Coculture System with an Organotypic Brain Slice and 3D Spheroid of Carcinoma Cells
Summary
The organotypic brain slice coculture with carcinoma cells enables visualizing morphological changes by fluorescence as well as bright field (video) microscopy during the process of carcinoma cell invasion of brain tissue. This model system also allows for cell exchange and replenishment approaches and offers a wide variety of manipulations and analyses.
Abstract
Patients with cerebral metastasis of carcinomas have a poor prognosis. However, the process at the metastatic site has barely been investigated, in particular the role of the resident (stromal) cells. Studies in primary carcinomas demonstrate the influence of the microenvironment on metastasis, even on prognosis1,2. Especially the tumor associated macrophages (TAM) support migration, invasion and proliferation3. Interestingly, the major target sites of metastasis possess tissue-specific macrophages, such as Kupffer cells in the liver or microglia in the CNS. Moreover, the metastatic sites also possess other tissue-specific cells, like astrocytes. Recently, astrocytes were demonstrated to foster proliferation and persistence of cancer cells4,5. Therefore, functions of these tissue-specific cell types seem to be very important in the process of brain metastasis6,7.
Despite these observations, however, up to now there is no suitable in vivo/in vitro model available to directly visualize glial reactions during cerebral metastasis formation, in particular by bright field microscopy. Recent in vivo live imaging of carcinoma cells demonstrated their cerebral colonization behavior8. However, this method is very laborious, costly and technically complex. In addition, these kinds of animal experiments are restricted to small series and come with a substantial stress for the animals (by implantation of the glass plate, injection of tumor cells, repetitive anaesthesia and long-term fixation). Furthermore, in vivo imaging is thus far limited to the visualization of the carcinoma cells, whereas interactions with resident cells have not yet been illustrated. Finally, investigations of human carcinoma cells within immunocompetent animals are impossible8.
For these reasons, we established a coculture system consisting of an organotypic mouse brain slice and epithelial cells embedded in matrigel (3D cell sphere). The 3D carcinoma cell spheres were placed directly next to the brain slice edge in order to investigate the invasion of the neighboring brain tissue. This enables us to visualize morphological changes and interactions between the glial cells and carcinoma cells by fluorescence and even by bright field microscopy. After the coculture experiment, the brain tissue or the 3D cell spheroids can be collected and used for further molecular analyses (e.g. qRT-PCR, IHC, or immunoblot) as well as for investigations by confocal microscopy. This method can be applied to monitor the events within a living brain tissue for days without deleterious effects to the brain slices. The model also allows selective suppression and replacement of resident cells by cells from a donor tissue to determine the distinct impact of a given genotype. Finally, the coculture model is a practicable alternative to in vivo approaches when testing targeted pharmacological manipulations.
Protocol
This new model is an adaptation of a previously published organotypic hippocampal brain slice approach9-12. Modifications and additions were introduced to optimize the cancer cell-brain tissue interactions and to guarantee reproducibility. The study was reviewed and approved by the local ethics committee. Animals were treated carefully according to the guidelines for animal care at the University Medicine Göttingen. The organotypic brain slice coculture can be subdivided into two steps. Step one is the preparation of the organotypic brain slice. Step two comprises the tumor cell preparation and deposition in the coculture model.
1. Organotypic Brain Slice
- Prepare the dissection medium consisting of minimum essential medium (MEM), supplemented with 0.2 mM glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin, and 4.5 mg/ml of glucose.
- Decapitate the mice from any mouse strain between postnatal day six and eight (P6-8).
- Remove the brain rapidly from the skull under aseptic conditions and transfer it to ice-cold dissection medium.
- Remove the frontal pole and the cerebellum from the whole brain section.
- Fix and stabilize the brain on a stage with cryo glue and 5% agarose.
- Slice the brain sections horizontally to a 350 μm thickness by using a vibratome.
- Collect four to six whole brain slices from a single mouse brain, depending on the species and age.
- Prepare the culture medium consisting of 50% MEM, 25% Hanks' balanced salt solution (HBSS), 25% normal horse serum (NHS), 0.2 mM glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin (Sigma, Munich, Germany), and 4.5 mg/ml glucose.
- Put each organotypic brain slice on a 0.4 μm polycarbonate transwell membrane insert in a six-well plate with 1 ml of cultivation medium in the lower well.
- Culture organotypic brain slices overnight in a humidified atmosphere with 5% CO2 at 37 °C incubator.
2. The Slice Coculture Model
- Embed 105 of GFP-transfected tumor or other cells (e.g. MCF-7-GFP cells) in 20 μl gel matrix, consisting of 15% RPMI medium and 85% ECM gel.
- Place the MCF-7- gel matrix mix into a sterile metallic spacer (3.8 mm diameter) directly adjacent to the cortical region of the organotypic brain slice and incubate for 2 hr.
- Remove the spacer and allow the 3D tumor spheroid to coculture with the organotypic slice for 24-96 hr.
- Change the culture medium every other day.
3. Immunofluorescence Staining of Astrocytes and Microglial in the Organotypic Brain Slice Coculture
- Fix the organotypic brain slice coculture with 4% paraformaldehyde for 8 hr at 4 °C.
- Wash the slice coculture with PBST (PBS with 0.5% Triton X-100) for 5 min.
- Block the samples with normal goat serum in PBST (1:20) at room temperature for 1 hr.
- Stain the astrocytes by incubating the brain slice coculture with anti-glial fibrillary acidic protein monoclonal antibody (GFAP, 1:200 in PBST) for 36 hr at 4 °C followed by goat anti-mouse-TRITC (1:100 in PBST) staining for 1 hr at room temperature.
- Wash the samples with PBST three times for 5 min.
- Stain the microglial cells with ILB4-Alexa Fluor 647 (1:100 in PBST) for 1 hr at room temperature.
- Counterstain the brain slice coculture with DAPI (1:1,000) for 3 min at room temperature.
- Mount and coverslip the brain slice coculture with DAKO fluorescent mounting medium.
- Evaluate the grade of tumor invasion based on the following scoring system: 0 = none of the cells; + < 1/3; ++ = 1/3 – 2/3; +++ ≥ 2/3 of the invaded cells (first measure the length of the contact section between the tumor plug and slice, then measure the fraction of contact detectable by the invading cells).
4. Live Imaging of the Interaction between Glial and Tumor Cells
- Perform the experiment under a Leica inverted DMI 6000B microscope at 10X magnification lens and a Leica DFC 350 FX CCD camera.
Representative Results
First, all used carcinoma cells (human: MCF-7 and murine: 410.4) invaded the organotypic mouse brain slice. The invasion was therefore species-independent (Figure 1A), which indicates a wide range of applications in a species-independent manner. Additionally, microglia as well as astrocytes accumulated at the interface as previously described in vivo and in patient samples13. Time-lapse imaging over an extended period of time not only revealed the viability of the slice but also offered a good platform to observe the cellular interactions. By time-lapse experiments, microglial cells were recorded to enter the 3D cell sphere while, in turn, cancer cells also invaded the brain slice (Figures 1B1-B2). Moreover, we have previously shown the ability of microglia to transport carcinoma cells by a still enigmatic mechanism to thereby assist in the carcinoma invasion. Using immunofluorescence labeling techniques, we described co-localizations of tumor cells and stromal cells (e.g. microglia and astrocytes), both in the brain tissue and in the tumor cell plug (Figures 1C-D), suggesting a tight interaction between these cells during the invasion process. Comparable results were obtained when mouse brain slices were cocultured either with human (Figures 1C1-C3) or murine (Figures 1D1-D3) carcinoma cells – showing the wide range of applications for studying cells from different species, genotypes and strains.
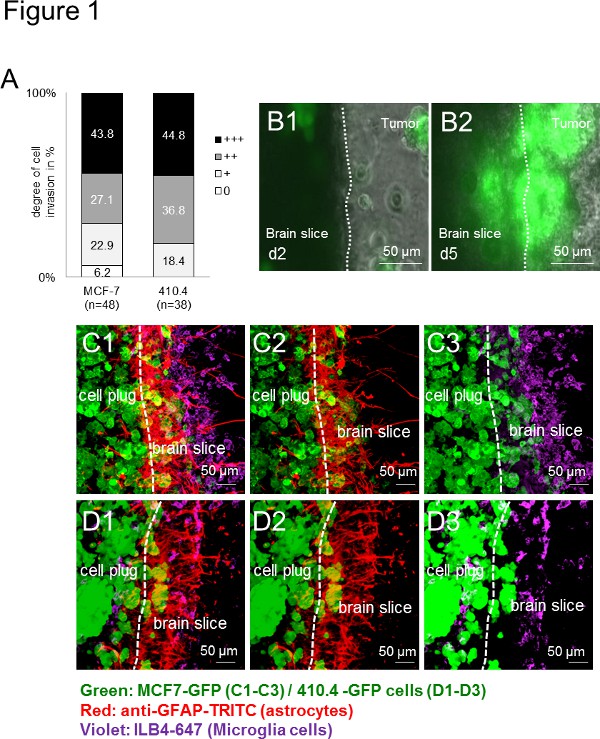
Figure 1. Brain slice coculture model with a 3D spheroid of mouse and human carcinoma cells. A) Quantification of cancer cell invasion in organotypic whole brain slice cocultures with a mouse breast cancer cell line 410.4 or a human breast cancer cell line MCF-7. Data represent the percentage of the degree of cell invasion into brain slice in each group with n ≥ 38. There was no significant difference between these two groups (Kruskal-Wallis test). B) Time-lapse images of organotypic whole brain slice cocultures with MCF-7-GFP cells and representative images from day2 (B1) and day 5 (B2) were shown. C-D) Confocal microscopy images showed cohorts of carcinomas, astrocytes and microglia. Double staining of astrocytes (anti-GFAP-TRITC, red) and microglia (ILB4-Alexa Fluor 647, violet) of the whole coculture with the 350 μm thick organotypic brain slice and the GFP-transfected tumor cell plug (green): 3D-MCF-7-GFP (C1-C3) and 3D-410.4-GFP (D1-D3). White dashes showed the edge of the brain slice and the tumor invasion front. Scale bars represent 50 μm. Microglia, astrocytes and tumors colocalizations (C1 and D1), astrocytes-tumors colocalizations (C2 and D2) and microglia-tumor colocalizations (C3 and D3) in the slice coculture. Click here to view larger image
Discussion
Previous histological investigations of cerebral metastasis demonstrated rapid and drastic changes of the resident glial cells, especially of astrocytes and microglia13. To study these changes and interactions with the carcinoma cells, this novel coculture system is well-suited. Other research fields already have long-standing experience with organotypic hippocampal brain slices. One advantage is that the organotypic hippocampal brain slice cultures are viable and effectively preserved for days to weeks, making it suitable for long-term experiments. Since Stoppini's introduction of the organotypic hippocampal slice system in 1991, it has widely been used, for example in research of degenerative diseases. Thus, this innovate coculture system represents a modification of a well-established approach with a range of applications in tumor biology9,11. The modification offered us a reproducible model to evaluate the grade of tumor invasion afterwards and cultures grown by the interface method are ideally suited for experiments that require a three-dimensional structure. Several techniques have been used to coculture organotypic hippocampal slices with other cells. These include an indirect system between macrophage cells and the organotypic brain slice14, and a direct coculture of two different slices from the hippocampal region15. Glioma aggregates have also been cocultured with brain slices16. These models can be used to analyze cellular and molecular events in the brain slices but do not allow direct, physiological contact between tumor cells, microglia and the brain parenchyma. Furthermore, this method allows the observation of microglia without contamination of bone marrow-derived peripheral monocytes/macrophages. The use of CCR2 and CX3CR1 transgenic mouse model is a critical improvement due to the fact that it is difficult to distinguish the resident microglia from invading monocytes based on their similar properties17,18,19. Though intracerebral injection of carcinoma cells allows investigating tumor progression, it cannot tell much as to whether the surrounded macrophage-like cells originate from the brain-resident microglia population or from bone marrow-derived peripheral monocytes/macrophages17. The team of Kettenmann introduced an organotypic brain slice model that involved inoculating glioma cells into brain slices with a micromanipulator 20. However, primary malignant gliomas differ in many regards from metastatic carcinomas. First, malignant gliomas are of mesenchymal origin, do not metastasize outside the nervous system, and migrate/invade as single cells with no border between tumor and brain tissue. By contrast, infiltrative growth is a typical pathological characteristic, and such carcinomas usually migrate/invade as cohorts. Second, carcinomas often try to rebuild epithelial structures in the brain, while glial cells try to separate the tumor from the brain tissue by a (pseudo) capsule. Considering biological and morphological features, malignant glioma and metastasis of carcinomas are not really comparable. For these reasons, we modified and developed a coculture system where we do not inject but coculture a carcinoma cell plug adjacent to the brain slice. Furthermore, we observed microglial and astrocytic accumulation at the border of the tumor plug, meaning that microglia enter the tumor plug and could be easily detected by bright field microscopy and confirmed afterwards by confocal microscopy. The cancer cells invade into the brain slice, which is comparable to the real in vivo situation and to an observation made by Baumert and colleagues, who found an infiltration zone in 63% of autopsy cases with brain metastases21.
Because of the missing blood perfusion, there are only resident macrophages/microglia in this culture. Moreover, because of the missing T cells and, therefore, absent allo-reactivity, human carcinoma cells could be used for coculture even with brain slices of immunocompetent mice or rats (NMRI, B6, or Wistar). This could serve as an alternative to the nude mouse model. Since four to five slices can be obtained from each mouse, significantly reduced numbers of animals are required, in comparison to the existing injection models. In addition, the animals do not suffer for a long period of metastatic disease and they do not undergo operative procedures repetitively22.
With this coculture system, we have demonstrated the activation of microglia by cancer cells and the capacity of promoting cancer cell invasion. Additionally, this is the first time, to our knowledge, microglia were found to actively transport carcinoma cells23.
Despite all these advantages, the coculture system has, indeed, also limitations. It is still an in vitro model missing the steps of metastasis prior to colonization. Because of the lack of perfusion, it is not possible to study the extravasation. Thus, the alternative way to study the extravasation is to use either the in vivo injection model or the modified Boyden chamber system with extracellular matrix, HUVEC and astrocytes to mimic the blood-brain barrier22,24. The brain slice coculture method is yet a reliable and reproducible model with many advantages and a potential for a wide variety of applications, such as analysis of colonization, in particular with a focus on the role of the resident cells. The combination with other established techniques (such as immunohistochemistry, confocal microscopy and time-lapse microscopy) supports the investigation of direct cell-to-cell interactions. It is an easy alternative and complementation of other techniques and offers access to the investigation of the cues and effects as imposed by the metastatic microenvironment.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Chalid Ghadban for his excellent technical assistance, Andreas Wodarz and Stephan Heermann for their technical advice regarding confocal and time-lapse microscopy. There is no conflict of interests for any of the authors. This work is funded by the German Research Council (DFG) in Project 2 of Forschergruppe 942 (FOR942 BI 703/3-1), by the Dres. Bayer-Stiftung (Baden Württembergischer Krebspreis, Germany) and by the Research Program of the Faculty of Medicine, Georg-August-University Göttingen, Germany.
Materials
| Material Name | Company | Catalogue number | Comment |
| Hank's balanced salt solution (HBSS) | Gibco, Darmstadt,Germany | 24020 | |
| Minimum essential medium (MEM) | Gibco, Darmstadt , Germany | 32360 | |
| RPMI-1640 | PAA Laboratories Inc., Cölbe, Germany | E15-840 | |
| Dulbecco's phosphate buffered saline (PBS) | Pan Biotech, Aidenbach, Germany | P04-36500 | |
| Normal horse serum (NHS) | Invitrogen, Karlsruhe, Germany | 16050-122 | |
| Fetal calf serum (FCS) | Invitrogen, Karlsruhe, Germany | 10091148 | |
| Glucose | B. Braum, Melsungen, Germany | ||
| L-Glutamine-Penicillin-Streptomycin solution | Sigma, Steinheim, Germany | G1146 | |
| Vibratome | Leica, Wetzlar, Germany | Leica VT1200S | |
| Microtome | Leica, Wetzlar, Germany | Leica SM 2000R | |
| Polycarbonate membrane | BD Falcon, Heidelberg,Germany | 353090 | transwell membrane insert (0.4 μm pore size) |
| ECM gel | Trevigen, R&D, Wiesbaden-Nordenstadt, Germany | 3432-005-01 | Basement membrane extract |
| Metallic spacer | Kig GmbG, Kirkel, Germany | DIN 433 | |
| Confocal microscope | Zeiss, Göttingen, Germany | LSM 510 | |
| Time-lapse microscope and camera | Leica, Wetzlar, Germany | DMI 6000B microscope and a DFC 350 FX CCD camera | |
| Anatomy microscope | Zeiss, Göttingen, Germany | Stemi SV11 | |
| Paraformaldehyde | Merck, Darmstadt, Germany | 1.04005.1000 | |
| Mouse monoclonal anti-glial fibrillary acidic protein (GFAP) antibody | Sigma, Steinheim, Germany | G3893 | |
| Goat anti-mouse IgG, F(ab')2– TRITC | Santa Cruz, Heidelberg, Germany | SC3796 | |
| Isolectin GS-IB4 from Griffonia simplicifolia, Alexa Fluor 647 conjugate | Invitrogen, Karlsruhe, Germany | 132450 | |
| DAPI (4',6'-diamidino-2-phenylindole dihyfrochloride) | Sigma, Steinheim, Germany | D8417 | |
| Fluorescent mounting medium | DAKO, Glostrup, Denmark | S3023 |
References
- Langley, R. R., Fidler, I. J. The seed and soil hypothesis revisited–the role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer. 128, 2527-2535 (2011).
- Joyce, J. A., Pollard, J. W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 9, 239-252 (2009).
- Mantovani, A., Allavena, P., Sica, A., Balkwill, F. Cancer-related inflammation. Nature. 454, 436-444 (2008).
- Fidler, I. J. The role of the organ microenvironment in brain metastasis. Semin. Cancer Biol. 21, 107-112 (2011).
- Rath, B. H., Fair, J. M., Jamal, M., Camphausen, K., Tofilon, P. J. Astrocytes enhance the invasion potential of glioblastoma stem-like cells. PloS one. 8, e54752 (2013).
- Steeg, P. S. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 12, 895-904 (2006).
- Eichler, A. F., et al. The biology of brain metastases-translation to new therapies. Nat. Rev. Clin. Oncol. 8, 344-356 (2011).
- Winkler, F., et al. Imaging glioma cell invasion in vivo reveals mechanisms of dissemination and peritumoral angiogenesis. Glia. 57, 1306-1315 (2009).
- Stoppini, L., Buchs, P. A., Muller, D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 37, 173-182 (1991).
- Kreutz, S., Koch, M., Ghadban, C., Korf, H. W., Dehghani, F. Cannabinoids and neuronal damage: differential effects of THC, AEA and 2-AG on activated microglial cells and degenerating neurons in excitotoxically lesioned rat organotypic hippocampal slice cultures. Exp. Neurol. 203, 246-257 (2007).
- De Simoni, A., Yu, L. M. Preparation of organotypic hippocampal slice cultures: interface method. Nat. Protoc. 1, 1439-1445 (2006).
- Fuller, L., Dailey, M. E. Preparation of rodent hippocampal slice cultures. CSH Protoc. 2007, pdb prot4848 (2007).
- Lorger, M., Felding-Habermann, B. Capturing changes in the brain microenvironment during initial steps of breast cancer brain metastasis. Am. J. Pathol. 176, 2958-2971 (2010).
- Brana, C., Biggs, T. E., Mann, D. A., Sundstrom, L. E. A macrophage hippocampal slice co-culture system: application to the study of HIV-induced brain damage. J. Neurosci. Methods. 90, 7-11 (1999).
- Kim, J. A., Yamada, M. K., Nishiyama, N., Matsuki, N., Ikegaya, Y. Mossy fiber pathfinding in multilayer organotypic cultures of rat hippocampal slices. Cell. Mol. Neurobiol. 23, 115-119 (2003).
- Matsumura, H., Ohnishi, T., Kanemura, Y., Maruno, M., Yoshimine, T. Quantitative analysis of glioma cell invasion by confocal laser scanning microscopy in a novel brain slice model. Biochem. Biophys. Res. Commun. 269, 513-520 (2000).
- Charles, N. A., Holland, E. C., Gilbertson, R., Glass, R., Kettenmann, H. The brain tumor microenvironment. Glia. , (2011).
- Mizutani, M., et al. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J. Immunol. 188, 29-36 (2012).
- Kettenmann, H., Hanisch, U. K., Noda, M., Verkhratsky, A. Physiology of microglia. Physiol. Rev. 91, 461-553 (2011).
- Markovic, D. S., Glass, R., Synowitz, M., Rooijen, N., Kettenmann, H. Microglia stimulate the invasiveness of glioma cells by increasing the activity of metalloprotease-2. J. Neuropathol. Exp. Neurol. 64, 754-762 (2005).
- Baumert, B. G., et al. A pathology-based substrate for target definition in radiosurgery of brain metastases. Int. J. Radiat. Oncol. Biol. Phys. 66, 187-194 (2006).
- Palmieri, D., Chambers, A. F., Felding-Habermann, B., Huang, S., Steeg, P. S. The biology of metastasis to a sanctuary site. Clin. Cancer Res. 13, 1656-1662 (2007).
- Pukrop, T., et al. Microglia promote colonization of brain tissue by breast cancer cells in a Wnt-dependent way. Glia. 58, 1477-1489 (2010).
- Bos, P. D., et al. Genes that mediate breast cancer metastasis to the brain. Nature. 459, 1005-1009 (2009).

