- 00:00Overview
- 01:05Principles of Accelerated Solvent Extraction
- 02:23Collection of Sample Materials and Preparation of ASE Cells
- 03:48Preparation of Collection Vials and Extraction
- 04:49Applications
- 06:18Summary
침전물에서 생물지표 추출-가속 용매 추출
English
Share
Overview
출처: 제프 살라컵 연구소 – 매사추세츠 대학교 애머스트
고고학 및 박테리아 의 제품군에 의해 생성 된 글리세롤 -dialkyl 글리세롤 테트라에테르 (GDGTs)라는 유기 바이오 마커의 그룹의 분포는 공기 또는 수온1,2에대한 응답으로 예측 가능한 방식으로 변화하는 현대 퇴적물에서 발견되었다. 따라서, 공지된 나이의 퇴적물의 순서로 이러한 바이오마커의 분포는 천년의 시간대에 데카달에 공기 및/또는 수온의 진화를 재구성하는 데 사용될 수있다(도 1). 고생물학이라고 불리는 과거의 기후의 긴 고해상도 기록의 생산은 수백, 아마도 수천 개의 샘플의 신속한 분석에 달려 있습니다. 초음파 처리 또는 Soxhlet과 같은 오래된 추출 기술은 너무 느립니다. 그러나, 새로운 가속 용매 추출 기술은 효율성을 염두에 두고 설계되었다.
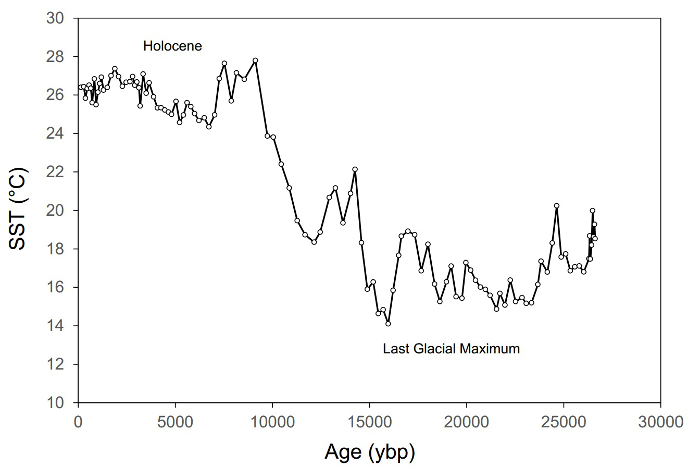
그림 1. 과거 ~27,000년3년동안 지중해 동부의 해수면 온도(SST)의 변화를 보여주는 고생물학 기록의예. 이 기록은 ~115개의 샘플로 구성되며 동위 상류GDGT 계의 TEX86 SST 프록시를 기반으로 합니다.
Principles
Procedure
Results
At the end of the extraction, there is a total lipid extract (TLE) for each sample. Each vial now contains the extractable organic matter from a sediment, soil, or plant tissue. These TLEs can be analyzed, and their chemical constituents identified and quantified.
Applications and Summary
The TLEs of the extracted samples contain a wide spectrum of different organic compounds, including the GDGTs to be used to reconstruct ancient temperatures. Glycerol-dialkyl glycerol-tetraethers are a large suite of biomarkers that show sensitivity to growth temperatures. There are two groups of GDGTs, branched and isoprenoid, which differ in the character of the branching patterns on the core alkyl groups (Figure 3). In the ocean, a cosmopolitan group of archaea, called Thaumarchaeota, produce isoprenoidal GDGTs4. Branched GDGTs are produced on land in soils5, lakes, and in lake sediments6 by as yet unidentified bacteria, likely Acidobacteria7. Both archaea and bacteria adjust the number of methyl branches and ring structures in the core alkyl group according to growth temperature, and because GDGTs are stable in sediments for millions of years, long high resolution records of climate change are generated using them.
The TEX86 paleo water temperature proxy is based on the ratio of certain isoprenoidal GDGTs, each containing 86 carbon atoms in its core alkyl group (Figure 3):
TEX86 = (GDGT-2 + GDGT-3 + GDGT-4') /
(GDGT-1 + GDGT-2 + GDGT-3 + GDGT-4')
Paleo water temperature is then inferred using a calibration, such as the original equation:
TEX86 = 0.015T + 0.28 (R2 = 0.92)
Proposed by Schouten et al.1, where T is paleotemperature. Several other regional and local calibrations have been developed to further refine the proxy for use in large lakes or in the tropics, for example.
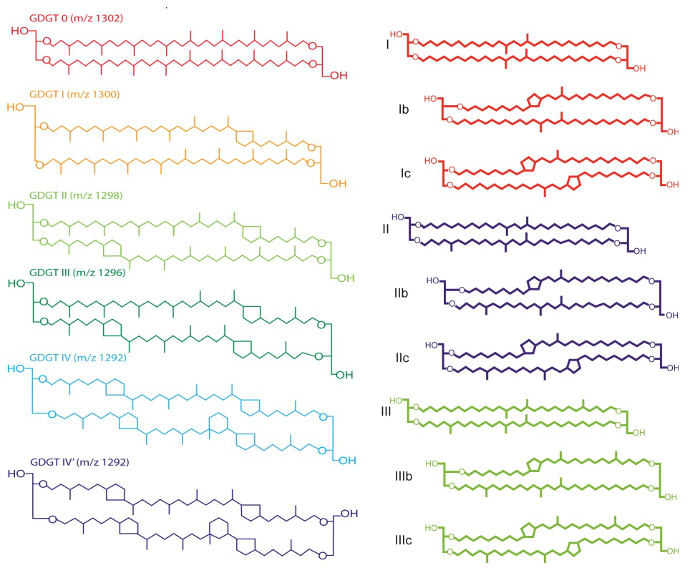
Figure 3. Chemical structures of isoprenoidal and branched GDGTs. Please click here to view a larger version of this figure.
References
- Schouten, S. et al. Distributional variations in marine crenarchaeotal membrane lipids: a new tool for reconstructing ancient sea water temperatures?, Earth and Planetary Science Letters, 204(1-2), 265-274 (2002).
- Weijers, J. W. H. et al. Environmental controls on bacterial tetraether membrane lipid distribution in soils, Geochimica et Cosmochimica Acta, 71(3), 703-713 (2007).
- Castaneda, I. S. et al. Millennial-scale sea surface temperature changes in the eastern Mediterranean (Nile River Delta region) over the last 27,000 years, Paleoceanography, 25, 13 (2010).
- Damste, J. S. S. et al. Crenarchaeol: the characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota, J Lipid Res, 43(10), 1641-1651 (2002).
- Hopmans, E. C. et al. A novel proxy for terrestrial organic matter in sediments based on branched and isoprenoid tetraether lipids, Earth and Planetary Science Letters, 224(1-2), 107-116 (2004).
- Tierney, J. E., Russell J. M. Distributions of branched GDGTs in a tropical lake system: Implications for lacustrine application of the MBT/CBT paleoproxy, Organic Geochemistry, 40(9), 1032-1036 (2009).
- Damste, J. S. S. et al. 13,16-Dimethyl Octacosanedioic Acid (iso-Diabolic Acid), a Common Membrane-Spanning Lipid of Acidobacteria Subdivisions 1 and 3, Appl Environ Microb, 77(12), 4147-4154 (2011).
Transcript
Accelerated solvent extraction is a fast, efficient, and high-throughput technique used to separate organic biomarkers from large numbers of geological sediment samples.
Traditionally, extraction methods such as sonication or Soxhlet are used, however, the drawback with these protocols is that they are too slow to process enough material for detailed paleoclimate reconstruction. A newer technique, the Accelerated Solvent Extraction, or ASE method, was developed with efficiency and high throughput in mind.
The ASE method uses a combination of high temperatures and high pressure to extract samples and can carry out multiple samples in a single, relatively fast preparation run.
This video is the third in a series detailing how to extract biomarkers from sediment. It will cover the procedure and provide insight into merits of ASE over sonication or Soxhlet extraction.
In accelerated solvent extraction, samples are loaded into steel cells that are then loaded onto a carousel. Collection vials for each sample cell are also loaded onto a separate carousel. The instrument loads a sample cell into an internal oven. Solvent is pumped from a solvent bottle through a series of valves until a sufficient pressure is reached.
This pressure is held for an amount of time dictated by the sample and analyte. Then, the solvent is flushed from the sample cell through a steel line into the corresponding collection vial. The process can be repeated a number of times. The temperature, pressure, and duration can all be customized for the sample.
The high temperature used increases the kinetics of the extraction while the high pressure keeps the solvent from volatizing. The collection vial now contains a total lipid extract, and what’s left in the sample cell is called a residue. It comprises non-organic material as well as organic material that is not solvent-extractable, called kerogen.
Now that we are familiar with the principles behind accelerated solvent extraction, let’s take a look at how this is carried out in the laboratory.
After collecting samples of interest; freeze-dry, homogenize, and decontaminate as demonstrated in another video in this series.
Once all samples have been prepared, assemble a cell for each one to be extracted, and one extra for a blank. To do this, screw an end cap onto the cell body. Using solvent-rinsed tweezers, place a combusted glass fiber filter on top of each one. Slowly and gently press the filter down with the plunger.
Label the cell bodies by number for each sample, and label the blank separately. Place a combusted weighing tin onto a laboratory scale and tare. Rinse a spatula with solvent, then use it to transfer 5 to 10 g of sample to the weighing tin, and record the mass.
Transfer all the material in the weighing tin to its corresponding cell. Place another glass fiber filter on top, and gently tamp until it reaches the top of the sample.
Add a dispersant, such as diatomaceous earth or sand, until it is almost full, being careful to avoid getting any debris into the cell body threads. Cap the top of the cell with another end cap. Repeat these steps for each sample and the blank.
Label each collection vial with the number of a corresponding sample cell or blank, and cap with a vial cap. Place each cell into a numbered slot on the upper ASE tray. Set the parameters for the extraction method using the keypad on the ASE to extract at 100 °C and 1,200 psi. Extract each sample 3 times with a static hold of 10 min and flush the cell body with 50% of its total volume between static holds.
Next, ensure that the solvent bottle contains enough to extract all of the samples. Rinse the instrument lines 3 times before starting the run. Finally, press start.
Remove the vial from the ASE. Now that the biomarkers have been extracted, they must be purified before analysis.
Accelerated Solvent Extraction is a versatile technique, which can be utilized for a variety of applications, some of which are explored here.
Accelerated solvent extraction can also be used on other types of sample, including food. Residue analysis to test for pesticide contamination is frequently carried out in regulatory and industrial facilities to ascertain the safety and quality of food products like fruit or vegetables. ASE can be used to extract organochlorine pesticides from food samples, and determine the types or levels of residue present in the produce. This information can then be used to establish whether produce is fit for human or animal consumption. For instance, dieldrin should be found within 0 to 0.1 parts per million on food, depending on the product.
Food nutritional components can also be extracted using ASE. For example, products like chocolate, which may have high gravimetric fat content, can be extracted. Using ASE with petroleum ether as a solvent, fat can be separated from chocolate samples and subjected to quantitative analysis to determine an accurate percentage fat content per known quantity of chocolate. Using this information, regulatory bodies can verify claims made by chocolate manufacturers, or manufacturers can obtain information to create accurate food labels.
You’ve just watched JoVE’s introduction to the extraction of lipid biomarkers using accelerated solvent extraction, or ASE. Methods for further processing and analysis can be found in subsequent videos.
Thanks for watching!
