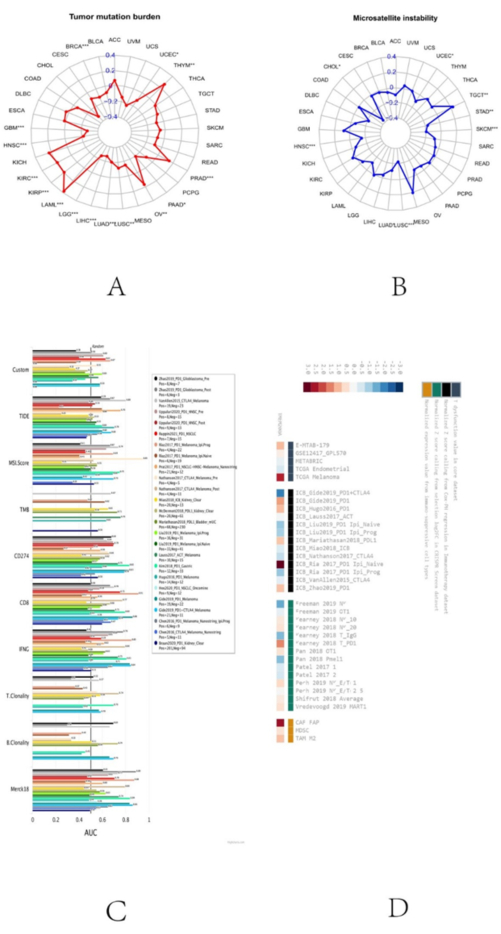
Figure 9: Analysis of immunotherapeutic response of TMEM200A in pancytopenia. (A) Correlation of TMEM200A expression in different cancers with TMB. (B) Correlation between MSI in pan-cancer and TMEM200A expression. (C) TMEM200A's potential as a biomarker for predicting immune checkpoint blockade response. (D) Weighted mean of TMEM200A's correlation with ICB survival result and logarithmic fold change (logFC) in CRISPR screening were used to rank it. Abbreviations: TMB = Tumor mutational burden; ICB = Immune Checkpoint Blockade; CRISPR =Clustered Regularly Interspaced Short Palindromic Repeats. Please click here to view a larger version of this figure.
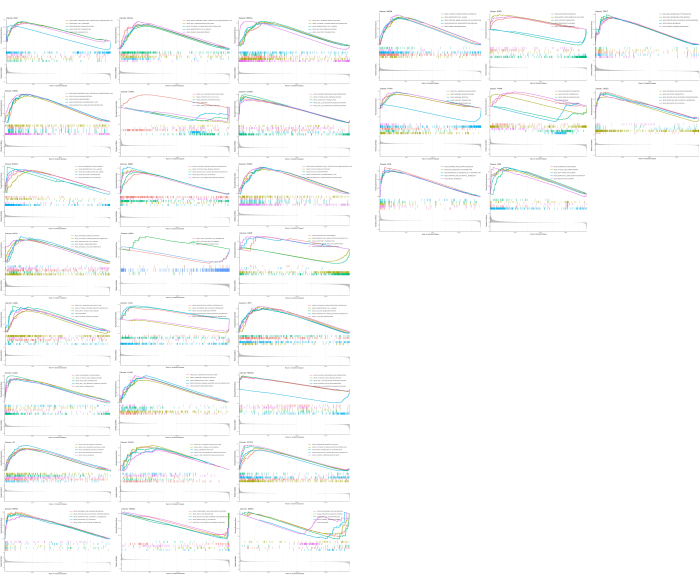
Figure 10: GSEA enrichment analysis of TMEM200A in different cancers. Top five pathways in which TMEM200A played a major functional role in regulating 32 cancers were identified by GSEA enrichment analysis. The left panel shows the results of GSEA enrichment analysis for TMEM200A in ACC, BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PCPG, PRAD, READ, and SARC. The right panel shows the results of GSEA enrichment analysis for TMEM200A in SKCM, STAD, TGCT, THCA, THYM, UCEC, UCS, and UVM. Abbreviations: GSEA =Gene Set Enrichment Analysis; ACC = Adrenocortical carcinoma; BLCA = Bladder Urothelial Carcinoma; BRCA = Breast invasive carcinoma; CESC = Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL =Cholangiocarcinoma; COAD = Colon adenocarcinoma; ESCA = Esophageal carcinoma; GBM = Glioblastoma multiforme; HNSC = Head and Neck squamous cell carcinoma; KICH = Kidney Chromophobe; KIRC = Kidney renal clear cell carcinoma; KIRP = Kidney renal papillary cell carcinoma; LAML = Acute Myeloid Leukemia; LGG = Brain Lower Grade Glioma; LIHC = Liver hepatocellular carcinoma; LUAD = Lung adenocarcinoma; LUSC = Lung squamous cell carcinoma; MESO = Mesothelioma; OV = Ovarian serous cystadenocarcinoma; PAAD =Pancreatic adenocarcinoma; PCPG = Pheochromocytoma and Paraganglioma; PRAD = Prostate adenocarcinoma; READ Rectum adenocarcinoma; SARC=Sarcoma; SKCM = Skin Cutaneous Melanoma; STAD =Stomach adenocarcinoma; TGCT =Testicular Germ Cell Tumors; THCA = Thyroid carcinoma; THYM = Thymoma; UCEC = Uterine Corpus Endometrial Carcinoma; UCS = Uterine Carcinosarcoma; UVM = Uveal Melanoma. Please click here to view a larger version of this figure.
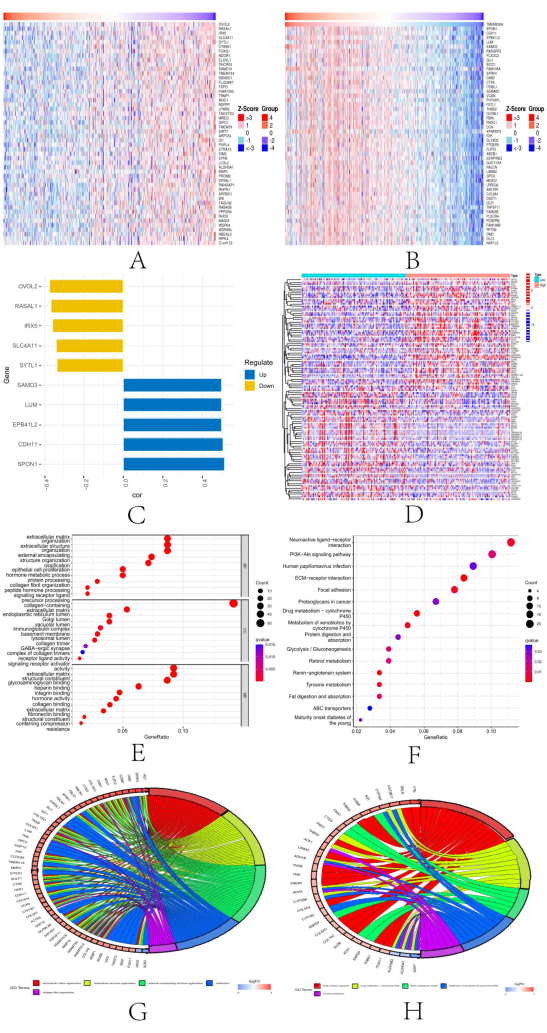
Figure 11: TMEM200A in STAD. (A) Top 50 genes found with a negative correlation with TMEM200A expression in STAD by the LinkedOmics database. (B) Top 50 genes in STAD that were positively connected to TMEM200A. (C) TMEM200A's top five positively and five negatively correlated genes in STAD. (D) Heat map of DEGs in STAD. (E–H) Investigation of enriched GO and KEGG pathways using the screened DEGs. Abbreviations: STAD=Stomach adenocarcinoma; DEGs= Differential expression of genes; KEGG = Kyoto Encyclopedia of Genes and Genomes; GO = Gene Ontology. Please click here to view a larger version of this figure.
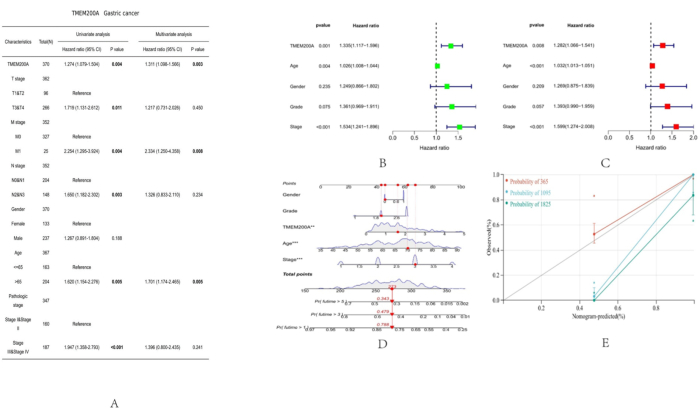
Figure 12: Correlation between the clinical features of STAD and TMEM200A expression level. (A) Analysis of TMEM200A expression levels and STAD clinical traits using logistic regression. (B,C) Cox analysis of STAD forest plots for single and multiple variables. (D) Column line plots predicting OS in patients with STAD based on sex, pathological grade, age, pathological stage, and TMEM200A expression. (E) Calibration plots showing the calibration of the column line graph model. Abbreviations: STAD= Stomach adenocarcinoma; OS=Overall Survival. Please click here to view a larger version of this figure.
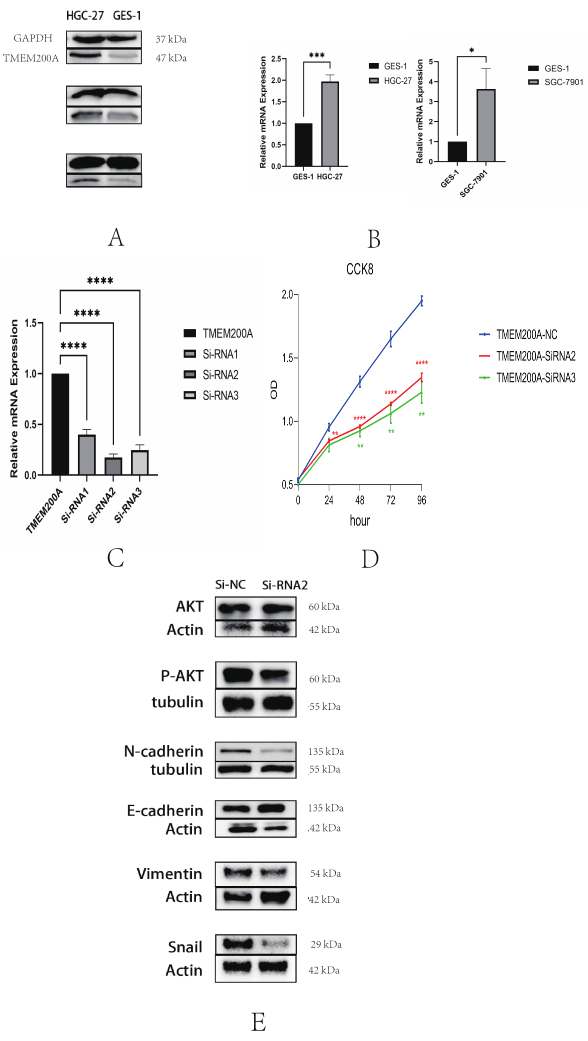
Figure 13: TMEM200A expression is upregulated in STAD and promotes proliferation of gastric cancer cells. (A) Detection of TMEM200A expression in gastric cancer cells HGC-27 and gastric mucosal epithelial cells GES-1 by western blotting. (B) Detection of TMEM200A expression in GC cells HGC-27 and SGC-7901, and gastric mucosal epithelial cells GES-1 by qRT-PCR. (C) The expression efficiency of TMEM200A was greatly reduced in the TMEM200A knockdown group compared to the non-knockdown group in GC cells HGC-27, as demonstrated by qRT-PCR. (D) TMEM200A knockdown significantly inhibited the proliferation of GC cells as measured by CCK8 assay. (E) The knockdown of TMEM200A significantly inhibited the related proteins in EMT and affected the phosphorylation of AKT in the PI3K/AKT signaling pathway. Abbreviation: GC = Gastric cancer; qRT-PCR: Quantitative real-time polymerase chain reaction; EMT=Epithelial-Mesenchymal Transition; AKT=Protein kinase B. Please click here to view a larger version of this figure.
Discussion
TMEM200A belongs to a family of TMEMs that is essential for cancer cells to proliferate38. The variable expression of TMEM200A in different malignancies has received less attention, and a thorough pan-cancer investigation is lacking. However, evidence continues to accumulate, showing that the TMEM transmembrane protein family may be important in keeping cancer cells malignant through interactions with several proteins, for example, activation of TMEM16A Ca2+-activated Cl– channels by ROCK1/moesin promotes BRCA metastasis39. Therefore, in the current work, our analyses focused on exploring the feasibility of TMEM200A as a therapeutic cancer target and tumor diagnostic marker. In particular, we investigated the expression levels of TMEM200A in 33 malignant tumors using RNA-seq data from the UCSC Xena database, and the results from the TIMER2.0 database successfully validated the analyses in the UCSC Xena database.
Subsequently, the Tumor Immune Single Cell Center (TISCH) web tool and CancerSEA database were used to analyze the expression levels of TMEM200A in different types of monocytes. The Sangerbox and TISIDB websites were used to help understand how TMEM200A affects immune infiltration. We also identified the signaling pathways in which TMEM200A plays a key function in each cancer by GSEA enrichment analysis to understand the major functional roles of TMEM200A in each malignancy.
TMEM200A may act as an adhesion molecule to control the immune environment of GC and promote the invasive spread of GC cells. Therefore, we identified 483 differentially expressed genes (DEGs) associated with the expression level of TMEM200A through the TCGA database and analyzed the GO and KEGG enrichment of these 483 DEGs. The enrichment results showed that the PI3K/AKT pathway plays a key role in cancer development, and the PI3K/AKT pathway may be one of the driving factors of cancer.
Further, we carried out cellular and molecular tests after learning more about the crucial function of TMEM200A in driving cancer development to confirm the correlation between TMEM200A expression and STAD cell activity. As expected, we discovered that downregulating TMEM200A in STAD cells greatly reduced cell proliferation, and that cancer cell lines expressed TMEM200A at considerably greater levels than normal cell lines. By combing the literature related to the transmembrane protein family in recent years, we found that transmembrane proteins, as cell adhesion molecules37, have a regulatory role in EMT.
Further, according to the results of the enrichment analysis of TMEM200A-related genes in GC, we found that TMEM200A may have a regulatory role in the PI3K/AKT signaling pathway in GC. Western blotting experiments revealed that TMEM200A knockdown could reduce Vimentin, N-cadherin, and Snai proteins and inhibit AKT phosphorylation, suggesting that TMEM200A regulates the tumor microenvironment by affecting the EMT, and that the PI3K/AKT signaling pathway may be involved in TMEM200A-mediated regulation of GC development. In recent years, some studies have found that EMT is the main cause of chemotherapy drug resistance in GC patients. Epithelial-mesenchymal plasticity (EMP) and tumor microenvironment have been described as limiting factors for effective treatment in many cancer types40. The occurrence of EMT can make GC cells lose their characteristic properties and show the characteristics of mesenchymal cells41. This feature not only reduces the sensitivity of GC patients to chemotherapeutic drugs but also enhances the invasive and migratory ability of GC cells, leading to tumor metastasis. Therefore, for the reasons mentioned above, our study aids the research and development of key regulatory points in EMT and the development of novel targeted therapeutic approaches, by exploring the specific mechanism of action of TMEM200A affecting EMT.
The use of bioinformatics for research has increased in recent years, and for this study, we accessed data from several internet databases. The use of these online databases in bioinformatics has streamlined and sped up the study of cancer, and they also provide researchers with an affordable means of validating their results.
The bioinformatics technique does, however, have certain drawbacks. When we utilize these databases for analysis, the same study may provide inconsistent or even conflicting findings in multiple databases since many online databases include data from several datasets. Furthermore, many databases do not get updated for a very long time, and because of copyright difficulties, their contents can never be enlarged; therefore, the analytical findings that researchers get from these databases are always constrained. Therefore, in this research, we employed different databases to examine the results collectively to overcome these constraints and ensure the correctness of the findings. To be sure that the findings we acquired were not altered considerably, we repeated the procedure when using different databases for analysis. The results of western blot experiments supported our bioinformatics prediction that TMEM200A may control the EMT in GC by regulating the PI3K/AKT signaling pathway, which would then affect the proliferation of GC cells. We predicted the mechanism of action of TMEM200A using bioinformatics in conjunction with related literature.
In summary, this work demonstrates how bioinformatics tools may significantly facilitate experimental design and be used to make many other types of scientific research predictions. Future cancer researchers are likely to adopt the primary paradigm of combining experimental validation with bioinformatics prediction, and this work serves as a good model for this objective.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82160550).
Materials
| Anti-AKT antibody | Proteintech Group, Inc | 60203-2-Ig | |
| Anti-E-cadherin antibody | Proteintech Group, Inc | 20874-1-AP | |
| anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody | Proteintech Group, Inc | 10494-1-AP | |
| Anti-N-cadherin antibody | Proteintech Group, Inc | 22018-1-AP | |
| Anti-P-AKT antibody | Proteintech Group, Inc | 66444-1-Ig | |
| Anti-snail antibody | Proteintech Group, Inc | 13099-1-AP | |
| Anti-Vimentin antibody | Proteintech Group, Inc | 10366-1-AP | |
| AxyPrepMultisourceTotalRNAMini- prep Kit |
Suzhou Youyi Landi Biotechnology Co., Ltd | UEL-UE-MN-MS-RNA-50G | |
| BCA Protein Assay Kit | Epizyme Biotech | ZJ101L | |
| CCK-8 reagent | MedChemExpress | HY-K0301-500T | |
| Fetal bovine serum (FBS) | CYAGEN BIOSCIENCES (GUANGZHOU) INC | FBSSR-01021 | |
| GAPDH primer | Sangon Biotech (Shanghai) Co., Ltd. | Forward primer (5’-3’): TGACATCAAGAAGGTG GTGAAGCAG; Reverse primer (5’-3’): GTGTCGCTGTTGAAG TCAGAGGAG |
|
| HighGene plus Transfection reagent | ABclonal | RM09014P | |
| HRP-conjugated Affinipure Goat Anti-Mouse lgG (H+L) | Proteintech Group, Inc | SA00001-1 | |
| HRP-conjugated Affinipure Goat Anti-Rabbit lgG (H+L) | Proteintech Group, Inc | SA00001-2 | |
| Human gastric mucosal epithelial GES-1 cells | Guangzhou Cellcook Biotech Co.,Ltd. | ||
| Human STAD HGC-27 cells | Procell Life Science&Technology Co.,Ltd | ||
| Human STAD SGC-7901 cells | Procell Life Science&Technology Co.,Ltd | ||
| MonAmp SYBR Green qPCR Mix (None ROX) | Mona (Suzhou) Biotechnology Co., Ltd | MQ10101S | |
| MonScript RTIII All-in-One Mix with dsDNase | Mona (Suzhou) Biotechnology Co., Ltd | MR05101M | |
| Omni-ECL Femto Light Chemiluminescence Kit | Epizyme Biotech | SQ201 | |
| PAGE Gel Fast Preparationb Kit | Epizyme Biotech | PG111 | |
| Penicillin-streptomycin (Pen-Strep) | Beijing Solarbio Science & Technology Co.,Ltd | P1400-100 | |
| Polyvinylidene difluoride (PVDF) membrane | Merck KGaA | IPVH00010-1 | |
| Protein Free Rapid Blocking Buffer | Epizyme Biotech | PS108P | |
| RIPA lysis solution | Beijing Solarbio Science & Technology Co., Ltd | R0010 | |
| RPMI 1640 complete medium | Thermo Fisher Scientific | C11875500BT | |
| Skimmed milk | Campina: Elk | ||
| TBST buffer solution | Beijing Solarbio Science & Technology Co., Ltd | T1082 | |
| The protein loading buffer | Epizyme Biotech | LT101S | |
| TMEM200A knockdown plasmid | MiaoLing Plasmid | ||
| TMEM200A primer | Sangon Biotech (Shanghai) Co., Ltd. | Forward primer (5’-3’): AAGGCGGTGTGGTGGTTCG; Reverse primer (5’-3’): GATTTTGGTCTCTTTGTCACGGTT | |
| TMEM200A SiRNA1 | MiaoLing Plasmid | Forward primer (5’-3’): ACAACTGATGATAAGACCAG; Reverse primer (5’-3’): TGTTGACTACTATTCTGGTC | |
| TMEM200A SiRNA2 | MiaoLing Plasmid | Forward primer (5’-3’): CGTGTGAATGTCAATGACTG; Reverse primer (5’-3’): GCACACTTACAGTTACTGAC | |
| TMEM200A SiRNA3 | MiaoLing Plasmid | Forward primer (5’-3’): ACAACCACAACATCTGCCCG; Reverse primer (5’-3’): TGTTGGTGTTGTAGACGGGC | |
| Transmembrane protein 200A Antibody | Proteintech Group, Inc | 48081-1 | |
| Equipment | |||
| CO2 cell culture incubator | Haier Group | PYXE-80IR | |
| Electrophoresis instrument | Bio-RAD | ||
| Fluorescence quantitative PCR instrument | Bio-RAD | ||
| Gel Imaging System (Tanon 5200) | Tanon Science & Technology Co., Ltd | LAB-0002-0007-SHTN | |
| Multifunctional Enzyme Labeler | Berthold |
Referências
- Torre, L. A., Siegel, R. L., Ward, E. M., Jemal, A. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomarkers Prev. 25 (1), 16-27 (2016).
- Long, X., et al. Economic burden of malignant tumors – Yichang City, Hubei Province, China, 2019. China CDC Wkly. 4 (15), 312-316 (2022).
- Harbeck, N., Gnant, M. Breast cancer. Lancet. 389 (10074), 1134-1150 (2017).
- Bagchi, S., Yuan, R., Engleman, E. G. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 16, 223-249 (2021).
- Lam, G. T., et al. Pitfalls in cutaneous melanoma diagnosis and the need for new reliable markers. Mol Diagn Ther. 27 (1), 49-60 (2023).
- Gromiha, M. M., Ou, Y. Y. Bioinformatics approaches for functional annotation of membrane proteins. Brief Bioinform. 15 (2), 155-168 (2014).
- Schmit, K., Michiels, C. TMEM proteins in cancer: a review. Front Pharmacol. 9, 1345 (2018).
- Marx, S., et al. Transmembrane (TMEM) protein family members: Poorly characterized even if essential for the metastatic process. Semin Cancer Biol. 60, 96-106 (2020).
- Fu, K., et al. Overexpression of transmembrane protein 168 in the mouse nucleus accumbens induces anxiety and sensorimotor gating deficit. PLoS One. 12 (12), e0189006 (2017).
- Cuajungco, M. P., et al. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 114 (7), 705-712 (2012).
- Zhang, S., et al. TMEM116 is required for lung cancer cell motility and metastasis through PDK1 signaling pathway. Cell Death Dis. 12 (12), 1086 (2021).
- Zhang, N., Pan, H., Liang, X., Xie, J., Han, W. The roles of transmembrane family proteins in the regulation of store-operated Ca(2+) entry. Cell Mol Life Sci. 79 (2), 118 (2022).
- Zhang, X., Zheng, P., Li, Z., Gao, S., Liu, G. The somatic mutation landscape and RNA prognostic markers in stomach adenocarcinoma. Onco Targets Ther. 13, 7735-7746 (2020).
- Jia, D., et al. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging (Albany NY). 10 (4), 592-605 (2018).
- Wang, S., et al. UCSCXenaShiny: an R/CRAN package for interactive analysis of UCSC Xena data. Bioinformatics. 38 (2), 527-529 (2022).
- Li, T., et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48 (W1), W509-W514 (2020).
- Thul, P. J., et al. A subcellular map of the human proteome. Science. 356 (6340), eaal3321 (2017).
- Li, Y., Ge, D., Lu, C. The SMART App: an interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin. 12 (1), 71 (2019).
- Chandrashekar, D. S., et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 25, 18-27 (2022).
- Tate, J. G., et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47 (D1), D941-D947 (2019).
- Gao, J., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 6 (269), pl1 (2013).
- Shen, W., et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 1 (3), e36 (2022).
- Ru, B., et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 35 (20), 4200-4202 (2019).
- Yuan, H., et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 47 (D1), D900-D908 (2019).
- Sun, D., et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 49 (D1), D1420-D1430 (2021).
- Warde-Farley, D., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214-W220 (2010).
- Zhu, Y., Feng, S., Song, Z., Wang, Z., Chen, G. Identification of immunological characteristics and immune subtypes based on single-sample gene set enrichment analysis algorithm in lower-grade glioma. Front Genet. 13, 894865 (2022).
- Mueller Bustin, S. A., R, Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci (Lond). 109 (4), 365-379 (2005).
- Sun, L., Zhang, H., Gao, P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 13 (12), 877-919 (2022).
- Ntontsi, P., Photiades, A., Zervas, E., Xanthou, G., Samitas, K. Genetics and epigenetics in asthma. Int J Mol Sci. 22 (5), 2412 (2021).
- Martínez-Reyes, I., Chandel, N. S. Cancer metabolism: looking forward. Nat Rev Cancer. 21 (10), 669-680 (2021).
- Chen, Y., et al. PremPS: Predicting the impact of missense mutations on protein stability. PLoS Comput Biol. 16 (12), e1008543 (2020).
- Li, M., Petukh, M., Alexov, E., Panchenko, A. R. Predicting the impact of missense mutations on protein-protein binding affinity. J Chem Theory Comput. 10 (4), 1770-1780 (2014).
- Hirsch, D., et al. Clinical responses to PD-1 inhibition and their molecular characterization in six patients with mismatch repair-deficient metastatic cancer of the digestive system. J Cancer Res Clin Oncol. 147 (1), 263-273 (2021).
- Poulogiannis, G., Frayling, I. M., Arends, M. J. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology. 56 (2), 167-179 (2010).
- Chintamani, J., et al. The expression of mismatched repair genes and their correlation with clinicopathological parameters and response to neo-adjuvant chemotherapy in breast cancer. Int Semin Surg Oncol. 4, 5 (2007).
- Deng, H., et al. High expression of TMEM200A is associated with a poor prognosis and immune infiltration in gastric cancer. Pathol Oncol Res. 29, 1610893 (2023).
- Stemmler, M. P. Cadherins in development and cancer. Mol Biosyst. 4 (8), 835-850 (2008).
- Bill, A., et al. ANO1/TMEM16A interacts with EGFR and correlates with sensitivity to EGFR-targeting therapy in head and neck cancer. Oncotarget. 6 (11), 9173-9188 (2015).
- De Las Rivas, J., et al. Cancer drug resistance induced by EMT: novel therapeutic strategies. Arch Toxicol. 95 (7), 2279-2297 (2021).
- Tian, S., et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway. Aging (Albany NY). 12 (4), 3574-3593 (2020).

