Methods to Identify the NMR Resonances of the 13C-Dimethyl N-terminal Amine on Reductively Methylated Proteins
Summary
Two methods for assigning the α- and ε-dimethylamine nuclear magnetic resonance signals of a reductively 13C-methylated N-terminal lysine are described. One method utilizes the pH-induced selectivity of the reductive methylation reaction, and the other uses aminopeptidase to selectively remove the N-terminal lysine.
Abstract
Nuclear magnetic resonance (NMR) spectroscopy is a proven technique for protein structure and dynamic studies. To study proteins with NMR, stable magnetic isotopes are typically incorporated metabolically to improve the sensitivity and allow for sequential resonance assignment. Reductive 13C-methylation is an alternative labeling method for proteins that are not amenable to bacterial host over-expression, the most common method of isotope incorporation. Reductive 13C-methylation is a chemical reaction performed under mild conditions that modifies a protein's primary amino groups (lysine ε-amino groups and the N-terminal α-amino group) to 13C-dimethylamino groups. The structure and function of most proteins are not altered by the modification, making it a viable alternative to metabolic labeling. Because reductive 13C-methylation adds sparse, isotopic labels, traditional methods of assigning the NMR signals are not applicable. An alternative assignment method using mass spectrometry (MS) to aid in the assignment of protein 13C-dimethylamine NMR signals has been developed. The method relies on partial and different amounts of 13C-labeling at each primary amino group. One limitation of the method arises when the protein's N-terminal residue is a lysine because the α- and ε-dimethylamino groups of Lys1 cannot be individually measured with MS. To circumvent this limitation, two methods are described to identify the NMR resonance of the 13C-dimethylamines associated with both the N-terminal α-amine and the side chain ε-amine. The NMR signals of the N-terminal α-dimethylamine and the side chain ε-dimethylamine of hen egg white lysozyme, Lys1, are identified in 1H-13C heteronuclear single-quantum coherence spectra.
Introduction
Nuclear magnetic resonance (NMR) spectroscopy is a valuable structure elucidation tool for proteins1. NMR spectroscopy can be used to determine the solution structure of a protein in its native state. To overcome the low natural abundance of stable magnetic isotopes, it is necessary to incorporate 13C and 15N into the protein of interest. The most common method employed is recombinant expression in a bacterial host2-3. However, two disadvantages of bacterial host over-expression are it cannot produce post-translational modifications and does not work for all proteins3-4. When bacterial expression is not a viable route for protein production, over-expression in nonbacterial hosts can be used, but isotopic labeling is difficult and expensive5. Alternative expression methods for incorporating 13C and 15N isotopes into proteins for NMR analysis include sparse labeling techniques using metabolic precursors for methyl labeling6 and single 13C, 15N amino acids7-9. A chemical approach to sparse labeling used herein is the well-established reductive 13C-methylation reaction (Figure 1), where the primary amino groups on a protein – the N-terminal α-amine and the lysine, side chain ε-amines – are methylated. Once monomethylamines are formed, the amine readily undergoes methylation again, due to the higher pKa value, to form dimethylamines.
Reductive methylation was first introduced as a method to chemically modify proteins by Means and Feeney10. The advantages of this reaction are its broad applicability and mild reactions conditions at buffered, physiological pH and low temperatures11,12. In the presence of formaldehyde and a reducing agent, such as dimethylamine borane complex (DMAB), the lysine ε-amino groups and the N-terminal α-amino group are selectively methylated to produce dimethylated amines. Although formaldehyde is known to cross-link proteins through the formation of methylene bridges, this process is blocked by the reducing agent13,14.
Reductive methylation has been successfully used to study proteins with both NMR and x-ray crystallography. Reductive methylation is used to facilitate the crystallization of otherwise intractable proteins15. Hen egg white lysozyme was the first protein crystallized in its dimethylated form. The root-mean square difference between the heavy atoms in the methylated and unmethylated lysozyme structures is 0.40 Å16. This comparison demonstrates that the protein structure can be maintained after reductive methylation, making the reaction a viable, labeling tool for structure elucidation.
By using 13C-labeled formaldehyde in the reductive methylation reaction, 13C-dimethylated amines are produced. The 13C-dimethylamines are NMR-detectable probes that have been widely used to study protein dynamics, structure, and function. NMR and reductive 13C-methylation have been used to study protein-ligand and protein-protein interactions for the β2 adrenergic receptor17, ribonuclease A18, lysozyme19, fd gene 5 protein20, and cytochrome c21. Similarly, structural and functional properties have been studied of reductively 13C-methylated ribonuclease A22, lysozyme23, fd gene 5 protein24, Clostridium pasteurianum ferredoxin25, Fc fragment of IgG26, apolipoprotein A-I27, and MIP-1α28 The dynamics of the 13C-dimethylamino groups have been studied on concanavalin A29-30 and calmodulin31.
Even though reductive 13C-methylation has been used widely to study proteins with NMR, the labeling method has always been limited by the difficulties of assigning the NMR resonances26. Most assignment strategies for reductively 13C-methylated proteins have relied on small numbers of sites,20,28 known structural properties11,19,23,26,31,32. or extensive genetic modifications33. None of these studies successfully assigned all the 13C-dimethylamine peaks except for the calmodulin study, where the peaks were assigned by site-directed mutagenesis of each lysine33. In the study of MIP-1α dimer formation, the use of mass spectrometry (MS) to aid in the NMR assignment of reductively 13C-methylated amines was first reported28. Matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) MS was used to identify the lysine at the interface of the dimer. The partially methylated lysines of human MIP-1α tryptic peptides were identified with MS and correlated with the appearance of mono- and dimethylamine signals of the intact protein observed in 2D 1H-13C HSQC NMR spectra28. Our group expanded on the use of MS and presented an assignment method that requires no prior knowledge of the protein's structure or properties other than the amino acid sequence34. This method is applicable to most proteins. One exception is when a protein has an N-terminal lysine because the MS isotopic profile of the N-terminal lysine α- and ε-13C-dimethylamines cannot be independently measured.
Here we present two methods, one chemical and one enzymatic, to identify the N-terminal α-dimethylamine and the side chain ε-dimethylamine sites of an N-terminal lysine residue. The first method was inspired by Córdova et al. who used pH to control the selectivity of protein acetylation35. The reaction favors the N-terminal α-amine at low pH and the side chain ε-amines at high pH, allowing the protein α- and ε-amino groups to be distinguished, in their studies, with capillary electrophoresis35. We demonstrate how high and low pH is used to alter the labeling of protein amino groups using the reductive 13C-methylation reaction and to allow application of the MS-assisted assignment strategy. The second method to assign the N-terminal α- and ε-13C-dimethylamines takes advantage of the selective removal of the N-terminal residue(s) using recombinant Aeromonas proteolytica aminopeptidase.
Protocol
1. Reductive Methylation of Lysozyme
- Prepare 500 ml of a 50 mM sodium phosphate buffer at pH 7.5. Store at room temperature.
- Label three 1 ml microcentrifuge tubes for fully labeled sample (FLS), low pH partially labeled sample (LpH), and high pH partially labeled sample (HpH).
- Weigh out 10 mg of lysozyme and dissolve in 2 ml of sodium phosphate buffer to create an aqueous solution with a concentration of 5 mg/ml. Cover the tubes with aluminum foil to protect the formaldehyde from degradation due light exposure. Add 500 μl of lysozyme solution to each tube.
- Prepare fresh 1.0 M solutions of dimethylamine borane complex (DMAB) and 13C-formaldehyde. Store at 0 °C.
- For the fully labeled sample, use a molar ratio of 1:10 (reactive amine: formaldehyde), and for the partially labeled samples, use a 1:5 ratio. Alternatively, the ratio can be optimized. For example, in order to see the dominance of the α-13C-dimethylamine peak at low pH, a 2:7 ratio was required.
- Add 6.1 μl of 1.0 M DMAB to FLS, add 3.2 μl of 1.0 M DMAB to LpH and HpH, and gently vortex. Then add 12.3 μl of 1.0 M 13C-formaldehyde to FLS and 6.1 μl of 1.0 M 13C-formaldehyde to LpH and HpH. Shake the reaction mixtures at 4 °C for 2 hr.
- Add another aliquot of 1.0 M DMAB and 13C-formaldehyde as in the previous step and shake the reaction mixture overnight at 4 °C for a total reaction time of 18-24 hr.
- Exchange the sample's buffer to phosphate at pH 7.5 to remove excess reagents and side products using a centrifugal filter (3 kDa molecular weight cut-off).
- For the partially labeled samples, reductively methylate the samples at pH 7.5 with natural abundance formaldehyde to create a homogeneously modified protein. Prepare new 1.0 M solutions of DMAB and natural abundance formaldehyde.
- Cover the sample tubes with foil, add 6.1 μl of DMAB to LpH and HpH, and gently vortex. Then add 12.3 μl of natural abundance formaldehyde. Shake the reaction mixtures at 4 °C for 2 hr.
- Add another aliquot of 1.0 M DMAB and natural abundance formaldehyde as in the previous step and shake the reaction mixtures overnight at 4 °C for a total reaction time of 18-24 hr. Then buffer exchange using steps 2.5-2.6 and store 50 μg samples at -20 °C for MS.
2. Prepare the Sample for NMR Analysis
- Prepare a 2.0 M sodium borate buffer solution at pH 8.5 in H2O.
- Add 375 μl of 2.0 M sodium borate buffer solution to four 15 ml conical tubes. Parafilm the tops and poke holes. Place the flask in a lyophilizing flask and freeze the sample.
- Place the frozen sample on a lyophilizer to dry. Remove when dry.
- Using a pipette, add 15 ml of D2O to each tube and mix.
- Label three 4 ml centrifugal filters (3 kDa molecular weight cut-off) with FLS, LpH, and HpH. Add the corresponding sample to the filter tubes (~500 μl). Add 3.0 ml of 50 mM sodium borate buffer at pH 8.5 to each tube. Make a balance tube for the third tube and concentrate the samples to 500 μl using a centrifuge with a 35° fixed angle rotor at 4 °C and 7,500 rcf.
- Empty the trap under each filter. Add another 3.0 ml of buffer to the samples and concentrate the samples again to 500 μl. Repeat step 2.5 4x.
- Using the bicinchoninic acid (BCA) protein assay36, determine the concentration of each sample. Dilute the lysozyme samples with 50 mM sodium borate buffer to make 150 μM samples.
- Transfer 530 μl to a 5 mm NMR tube or 250 μM to a 5 mm magnetic susceptibility matched tube.
- Prepare an 80 mM stock solution of 1,2-dichloroethane-13C2 in D2O. Store this stock solution at -80 °C. Dilute to 24 mM in 80 μl and transfer to coaxial insert tube to use as a reference in NMR analysis. Analyze the samples with NMR. Note: Run a 1D 1H-13C HSQC and integrate the peak areas for each resonance. Peak areas should be the same for fully labeled sample.
3. Aminopeptidase Degradation of Lysozyme
- Prepare a 5 mg/ml solution of lysozyme in ultrapure water.
- Add 250 μl of the lysozyme solution to a clean, 1 ml centrifuge tube.
- Add 25 μl of a 0.1 M tricine solution, pH 8.0, and 6 μl of a 2.16 μg/ml Aeromonas proteolytica aminopeptidase solution.
- Incubate at 37 °C for 6 hr.
- Add an additional 6 μl of the 2.16 μg/ml aminopeptidase solution to the sample for a 1:50 molar ratio of aminopeptidase to lysozyme. Incubate for an addition 20 hr at 37 °C.
- Desalt 30 μl of the sample using a C18 spin column, (follow manufacturer's protocol). Store at -20 °C for MALDI MS analysis.
- Prepare the remaining protein sample for NMR as described in protocol 2).
4. Prepare Samples for Mass Spectrometry
- Using the 50 μg of sample reserved in step 2.3, exchange the buffer to 100 mM ammonium bicarbonate, pH 8.0, and concentrate to approximately 50 μl using a 1 ml centrifugal filter (3 kDa molecular weight cut-off). Transfer the sample to a 1 ml centrifuge tube and wrap in aluminum foil.
- Add 2.0 μl of 500 mM dithiothreitol to reduce the disulfide bonds. Incubate the solution at 60 °C for 1 hr.
- Add 2 μl of freshly prepared 500 mM iodoacetamide to alkylate the thiol groups. Incubate at room temperature for 20 min.
- Remove the foil and add 1.0 μl of the 500 mM dithiothreitol solution to quench the reaction.
- Add 2.0 μg of sequencing grade trypsin to digest the protein. Incubate the mixture at 37 °C for 16-24 hr.
- Add TFA until the pH is less than 2 to quench the digestion. (Formic and acetic acids can be used as well)
- Desalt the sample on a C18 spin column and elute the protein in 80% acetonitrile/0.1% TFA. Dry the sample using a vacuum concentrator.
- Reconstitute the sample in 5.0 μl of 50% acetonitrile/0.1% TFA for MALDI MS analysis. Mix equal volumes of the reconstituted sample and a saturated solution of 2,5-dihydroxybenzoic acid matrix solution on the MALDI target plate. Analyze the samples on a MALDI MS. Note: Use tandem MS to confirm the identity of peptide peaks. Smooth the mass spectra and analyze. For each dimethylamine-containing peptide, determine the peak intensity and peak area of each isotope in the profile. Calculate the percentage of 13C-incorporation as described previously34.
Representative Results
Reductive Methylation and pH
A pH induced chemoselective reductive 13C-methylation reaction has been demonstrated on lysozyme. At low pH, the reaction prefers the N-terminal α-amine over the side chain ε-amines and vice versa at high pH. In solution the protein's amino groups exist in equilibrium between the free amine and the conjugate acid, which is tunable with pH. The optimum pH range for this reaction depends on the reducing agent used. In this case, dimethylamine borane has an optimum pH of 7.0-9.0. Working outside the optimum range of the reducing agent does make the reaction less efficient, which becomes an advantage in this case. The reductive methylation reaction preferentially modifies the N-terminal α-amine at pH 6.0 due to the lower acid dissociation constant (pKa) of the α-amine (~7)23 versus the ε-amines (~10)32. At pH 6.0 the equilibrium is drastically shifted to favor the conjugate acid; the α-amine has the highest concentration of free amine, the reactive form in the reductive methylation reaction. At pH 10.0, the equilibrium favors the free amine. Even though the α-amine with its lower pKa exists nearly entirely in the free amine form, the ε-amines are stronger bases and more reactive. In the presence of limiting 13C-formaldehyde, we are able to take advantage of the differing rates of reaction between the ε-amines and the α-amine. Reductive 13C-methylation of lysozyme at pH 10.0 with a molar ratio of 1:5 (moles of reactive amine: moles of 13C-formaldehyde) achieved methylation at each of the side chain ε-amines but not at the α-amine site, due to the weaker nucleophilic nature of the α-amine. As illustrated in Figure 2, peak 7 is absent for the lysozyme sample reacted at pH 10.0, indicating that peaks 1-6 belong to ε-dimethylamino groups and peak 7 is the N-terminal α-dimethylamino group. α-Amine favored reductive 13C-methylation at pH 6.0 was observed when the amount of 13C-formaldehyde used was reduced to a molar ratio of 2:7 and the sample was not further reacted with natural abundance formaldehyde. As illustrated in Figure 3, the methylation at low pH is a mixture of monomethylamines (13C-chemical shift ~32-34 ppm) and dimethylamines (13C-chemical shift ~43-44 ppm). Three amino groups show 13C-dimethylation, indicating that these groups reacted faster than the groups showing 13C-monomethylation. Of the 3 13C-dimethylamines, peak 7 has the highest intensity and was assigned as the N-terminal α-dimethylamino group, corroborating the assignment of peak 7 using the sample reacted at high pH.
With the ability to alter the rate of reductive methylation of the N-terminal α-amino group, the MS-assisted method for assigning the NMR resonances can be applied34. The lysozyme sample that was reductively 13C-methylated at pH 10.0 with a 5:1 ratio was digested with trypsin and analyzed with MALDI-TOF MS. Because the α-amino group was not reductively 13C-methylated under these conditions, as shown in Figure 2, the MS isotopic profile of the tetramethylated, N-terminal peptide can be used to quantify the percentage of 13C-incorporated at the N-terminal ε-dimethylamino group. The 13/12C-dimethylated N-terminal peptide, K(13/12CH3)4VFGR, with a mass-to-charge ratio (m/z) of 662, gave a scaled percentage of 13C-incorporation of 27%. NMR analysis of the same lysozyme sample gave a percentage of 13C-incorporation of 33% for peak 6, indicating that peak 6 is the N-terminal Lys1 ε-13C-dimethylamine.
Aminopeptidase and Reductive Methylation
MALDI-TOF mass spectra of untreated- and aminopeptidase-treated lysozyme samples are shown in Figure 4. Insulin, which was used as a positive control, shows a shift to lower average m/z compared to its native form, 5731.2 to 5242.7. The treated lysozyme shows a shift from 14305.8 to 13930.5, corresponding to the loss of the first three N-terminal residues of lysozyme, Lys1-Val2-Phe3. Figure 5 shows the NMR spectra of the untreated- and aminopeptidase-treated reductively 13C-methylated lysozyme samples. Compared to the control spectrum (Figure 4a), the spectrum of the aminopeptidase-treated sample (Figure 4b) shows a decrease in intensity for peaks 6 and 7 and an additional peak labeled 7a. From this data, peaks 6 and 7 can be assigned pairwise as Lys1 α- and ε-dimethylamine.
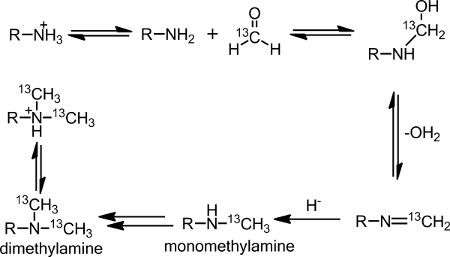
Figure 1. Reaction scheme of the reductive methylation reaction.
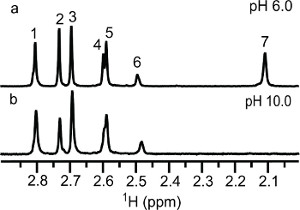
Figure 2. 1D 1H-13C HSQC NMR spectra obtained at 25 °C and pH 8.5 of reductively 13C-methylated lysozyme at reaction conditions of a 1:5 molar ratio (amine: 13C-formaldehyde) and (a) pH 6.0 and (b) pH 10.0 followed by reductive methylation with natural abundance formaldehyde in a ratio of 1:10 at pH 7.5.
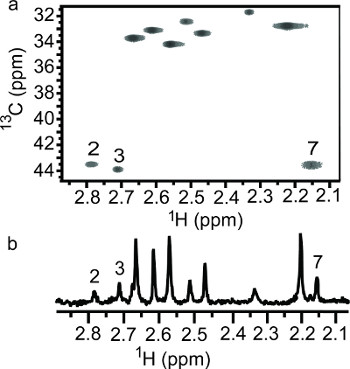
Figure 3. NMR spectra of lysozyme reductively 13C-methylated with a 2:7 molar ratio (amine: 13C-formaldehyde). (a) 2D 1H-13C HSQC and (b) 1D 1H-13C HSQC.
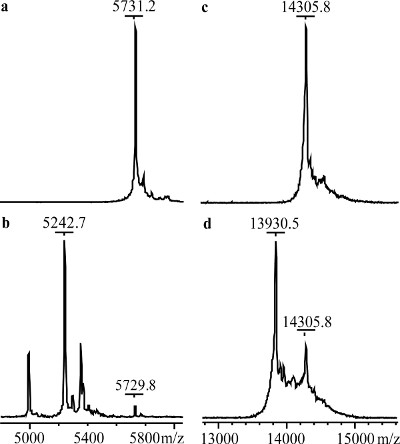
Figure 4. MALDI-TOF mass spectra. (a) Bovine pancreas insulin, (b) aminopeptidase-treated bovine pancreas insulin, (c) lysozyme, and (d) aminopeptidase-treated lysozyme.
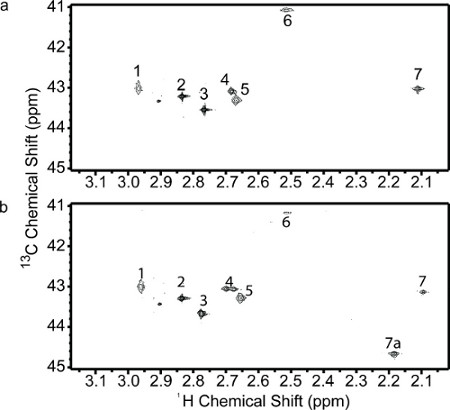
Figure 5. 2D 1H-13C HSQC NMR spectra. (a) Reductively 13C-methylated lysozyme and (b) Aminopeptidase-treated reductively 13C-methylated lysozyme.
Discussion
Assigning the NMR signals of reductively 13C-methylated proteins is necessary to fully utilize this isotopic labeling method. The use of MS to aid in the assignment of the NMR signals thru correlation of the partial 13C-incorporation data is a promising technique. The advantage of this technique over other assignment methods is that only the primary amino acid sequence is needed. The MS-assisted assignment method is limited when the N-terminal amino acid is a lysine because the 13C-incorporation at the α-dimethylamine and ε-dimethylamine cannot be measured separately with MS. Titration experiments usually show a pH dependence of the α-dimethylamine chemical shift, but salt bridges and other structural contributions make the chemical shift an unreliable assignment method. Using the methods presented not only solves this problem, but also circumvents the problem of measuring individual 13C %-incorporation for the α- and ε-dimethylamino peptide with MS. Since one site is labeled, the 13C %-incorporation can be directly measured with MS. The previous assignments of Lys1 α- and ε-dimethylamine were ambiguous because only the average 13C %-incorporation was measured34. With the pH and aminopeptidase methods, the α- and ε-dimethylamine peaks can be assigned directly and agree with assignments made by others23,37,38. Controlling the pH of the reductive 13C-methylation and using aminopeptidase provide two, complementary methods to distinguish the NMR signals of the N-terminal α- and ε-dimethylamine groups.
The reductive methylation reaction is more efficient at higher protein concentrations. Reductive methylation of lysozyme was carried out at a 5 mg/ml protein concentration. The concentration used depends on the solubility and aggregation tendencies of the protein of interest. For example, if dimerization of the protein occurs and one of the lysine residues sits at the interface of the dimer, then complete methylation may be inhibited as demonstrated by Fisher et al. with human MIP-1α28. In this case, the protein concentration was used to control the selectivity of the reductive methylation reaction. To prevent aggregation or precipitation, lower protein concentrations and/or different buffers should be tested, keeping in mind that the buffer cannot have reactive amino groups.
When comparing different reductive methylation conditions, it is important to minimize variability and analyze replicate samples. This is mainly achieved by using stock solutions from which each subsequent sample is produced. For example, a stock protein solution at the proper concentration can be divided into smaller aliquots appropriate for the reaction. Stock solutions of the reagents, DMAB and formaldehyde, should be prepared to aliquot the appropriate molar ratio. It is important to make the DMAB and formaldehyde solutions fresh before use. To reduce the loss of protein through precipitation, protein samples should not be concentrated beyond the reaction concentration when exchanging the buffer. Using a standard BCA assay to determine reductively methylated protein final concentration, prepare 50-200 μM samples to be analyzed with NMR. Depending on the NMR instrument, even lower concentrations can be used, making the method amenable to proteins in limited supply.
The NMR spectra presented were obtained at 25 °C in a borate buffer at pH 8.5. The relative chemical shifts of the dimethylamine resonances depend on the type of buffer, pH, and temperature of the sample. Chemical shifts presented here are slightly different from previous literature due to different buffer conditions. Dichloroethanewas used as an external reference in these experiments for two reasons. The initial reason was to normalize the peak areas between samples for the MS-assisted assignment method. Since the dichloroethane peak does not overlap with the dimethylamine peaks, we used it as a chemical shift reference as well; however, DSS and chemical shift referencing ratios can be used instead.
The reaction conditions for the aminopeptidase digestion should be optimized for the protein of interest. A molar ratio of 1:100 was needed to digest lysozyme, while a 1:50 ratio was sufficient for the control protein, insulin. The buffer may also need to be optimized as was the case for lysozyme. The digestion did not work for lysozyme in phosphate buffer, but was successful in a tricine solution. The zwitterionic tricine is known to stabilize the charged intermediates formed during aminopeptidase digestion39. Although this protocol calls for pH 8, the aminopeptidase used has a pH range of 6-9. Also, there are a number of other aminopeptidases that work at varying pH ranges and are specific to certain residues. For those proteins not soluble at high pH, this method is still applicable through a combination of the low pH reaction and aminopeptidase treatment. It should be noted that some proteins cannot be digested with aminopeptidase while they are in their native structure because the N-terminal residue(s) may be buried and inaccessible. If the protein can be denatured (fully or partially) and refolded, then aminopeptidase can be used to digest the denatured protein. The converse could also be a problem, when the protein structure changes significantly upon truncation of the N-terminus. In this case, limiting the aminopeptidase concentration and time of the reaction should be used to optimize the digestion. The aminopeptidase method is useful for identifying the N-terminal α-dimethylamine and lysine, ε-dimethylamines near the N-terminus.
For the MS-assisted assignment method, the reductively methylated protein is digested with a protease to produce peptides. Trypsin was used with reductively methylated lysozyme, but other proteases can be used. The choice of protease(s) depends on the primary amino acid sequence of the protein and the need to isolate each dimethylamine to a peptide. Trypsin specifically cleaves the peptide bonds on the C-terminal side of arginine and lysine residues, but not of dimethyl-lysine residues40. Trypsin has good activity in ammonium bicarbonate, pH 8 buffer. The choice of buffer and trypsin concentration minimizes interferences during MS analysis. Reduction and alkylation of the cysteines reduce the complexity of the peptide mixture be preventing the formation of random disulfide bridges between peptides. Breaking the disulfide bonds of lysozyme also allows better access of trypsin and reduces the number of missed cleavages.
The procedures described herein utilize both NMR and MS to quantify isotopic labeling of the N-terminal residue of proteins. These incorporated isotopes can then be used for structural and functional studies of the protein. Careful attention must be paid to the details of this method to achieve the desired results. The procedure is tunable and can be optimized for different proteins of interest. These methods are beneficial because they do not require extensive prior knowledge of the target protein.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by Award Number R00RR024105 from the National Center For Research Resources, National Institute of Health.
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| Acetonitrile | Sigma | 34851 | |
| Amicon Ultra 4 ml centrifugal filter (3k MWCO) | Fisher Scientific | UFC800308 (8pk) | |
| Aminopeptidase | Creative Biomart | Aminopeptidase-86A | |
| Ammonium bicarbonate | Sigma | A6141 | |
| Ammonium sulfate | Sigma | A5132 | |
| Borane-dimethylamine complex | Sigma | 180238 | Make fresh 1 M solution before use. |
| Boric acid | Sigma | B6768 | |
| Bovine pancreas insulin | Sigma | I5500 | |
| C18 Spin Columns | Thermo Scientific | 89870 | |
| Deuterium oxide | Cambridge Isotope Laboratories | DLM-4-100 | |
| 1,2-dichloroethane-13C2 | Sigma | 714321 | 80 mM stock solution stored at -80 °C. Used as reference in NMR spectra at 24 mM. |
| 2,5-dihydroxybenzoic acid | Acrōs Organics | 165200050 | |
| Formaldehyde (36.5% wt/wt) | Sigma | 33220 | Make fresh 1 M solution before use. |
| 13C-formaldehyde (99%) | Cambridge Isotope Laboratories | CLM-806-1 | Make fresh 1 M solution before use. |
| Lysozyme (hen egg white) | Sigma | L6376 | |
| Sodium phosphate (dibasic heptahydrate) | Sigma | S9390 | |
| Sodium phosphate (monobasic) | Sigma | S9638 | |
| Tricine | Sigma | T0377 | |
| Trifluoroacetic acid | Sigma | 302031 | |
| 700 MHz Varian VNMRS with 5 mm HCN 5922 probe | Agilent Technologies | ||
| Bruker UltrafleXtreme MALDI TOF/TOF | Bruker |
References
- Bax, A., Grzesiek, S. Methodological Advances in Protein NMR. Accounts of Chemical Research. 26 (4), 131-138 (1993).
- Shimba, N., Yamada, N., Yokoyama, K., Suzuki, E. Enzymatic Labeling of Arbitrary Proteins. Analytical Biochemistry. 301 (1), 123-127 (2002).
- Palomares, L., Estrada-Moncada, S., Ramírez, O., Balbás, P., Lorence, A. Production of Recombinant Proteins. Recombinant Gene Expression. 267, 15-51 (2004).
- Graslund, S., et al. Protein Production and Purification. Nature Methods. 5 (2), 135-146 (2008).
- Terwilliger, T. C., Stuart, D., Yokoyama, S. Lessons from Structural Genomics. Annual Review of Biophysics. 38, 371-383 (2009).
- Goto, N. K., Kay, L. E. New Developments in Isotope Labeling Strategies for Protein Solution NMR Spectroscopy. Curr. Opin. Struct. Biol. 10 (5), 585-592 (2000).
- Lian, L., Middleton, D. Labelling Approaches for Protein Structural Studies by Solution-State and Solid-State NMR. Progress in Nuclear Magn. Reson. Spectroscopy. 39 (3), 171-190 (2001).
- Mason, A., Siarheyeva, A., Haase, W., Lorch, M., van Veen, H., Glaubitz, C. Amino Acid Type Selective Isotope Labelling of the Multidrug ABC Transporter LmrA for Solid-State NMR Studies. Febs Lett. 568 (1-3), 117-121 (2004).
- Chen, C., et al. Preparation of Amino-Acid-Type Selective Isotope Labeling of Protein Expressed in Pichia pastoris. Proteins:Structure Function and Bioinformatics. 62 (1), 279-287 (2006).
- Means, G., Feeney, R. Reductive Alkylation of Amino Groups in Proteins. Biochem. 7 (6), 2192 (1968).
- Jentoft, J. E., Jentoft, N., Gerken, T. A., Dearborn , D. G. C-13 NMR-Studies of Ribonuclease-A Methylated With Formaldehyde-C-13.. J. Biol. Chem. 254 (11), 4366-4370 (1979).
- Rayment, I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Macromolecular Crystallography, Pt A. 276 (97), 171-179 (1997).
- French, D., Edsall, J. T. The Reactions of Formaldehyde with Amino Acids and Proteins. Adv. Protein Chem. 2, 277-335 (1945).
- Jentoft, N., Dearborn, D. G. Labeling of Proteins by Reductive Methylation Using Sodium Cyanoborohydride. J. Biol. Chem. 254 (11), 4359-4365 (1979).
- Walter, T., et al. Lysine Methylation as a Routine Rescue Strategy for Protein Crystallization. Struct. 14 (11), 1617-1622 (2006).
- Rypniewski, W., Holden, H., Rayment, I. Structural Consequences of Reductive Methylation of Lysine Residues in Hen Egg-White Lysozyme – An X-ray Analysis at 1.8-Angstrom Resolution. Biochem. 32 (37), 9851-9858 (1993).
- Bokoch, M. P., et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nat. 463 (7277), (2010).
- Jentoft, J., Gerken, T., Jentoft, N., Dearborn, D. . C-13] Methylated Ribonuclease-A – C-13 NMR Studies of the Interaction of Lysine 41 with Active Site. 256 (1), 231-236 (1981).
- Sherry, A., Teherani, J. Physical Studies of C-13-Methylated Concanavalin A – pH and Co2+-Induced Nuclear Magn. Reson. Shifts.. J. Biol. Chem. 258 (14), 8663-8669 (1983).
- Dick, L., Sherry, A., Newkirk, M., Gray, D. Reductive Methylation and C-13 NMR Studies of the Lysyl Residues of fd Gene 5 Protein – Lysines 24, 46, and 69 May be Involved in Nucleic Acid Binding.. J. Biol. Chem. 263 (35), 18864-18872 .
- Moore, G. R., et al. N-epsilon,N-epsilon-dimethyl-lysine cytochrome c as an NMR probe for lysine involvement in protein-protein complex formation. Biochem. J. 332, 439-449 (1998).
- Jentoft, J. E., Gerken, T. A., Jentoft, N., Dearborn, D. G. Studies of the Active-Site Lysine of Ribonuclease-A By C-13 NMR.. Biophys. J. 25 (2), 56 (1979).
- Gerken, T., Jentoft, J., Jentoft, N., Dearborn, D. Intramolecular Interactions of Amino Groups in C-13 Reductively Methylated Hen Egg-White Lysozyme. J. Biol. Chem. 257 (6), 2894-2900 (1982).
- Dick, L. R., Geraldes, C., Sherry, A. D., Gray, C. W., Gray, D. M. C-13 NMR of Methylated Lysines of fd Gene-5 Protein – Evidence for A Conformational Change Involving Lysine-24 Upon Binding of A Negatively Charged Lanthanide Chelate. Biochem. 28 (19), 7896-7904 (1989).
- Gluck, M., Sweeney, W. V. C-13-NMR of Clostridium-Pasteurianum Ferredoxin After Reductive Methylation of the Amines Using C-13 Formaldehyde. Biochimica Et Biophysica Acta. 1038 (2), 146-151 (1990).
- Jentoft, J. E. Reductive Methylation and C-13 Nuclear-Magnetic-Resonance in Structure-Function Studies of Fc Fragment and Its Subfragments. Methods in Enzymology. 203, 261-295 (1991).
- Sparks, D., Phillips, M., Lundkatz, S. The Conformation of Apolipoprotein A-I in Discoidal and Spherical Recombinant High Density Lipoprotein Particles – C-13 NMR Studies of Lysine Ionization. J. Biol. Chem. 267 (36), 25830-25838 (1992).
- Ashfield, J., et al. Chemical Modification of a Variant of Human MIP-1 Alpha, Implications for Dimer Structure.. Protein Science. 9 (10), 2047-2053 (2000).
- Goux, W., Teherani, J., Sherry, A. Amine Inversion in Proteins – A C-13-NMR Study of Proton Exchange and Nitrogen Inversion Rates in N-Epsilon,N-Epsilon,N-Alpha,N-Alpha-[C-13]Tetramethyllysine, N-Epsilon,N-Epsilon,N-Alpha,N-Alpha[C-13]Tetramethyllysine Methyl-Ester, and Reductively Methylated Concanavalin-A.. Biophysical Chemistry. 19 (4), 363-373 (1984).
- Sherry, A. D., Keepers, J., James, T. L., Teherani, J. Methyl Motions in C-13 Methylated Concanavalin-as Studied by C-13 Magnetic-Resonance Relaxation Techniques. Biochemistry. 23 (14), 3181-3185 (1984).
- Huque, M., Vogel, H. C-13 NMR Studies of the Lysine Side Chains of Calmodulin and Its Proteolytic Fragments. Journal of Protein Chemistry. 12 (6), 695-707 (1993).
- Brown, L., Bradbury, J. Proton-Magnetic-Resonance Studies of Lysine Residues of Ribonuclease-A. Eur. J. Biochem. 54 (1), 219-227 (1975).
- Zhang, M., Vogel, H. Determination of the Side Chain pKa Values of the Lysine Residues in Calmodulin. J. Biol. Chem. 268 (30), 22420-22428 (1993).
- Macnaughtan, M., Kane, A., Mass Prestegard, J. Spectrometry Assisted Assignment of NMR Resonances in Reductively C-13-Methylated Proteins. J. Am. Chem. Soc. 127 (50), 17626-17627 (2005).
- Cordova, E., Gao, J., Whitesides, G. M. Noncovalent Polycationic Coatings for Capillaries in Capillary Electrophoresis of Proteins. Anal. Chem. 69 (7), 1370-1379 (1997).
- Smith, P. K., et al. Measurement of Protein Using Bicinchoninic Acid. Anal. Biochem. 150 (1), 76-85 (1985).
- Larda, S. T., Bokoch, M. P., Evanics, F., Prosser, R. S. Lysine methylation strategies for characterizing protein conformations by NMR. J. Biomolec. NMR. 54 (2), 199-209 (2012).
- Bradbury, J. H., Brown, L. R. Determination of Dissociation Constants of Lysine Residues of Lysozyme by Proton-Magnetic-Resonance Spectroscopy. Eur. J. Biochem. 40 (2), 565-576 (1973).
- Frey, S. T., Guilmet, S. L., Egan, R. G., Bennett, A., Soltau, S. R., Holz, R. C. Immobilization of the Aminopeptidase from Aeromonas proteolytica on Mg2+/Al3+ Layered Double Hydroxide Particles. Acs Appl. Mater., & Interfaces. 2 (10), 2828-2832 (2010).
- Poncz, L., Dearborn, D. G. The Resistance to Tryptic Hydrolysis of Peptide-Bonds Adjacent to N-epsilon,N-Dimethyllysyl Residues. J. Biol. Chem. 258 (3), 1844-1850 (1983).

