Scalable High Throughput Selection From Phage-displayed Synthetic Antibody Libraries
Summary
A method is described with visual accompaniment for conducting scalable, high throughput selections from phage-displayed combinatorial synthetic antibody libraries against hundreds of antigens simultaneously. Using this parallel approach, we have isolated antibody fragments that exhibit high affinity and specificity for diverse antigens that are functional in standard immunoassays.
Abstract
The demand for antibodies that fulfill the needs of both basic and clinical research applications is high and will dramatically increase in the future. However, it is apparent that traditional monoclonal technologies are not alone up to this task. This has led to the development of alternate methods to satisfy the demand for high quality and renewable affinity reagents to all accessible elements of the proteome. Toward this end, high throughput methods for conducting selections from phage-displayed synthetic antibody libraries have been devised for applications involving diverse antigens and optimized for rapid throughput and success. Herein, a protocol is described in detail that illustrates with video demonstration the parallel selection of Fab-phage clones from high diversity libraries against hundreds of targets using either a manual 96 channel liquid handler or automated robotics system. Using this protocol, a single user can generate hundreds of antigens, select antibodies to them in parallel and validate antibody binding within 6-8 weeks. Highlighted are: i) a viable antigen format, ii) pre-selection antigen characterization, iii) critical steps that influence the selection of specific and high affinity clones, and iv) ways of monitoring selection effectiveness and early stage antibody clone characterization. With this approach, we have obtained synthetic antibody fragments (Fabs) to many target classes including single-pass membrane receptors, secreted protein hormones, and multi-domain intracellular proteins. These fragments are readily converted to full-length antibodies and have been validated to exhibit high affinity and specificity. Further, they have been demonstrated to be functional in a variety of standard immunoassays including Western blotting, ELISA, cellular immunofluorescence, immunoprecipitation and related assays. This methodology will accelerate antibody discovery and ultimately bring us closer to realizing the goal of generating renewable, high quality antibodies to the proteome.
Introduction
With the onset of the post-genomic age, the availability of high quality binding reagents to characterize and modulate proteins is essential to open new research and therapeutic avenues. Antibodies continue to be critical to both academic and industrial researchers as basic research and diagnostic tools and potential therapeutics. Not surprisingly, there has been an impressive growth of contract antibody development firms, most of which rely upon conventional hybridoma technologies to generate custom antibodies. Nevertheless, in vitro selection using phage-displayed antibody libraries is becoming a powerful alternative technology that can offer unique advantages and success where conventional technologies can face limitations1, 2.
In light of the considerable demand for high quality antibodies as research tools, two primary challenges for generating renewable antibodies are 1) selection throughput and 2) antigen availability. A number of groups have now described in vitro selection pipelines aimed at increasing throughput and the rate of antibody identification. These descriptions detail a variety of viable approaches that include selecting upon either full-length targets3,4, or structurally related domains5,6,7, using either bead-based6,8 or plate-based3,4 antigen immobilization schemes. In addition, the growing adoption of gene synthesis technologies9 has made systematic antigen generation, particularly of isolated domains, reasonably cost effective and can potentially alleviate the difficulty in obtaining sufficient quantities of purified, full-length antigen. By using the two technologies in tandem, a self-contained and scalable antigen generation and antibody selection pipeline was devised that would enable the parallel isolation of antibodies for large sets of expressed antigen domains and facilitate the development of reagents for characterizing entire classes of structurally or functionally related proteins.
Toward this aim, an integrated pipeline that couples in silico identification of expressible antigen domains, gene synthesis, high-throughput bacterial expression of antigens and scalable phage-displayed antibody selections has been developed. This pipeline requires only basic infrastructure available to most life science laboratories (including antibody libraries which are increasingly available through license or material transfer agreement), but is also amenable to automation for use on an industrial scale. Using this protocol, it is possible to generate hundreds of affinity-tagged antigen domains, and routinely isolate highly specific antibody fragments to many of these antigens.
Phage display technology has shown demonstrated compatibility with a wide variety of recombinant affinity reagent formats including Fab, scFv, and autonomous Fv domains and a growing array of small ‘alternative frameworks’ (designed ankyrin repeat proteins (DARPINS), fibronectin (Fn), lipocalin domains and more10). Discussion is restricted in this example to the isolation of Fab antibody fragments, although it is presumed these methods can be adapted to other types of libraries. Using this technology, Fabs with low nanomolar affinity to small, tagged protein domains including transcription factor domains, SH2 domains, RNA-binding proteins and others have been successfully selected, many of which bind full-length protein and are functional in immunoassays such as immunofluorescence, immunoprecipitation and immunohistochemistry. Importantly, recombinant binding clones are fully renewable and can be re-generated from expression constructs via bacterial production offering increased consistency, reproducibility and cost-effectiveness, thus justifying the expense of rigorous clone validation.
In this protocol and accompanying video, basic methods for antibody selection from phage-displayed libraries using immobilized antigen domains are demonstrated. This particular method employs GST-tagged protein domains immobilized by passive adsorption in microwell plates, although other tags11,12,13 and selection formats13,14,2 have also been used successfully. Critical considerations for the setup and conduct of selections with parallel monitoring of selection parameters aimed at identifying and isolating specifically enriched clonal antibodies for validation are detailed.
Protocol
1. Antigen Generation
NOTE: Antigen domains can be synthesized and cloned by a variety of commercial vendors into an appropriate IPTG-inducible expression construct.
- Transform expression constructs into chemically competent, T1-phage resistant, BL21 E. coli cells by mixing 10 ng of encoding DNA in 20 µl of 1X KCM on ice in a 96 well PCR plate followed by the addition of 20 L of chemically competent cells.
- Incubate the cell/DNA mixture on ice for 20 min, at RT for 10 min and then on ice again for 2 min before rescuing with 100 µl of pre-warmed super optimal broth with glucose (SOC) media, covering with a breathable plate seal and shaking at 37 °C for 1 hr at 200 rpm in a 2.5 cm orbital shaker.
- Plate 5 µl of transformed cells on to Luria-Bertani (LB)/agar plates supplemented with carbenicillin (100 g/ml) and grow O/N to obtain single colonies used for generating glycerol stocks.
- Inoculate 1 ml of 2YT/carb media with 5 µl glycerol stock and grow for 12 hr at 37 °C in a 96 well deep-well block with shaking at 200 rpm in a 2.5 cm orbital shaker.
- Inoculate NZY media15 (supplemented with 0.05 % glucose, 2% lactose, and 100 g/ml carbenicillin) with a 1:40 dilution of this culture, then shake for 6-8 hr at 30 °C and 200 rpm in a 2.5 cm orbital shaker.
- Pellet bacteria and purify antigenic proteins in high-throughput in a 96-well filter plate containing 100 µl of Ni-NTA resin as per previously published protocols16,17.
- Determine the concentration of purified proteins by Bradford dye-binding assay18 and characterize for size and purity by separation and visualization of 10-50 µg with Coomassie stain on an SDS-page gel (See Figure 2).
NOTE: At this stage proteins can be further characterized by methods that assess protein aggregation19 or folding20 depending on the nature of the antigen and the requirements of the selection pipeline.
2. Preparation and Titration of the Phage Library
- Pick a single colony of T1 phage-resistant E. coli cells into 1 ml of 2YT media supplemented with 50 μg/ml tetracycline.
- Once cell growth is visually established, dilute the culture in a larger volume of 2YT supplemented with 50 μg/ml tetracycline to ensure a sufficient volume of cells for infection (generally 45 µl per library dilution, see below.).
- Prepare the library for use by precipitating the phage from storage buffer with 1/5th volume of autoclaved 20% PEG-8000, 2.5 M NaCl (PEG-NaCl) incubating on ice for 30 min and pelleting precipitated phage by centrifugation at 12,000 x g.
- Discard the supernatant and resuspend precipitated phage in an appropriate volume of 1x PBS pH 7.4, 0.2% BSA and 0.05% Tween (PBT), adjusting to an appropriate phage concentration for selections (if necessary) to ensure adequate library coverage. See Step 2.9 and Discussion.
- Dilute a 5 μl phage aliquot into 45 μl of 1x PBS pH 7.4 in a sterile multi-well plate, mix, and create a series of 10 fold dilutions in the same buffer.
- Add 5 µl of each phage dilution to 45 µl of T1 phage-resistant E . coli cells in log phase growth (OD600 = 0.4-0.8) in another multiwell plate, cover with a breathable seal and incubate at 37 °C with shaking at 200 rpm for 30 min in a 2.5 cm orbital shaker.
- Plate 5 μl of infected cells from each of the wells on to a pre-warmed agar plate supplemented with 100 g/ml carbenicillin and grow O/N at 37 °C until colonies are visible.
- Calculate the total phage/ml as follows:
Phage/ml = number of colonies on a carbenicillin plate in the most dilute sample x 200 x 10i (where i = the maximum number of dilutions in which colonies are apparent). - To determine the number of coated antigen wells to ensure coverage of the library diversity, calculate as follows:
Number of coated antigen wells required =
Desired fold coverage * library diversity
# of phage per 100 µl
3. Antibody Selection from Phage-displayed Libraries
3.1) Antigen Immobilization
- To avoid freeze-thaw cycles that can affect antigen quality and to facilitate dilutions, prepare a stock antigen protein plate normalized to 100x working concentrations (e.g., 500 g/ml) and aliquot in to non-binding 96-well plates.
- Prepare antigen-coated plates from stock protein solutions one day prior to each round of selections by diluting from stock plates with PBS.
- Coat 96-well high protein-binding polystyrene plates with 100 μl of GST-tagged antigen protein or negative control protein (GST, BSA, neutravidin, etc.) at 5 μg/ml in PBS. Shake at 250 rpm 4 °C O/N on a microplate shaker.
- Remove protein solution from the coated microplates, block the antigen-coated and negative selection plates with 200 μl blocking buffer (0.2% BSA in 1x PBS pH 7.4) and shake at 250 rpm at RT for 1-2 hr.
- After blocking, wash the blocked protein-coated plates four times with 200 µl of 1x PBS pH 7.4 with 0.05% Tween (PT) buffer either manually or using an automated microplate washer.
3.2) Library Pre-clearance and Antigen-phage Incubation
- Deplete the library of non- (or tag-) specific binding phage-displayed antibody fragment clones via transfer of 100 µl (1012 phage) of prepared phage library in PBT buffer to a number of wells coated with negative control protein or free affinity tag protein (e.g., GST) equivalent to the number of targets and incubate phage in wells for 1-2 hr at RT with shaking at 250 rpm.
NOTE: As an alternate or additional means of negative selection, incubation can also be performed in the presence of excess (5-25 μM) soluble protein affinity tag (e.g., GST) to further remove tag-binding clones and enhance recovery of specific target binding clones. - Transfer phage supernatant from negative selection to antigen-coated plates and incubate for 1–2 hr at RT with shaking at 250 rpm for capture of specific binding clones.
- Remove unbound phage and wash wells of the microplate 8-15 times with PT buffer either manually or using a microplate washer. Remove excess wash buffer just prior to elution.
3.3) Elution, Infection and Phage Amplification
- Early on the day of selections, pick a single colony of T1 phage-resistant E. coli cells into 1 ml of 2YT media supplemented with 50 μg/ml tetracycline and grow at 37 °C with shaking at 200 rpm in a 2.5 cm orbital shaker or equivalent.
- Once cell growth is established and can be seen by eye, increase the volume of media with 2YT supplemented with 50 ug/ml tetracycline to ensure a sufficient volume of cells for infection (generally 100 µl per selection well).
- Grow the cells until an OD600 of 0.4–0.8 is reached (approximately 5-7 hr), then to elute bound phage add 100 μl of cells to the washed antigen plate and shake at 37 °C for 30 min.
- Add M13K07 helper phage to a final concentration of 1 x 1010 cfu/ml and incubate at 37 °C for 45 min with shaking at 200 rpm.
- Transfer cells to 1.2 ml of 2YT supplemented with 150 μg/ml carbenicillin and 75 μg/ml kanamycin in a 96-well deep well block with V-bottom and grow O/N at 37 °C with shaking at 200 rpm in a 2.5 cm orbital shaker.
3.4) Phage Preparation and Repeated Rounds of Selection
- Pellet bacteria by centrifugation at 4,000 x g for 15 min at 4 °C.
- Mix equal volumes of phage supernatants from replicate plates in a mini-tube 96-well sterile microplate blue box, neutralize with 1/10th volume of 10X PBT and mix. Repeat rounds of selection (starting from Step 3.1.1) for 3-4 rounds or until enrichment is observed (See Sections 4. and 7.) compared to negative control protein.
NOTE: In the early rounds of selection when significant / detectable enrichment is not expected (i.e., rounds 1 and 2), quantification of input and output phage titres (described in Section 7 and representative results shown in Figure 3) may be helpful in ensuring minimum phage concentrations for successful selections (described in Discussion) and are more appropriate than pooled ELISAs.
4. Characterization by Pooled ELISA
- Coat 96-well high protein-binding polystyrene plates with 100 μl of GST-tagged antigen protein or negative control protein (GST, BSA, neutravidin etc.) at 2 μg/ml as per Section 3.1 and shake at 4 °C O/N.
- Remove excess antigen, block the plate with 200 μl of blocking buffer with shaking for 1–2 hr at RT at 250 rpm on a microplate shaker and wash four times with 200 μl PT buffer either by hand or by automated plate washer.
- Apply 100 μl of phage supernatant (diluted 1:10-1:20 in PBT buffer), shake at 250 rpm for 15 min at RT, and wash the plate eight times with 200 μl PT buffer.
- Add 100 μl of anti-M13-HRP (1:5,000 in PBT buffer). Shake at 200 rpm for 30 min at RT, then wash the plate six times with 200 μl PT buffer and twice with 200 μl of PBS.
- Apply 100 μl of 1:1 TMB substrate and develop for 5–15 min (or until substantial color has developed), then stop the reaction by addition of 100 μl of 1 M H3PO4 and read the plate at 450 nm.
- In general, enrichment in phage pools can be observed in rounds 3–4 as a ratio of specific binding versus non-specific binding of >2 (See Figure 4)
NOTE: It is informative to note that high absolute binding signal on the affinity tag protein indicates enrichment of tag-specific binders (rather than target binders), and that high absolute binding signal on the negative control protein indicates enrichment of non-specific or “sticky” binders.
5. Clone Selection, Sequencing and Characterization
5.1) Isolation of Single Fab-phage Clones
- To isolate individual clones for sequencing and characterization, infect 1/10th volume of the amplified phage pool in to T1 phage-resistant E. coli cells in log phase of growth (OD600 = 0.4-0.8) in a round-bottom microtiter plate, cover with a breathable plate seal and incubate with shaking at 200 rpm for 30 min at 37 °C.
- Prepare 10-fold serial dilutions in 2YT media and plate out on separate agar plates supplemented with 100 g/ml carbenicillin.
- Pick single colonies in to 450 µl of 2YT/carb media supplemented with 1 x 109 M13K07 phage/ml and grow O/N in a mini-tube 96 well sterile microplate blue box incubating at 37 °C with shaking at 200 rpm.
- Pellet bacteria by spinning at 4,000 x g for 10 min at 4 °C.
- Clonal phage supernatant can be used for sequencing of Fab-phage clones (as described in Section 5.4) or for clonal direct-binding ELISAs to determine specificity against immobilized antigens (as described in Sections 5.3 and 21) for which representative results are shown in Figure 5.
5.2) Expression of Fab clones for characterization
- Following cloning in to an appropriate expression vector (see Discussion), pick single E. coli colonies transformed with expression constructs in to 1 ml of 2YT/carb media in a 96 well deep-well plate using a sterile toothpick or pipette tip and grow for 12-16 hr at 37 °C with shaking at 200 rpm. This can be done either manually or with an automated colony picker.
- Transfer 40 µl of growing culture to 1.5 ml of NZY media supplemented with glucose/lactose/glycerol in a 96 well deepwell block as per the methodology of Studier et al.15
NOTE: Alternately, cells can be grown for 3-4 hr (OD 0.8-1.0) in non-supplemented and protein expression induced by the addition of 1 mM IPTG. - Cover the culture block(s) with a breathable seal and express Fab clones for 6-8 hr at 30 °C with shaking at 200 rpm. Spin plates at 4,000 x g for 15 min to pellet bacterial cells, remove the supernatant, cover plates with a foil seal and freeze pellets at -20 °C until lysis.
- Liberate expressed Fabs from collected pellets with 100 µl of lysis buffer (Tris HCl 50 mM, 200 mM NaCl, 1.25% Triton X-100 pH 8.0 with 1mg/ml lysozyme and 10 U benzonase/ml culture and protease inhibitors) and shaking for 2 hr at 4 °C.
- Remove cell debris by centrifugation at 4,000 x g for 15 min at 4 °C and collect crude lysate supernatant for use in initial characterization of specificity by ELISA plates (see Section 5.3).
5.3) Clone Characterization by Direct Binding Clonal Fab ELISA
- Immobilize antigens as described in Section 3.1 instead aliquotting 30 µl of 2 µg/ml antigen in PBS to wells of a 384 well plate. Quad-based antigen layouts in the plate can facilitate reagent transfer when using 96 channel-based dispensing heads. Wash and block ELISA plates as per Section 3.1.
- Cleared lysates collected from Section 5.2 can be applied directly to immobilized antigen for evaluation of binding. Incubate 30 µl of lysate per well for 15 min at RT with shaking at 200 rpm.
NOTE: Purified Fab clone protein can alternately be used by diluting to 10 µg/ml in PBT buffer and applying 30 µl of each clone to blocked wells containing antigen or negative control protein (GST, BSA etc.) and developing in the same manner described below. Alternately, Fab-phage can also be used by diluting 10 µl of phage supernatant (described in Section 5.1) in 20 µl PBT buffer (1:3) and detecting with anti-M13-HRP antibodies (described in Steps 4.4 – 4.5) - Remove excess lysate and wash wells either manually or using an automated plate washed 8x with 100 µl of PT buffer and remove excess buffer.
- Incubate 30 µl of a 1:5,000 dilution of secondary anti-FLAG antibody in PBT buffer for 30 min at RT with shaking at 200 rpm.
- Wash, develop and quantify as per Sections 4.3 – 4.5.
NOTE: Representative results illustrating both specific and non-specific binding clones are shown in Figure 5.
5.4) Clone Sequencing
- Prepare a PCR master mix as indicated in Table 2.
- Add 2 µl of the appropriate PCR template (either phage supernatant or re-suspended single phagemid-containing bacterial colony) to 20 µl of PCR master mix prepared with the appropriate primers in wells of a PCR plate.
NOTE: Separate master mixes are required for sequencing of both the heavy and light chain genes. - Run the PCR reaction using the parameters listed in Table 2 – PCR Settings.
- Verify success of the PCR reaction by mixing 5 µl of the completed reaction with loading dye on a 1% agarose gel supplemented with DNA visualization stain and imaging on a 300 nm transilluminator.
- Add 2 µl of PCR product to 10 µl of sterile water containing 0.2 µl each of exonuclease and shrimp alkaline phosphatase and incubate at 37 °C for 20 min followed by enzyme inactivation at 80 °C for 10 min to remove excess primers in preparation for sequencing.
6. Scaling for Automated Selections
6.1) Pipette-based Liquid Handling
- Prepare a 1% (w/v) bromophenol blue in water and remove insoluble particles using a 0.45 ml filter.
- To determine the linear range of absorbance on the plate reader, prepare a 1:1,000 dilution of the 1% bromophenol blue solution in water, and continue with two-fold serial dilutions (16 dilutions total).
- Transfer 100 µl of each dilution to a flat bottom 96 well plate and read the absorbance at 590 nm. Plot absolute absorbance versus dilution factor and choose a dilution at the mid to high end of the linear range for subsequent tests.
- Add bromophenol blue to water or to the actual solution that will be pipetted based on the dilution chosen in the previous step in sufficient volume for pipetting to three 96 well plates, plus the dead volume in the reservoir.
- Calibrate a single-channel pipettor to deliver the test volume by pipetting water onto an analytical balance (100 µl of water weighs 100 mg at RT.)
- Using the calibrated pipettor, transfer the test volume of the dyed solution to at least three wells of a flat bottom 96 well plate and read their absorbances at 590 nm, in triplicate.
- Weigh three empty flat-bottom 96 well plates on an analytical balance and record their masses.
- Pipette test volume from reservoir to each of three 96 well plates and measure their absorbances at 590 nm, in triplicate.
- Weigh each plate and calculate the difference in mass.
- First, evaluate whether the absorbances in each well are uniform within the range of tolerance (e.g., +/- 5%) and whether they are consistent with the absorbances obtained by hand-pipetting with the calibrated pipetman. If particular channels are consistently over- or under-delivering (for example see Figure 6), follow repair procedures and repeat the evaluation procedure until all channels deliver uniform volumes.
- Second, once all channels are confirmed to deliver uniform volumes, check the accuracy of that volume by calculating the average mass difference, per well. For instance, a perfectly calibrated liquid handler set for 100 µl will deliver 9.60 g of water per plate, or 0.10 g of water per well. If the real volume delivered is consistently over or under the intended volume, then a correction factor can be applied, i.e., programming 110 µl in order to actually deliver 100 µl.
NOTE: For small volumes, e.g., 10 µl, use ten-fold more bromophenol blue in the dyed solution, and pre-aliquot 90 µl of water to the 96 well plates by hand, to ensure that the absorbances are measureable and fall in the linear range.
6.2) Automated Plate Washing
- Immobilize positive target and negative control proteins in separate wells of 96 well microplates (two plates for each condition to be tested) as described in Section 3.1.
- Block non-specific binding sites by incubation with blocking solution for one hr at RT.
- Add identical 100 µl aliquots of a 1013 cfu/ml target-binding Fab-phage solution in PBT to all wells in all plates and incubate with gentle agitation at RT for 2 hr.
- Wash two plates according to each condition, one plate for infection and titration and one plate for ELISA.
- Evaluate the efficacy of the wash by direct infection into bacteria and titration (as described in Section 3.3 (without M13K07 addition) and 7.2.1) or development with anti-M13 HRP antibody and colorimetric substrate (as described in Section 4).
- Phage titres from specific clones will often yield in the range of 105–107 phage/ml from an immobilized protein against which the clone binds specifically. Some residual binding to negative control protein (e.g., BSA) is to be expected and generally 102-105 phage/ml are observed, however very low titres (i.e., <101) can signal overly stringent washing, which can compromise a selection.
- For phage binding determined by ELISA, compare the absolute binding signals for positive and negative control proteins to determine if robotic washing is equivalent to established hand washing results (Figure 7).
6.3) Plate Washer Decontamination
- To start, run the decontamination protocol to be tested.
NOTE: We recommend using at least double the total line volume, and prime and soak the washer head for 5 min in 0.5% sodium hypochlorite, freshly diluted from commercial bleach (typically 6% sodium hypochlorite). - Prime and soak the washer head for 5 min in sterile (autoclaved) water and finally, prime the system until the lines are full of air.
- Prime the lines with sterile water or wash buffer.
- Wash a sterile 96 well microplate once, remove from washer and seal with sterile plate seal. This is the pre-contamination control plate.
- Aliquot 100 µl of O/N amplified phage culture supernatant to all wells of a 96 well plate.
- Wash the phage-containing plate once, then remove from the washer and seal with a plastic or foil plate seal. This is the contamination control plate.
- Carry out the decontamination protocol being tested. In this example, repeat steps 6.3.1 and 6.3.2.
- Wash sterile microplate once, then remove from washer and seal. This is the decontamination test plate.
- Plate 50 µl of log phase E. coli to titration plates; this is the pre-infection control plate.
- Unseal pre-contamination, contamination and decontamination 96-well plates and add 100 µl of E. coli in the log-phase growth stage to all wells, using new tips for each well.
- Seal and incubate at 37 °C for 30 min at 200 rpm.
- Plate 50 µl of infected cells from three random wells of each 96-well plate to 10 cm 2YT/carb plates and incubate at 37 °C O/N.
- Transfer 50 µl of infected cells from all wells of each plate to deep well plates containing 1 ml 2YT plus 100 g/ml carbenicillin per well and grow O/N at 37 °C.
- Repeat the decontamination protocol and leave the washer lines dry.
NOTE: No O/N growth on plates or in liquid medium should be observed on the pre-infection control, pre-contamination control or decontamination test plates. Many colonies and growth should be observed on the contamination control plate; if not, no conclusions about the efficacy of the decontamination protocol can be made. Ideally, the decontamination plates will yield no colonies and no O/N growth in any of the wells in the O/N plates. The stringency of the decontamination protocol can be increased with higher bleach concentrations, longer soak times and additional cycles.
7. Input/Output Titrations
7.1) Input Phage Titrations
- Dilute a 5 µl aliquot of phage to 50 µl with sterile-filtered PBS and mix.
- Prepare serial 10-fold dilution in sterile-filtered PBS to 1010 using multi-channel pipette or a 96 channel pipettor.
- Inoculate 5 µl of diluted phage in to 45 µl of T1 phage-resistant E. coli cells in log-phase of growth (OD 0.6-0.8) and incubate covered with a breathable seal for 30 min at 37 °C.
- Plate 5 µl of infected cells on to an LB/agar plate supplemented with 100 µg/ml carbenicillin and incubate O/N at 37 °C.
- The phage concentration can be estimated from the most dilute sample in which colonies appear according to the calculations detailed in Section 2.8.
7.2) Output Phage Pitrations
- Following elution of plate-bound phage by the addition of E. coli cells, dilute a 5 µl aliquot of phage-infected cells to 50 µl with autoclaved 2YT media
- Prepare serial 10-fold dilution in autoclaved 2YT media to 106 using a multi-channel pipette or 96 channel pipettor.
- Plate 5 µl of infected cells on to an agar plate supplemented with 100 µg/ml carbenicillin and incubate O/N at 37 °C.
- The phage concentration can be estimated from the most dilute sample in which colonies appear according to the calculations detailed in Section 2.8.
NOTE: In both cases (i.e., input and output phage titres, comparison with colony numbers on both tetracycline (total cell number) and kanamycin (helper phage titre) can also be informative.
Representative Results
The protocol described herein has been used to isolate antibody fragments to a variety of both structurally- and functionally-related antigen domains from combinatorial phage-displayed Fab libraries in parallel. Issues related to antigen availability can be circumvented, in many cases, by in silico identification of expressible antigen domains that are suitable for antibody selection23. By coupling antigen expression to antibody selection within the same laboratory (Figure 1) it becomes feasible to construct an integrated pipeline, in which antigen parameters critical to the selection can be more easily controlled. In most cases, careful determination of domain boundaries enables the expression and isolation of adequate quantities of sufficiently pure antigen from high-throughput expression in 96 or 24 well deepwell plates (Figure 2) for successful use in antibody selections. During successive rounds of the selection process, eluted phage concentrations and enrichment of phage that specifically bind antigen can be monitored by either phage titration (Figure 3) or with phage pools in an ELISA assay (Figure 4). A binding ratio (>2) generally indicates the presence of specific binding clones. Conversely, a low ratio (<2) can help determine the cause of a selection failure. For instance, a ratio <2 with high non-specific OD can indicate insufficient removal of non-specific clones (or sticky binders) and may warrant further optimization of the selection protocol. Nevertheless, once enrichment is detected, individual high affinity clones can then be isolated for sequencing and characterization. Clone binding can be characterized in a colorimetric immunoassay and enables the comparison of binding of the Fab-phage or Fab to immobilized antigen versus negative control protein, yielding a measure of enrichment (ratio of binding to target versus negative control protein or affinity tag protein) (Figure 5).
During scaling and automation of this method, assessment of key functions of the liquid handling unit can be helpful to ensure proper and accurate operation with solutions similar to those used during actual selections. The use of absorbing chromophores in solutions can help to detect when specific channels in the fluidics (aspiration or dispensing) are either clogged or have lost seal resulting in partial volume delivery as illustrated in Figure 6. Automated wash units can also be a source of variability and can require attention. The use of known binding clones enables comparison of binding between target control protein to ensure that target binding is maintained while background binding removed (Figure 7) when wash parameters such as dispensing speed, wash volumes and the number of washes are being optimized. Following the use of an automated plate washer with phage solutions, effective decontamination protocols are necessary to ensure the absence of cross-contamination from successive rounds of selections or different selection procedures and a troubleshooting/interpretation guide is provided in Table 3.
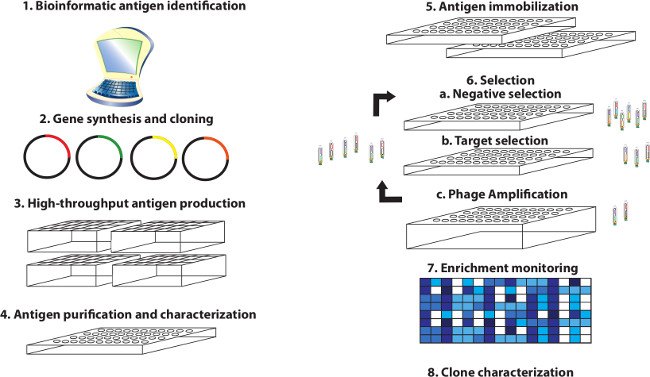
Figure 1. Overview – High-throughput antigen production and antibody selection pipeline. This overview shows a step-by-step visualization of a representative antigen generation and antibody selection pipeline. The in silico identification of antigen domain boundaries enables gene synthesis, expression and selection upon domains for proteins that may be poorly characterized. Expressed antigen domains are immobilized and used to isolate pools of specific high affinity antibody clones from a combinatorial antibody library displayed on the surface of filamentous phage. Pools of antibody-phage eluted and amplified from individual antigens can then be used to monitor round-to-round enrichment of specific binding clones in ELISA-based assays comparing immobilized target versus negative control protein prior to isolation of individual binding clones for characterization. Please click here to view a larger version of this figure.
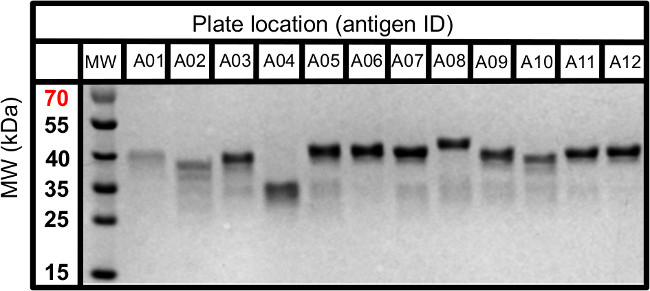
Figure 2. SDS-PAGE visualization of affinity-tagged antigens. A representative SDS-PAGE of expressed and purified 6X-HIS-GST-tagged antigen domains (5-10 kDa domain + 26 kDa GST) originally identified by in silico analysis of antigen domain boundaries. Protein domains expressed and isolated by affinity purification are characterized by SDS-PAGE prior to selections to ensure size-based identity, adequate yields and purity to enable use in phage-antibody selections. Please click here to view a larger version of this figure.
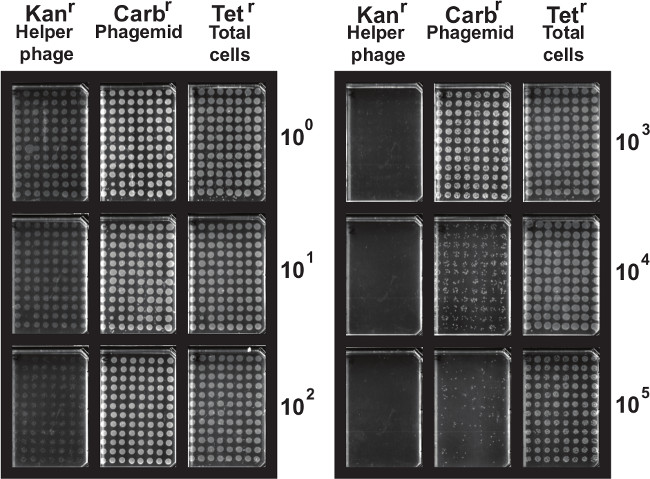
Figure 3. Visualization of phage output titres. Phage output titres are determined by plating out cells infected with phage over a 10-fold dilution series on agar media supplemented with various antibiotics (tetracycline, carbenicillin and kanamycin) to determine the lowest dilution in which colonies can be observed and estimate the number of phage bound to target antigen and eluted. Please click here to view a larger version of this figure.
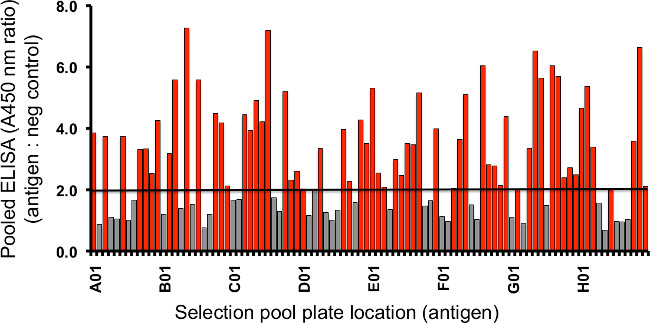
Figure 4. Binding ratio of target to negative control protein by pooled ELISA. The progress of enrichment can be evaluated following rounds of selection by screening individual amplified eluents against both the specific targets and negative control proteins and/or isolated affinity tag. Shown in the plot are the enrichment ratios for binding of 96 individual phage-antibody pools against their 96 respective antigens versus negative control protein in parallel. Each ratio value is derived from two absorbance measurements that quantify binding of the phage pool to either target or negative control protein using a HRP-fused antibody specific for M13 phage coat protein. A ratio threshold of 2 or greater (often ranging from 2–10 or more as depicted in red) is used as an indication of enrichment and the likely presence of specific binding clones. Lesser ratios can signify failure of or specific issues with the selection that may warrant further optimization of the protocol. Please click here to view a larger version of this figure.
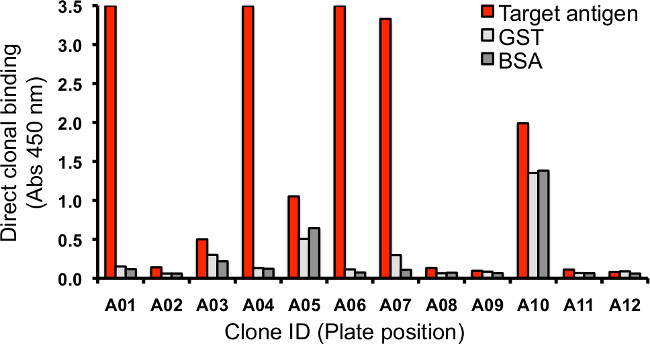
Figure 5. Direct binding clonal Fab-phage or Fab ELISA. The binding of single clones (whether in Fab-phage or Fab format) can be evaluated by ELISA-based assay versus immobilized protein in 384 well plates. Binding is detected using a secondary HRP-fused antibody against the M13 coat protein on Fab-phage or an affinity tag on the Fab. Raw absorbance values for binding of clonal Fab-phage to target antigen, negative control protein and antigen affinity tag (GST) are depicted illustrating specific binders (A01, A04, A06), and sticky binders (A05).
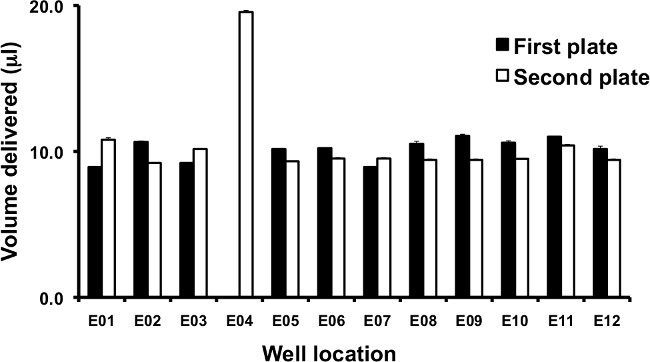
Figure 6. Evaluation of dispensed volumes by automated liquid handling unit. Consistent and accurate delivery of intended volumes by robotic or manual 96-well pipetting systems can be evaluated using dyed solutions and a plate reader. In this example, 10 µl of a solution containing bromophenol blue is pipetted sequentially to all wells of two 96 well plates using a single tip box. Each well contains 90 µl/well dH2O, analogous to the addition of helper phage to infected E. coli. Each plate’s absorbances at 590 nm are read in triplicate, and the volumes are interpolated from a standard curve prepared using a calibrated single channel pipetman. Volumes delivered to a single row on the first and second plate are shown. Note that on the first plate, well E04 receives no dyed solution, and on the second plate, it receives a double volume. This indicates that the tip touch is not sufficient for consistent delivery, and requires further optimization. 95% confidence intervals for triplicate absorbance measurements are shown.
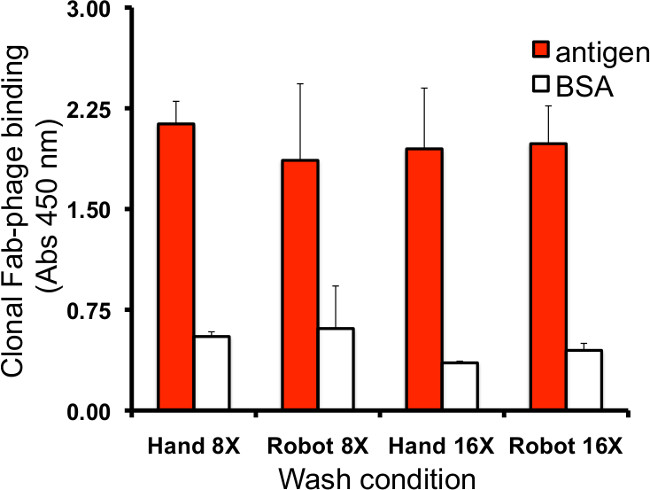
Figure 7. Evaluation of automated plate washing. Effective removal of unbound phage can be assessed using an ELISA to measure Fab-phage binding to immobilized positive (anti-FLAG antibody) and negative (BSA) control proteins while varying the wash conditions. In this example, input phage analogous to naïve library (1013 cfu/ml in PBT) is removed using eight or sixteen washes, by hand or using the robot. Residual phage binding to BSA is the same across all conditions, as is specific binding to immobilized anti-FLAG antibody, indicating that at this low flow rate, both methods are equivalent and sufficiently stringent. 95% confidence intervals from duplicate wells are shown.
| SOLUTIONS | |||
| 2YT media – Add water to 1 L, adjust pH to 7.0, and autoclave. | yeast extract | 10 g | |
| tryptone | 16 g | ||
| NaCl | 5 g | ||
| NZY media (1 L) | N-Z Amine | 10 g | |
| yeast extract | 5 g | ||
| 50X ZYM-5052 | 20 ml | ||
| 20 salt stock | 50 ml | ||
| 50x ZYM-5052 sugar stock (1 L) | glucose | 25 g | |
| lactose | 100 g | ||
| glycerol | 250 ml | ||
| 20x salt stock (1 L) | Na2HPO4 | 500 mM | |
| KH2PO4 | 500 mM | ||
| NH4Cl | 1 M | ||
| Na2SO4 | 100 mM | ||
| Carbenicillin (Carb): | 100 mg/ml in water. Filter-sterilize. | ||
| Kanamycin (Kan): | 25 mg/ml in water. Filter-sterilize. | ||
| Tetracycline (Tet): | 10 mg/ml in water. Filter sterilize. | ||
| PEG-NaCl | PEG | 20% | |
| NaCl | 2.5 M | ||
| 1x PBS (pH7.2) | NaCl | 137 mM | |
| KCl | 3 mM | ||
| Na2HPO4 | 8 mM | ||
| KH2PO4 | 1.5 mM | ||
| 1X KCM | KCl | 500 mM | |
| CaCl2 | 150 mM | ||
| MgCl2 | 250 mM | ||
| SOC Media | yeast extract | 5 g/L | |
| tryptone | 20 g/L | ||
| NaCl | 10 mM | ||
| KCl | 2.5 mM | ||
| MgSO4 | 20 mM | ||
| Glucose | 20 mM | ||
| Blocking buffer | PBS | 1x | |
| BSA | 0.20% | ||
| PBT buffer | PBS | 1x | |
| BSA | 0.20% | ||
| Tween | 0.05% | ||
| PT buffer | PBS | 1x | |
| Tween | 0.05% | ||
| Phosphoric acid | H3P04 | 1 M | |
Table 1. Media and Solutions.
| PCR MASTER-MIX SET-UP | |||
| Component | µl per reaction | ||
| Water | 19.45 | ||
| 10x Taq buffer | 2.5 | ||
| dNTPs (10 mM stock) | 0.675 | ||
| DMSO | 0.75 | ||
| Taq enzyme | 0.125 | ||
| Forward Primer (100 uM stock) | 0.25 | ||
| Reverse primer (100 uM stock) | 0.25 | ||
| Heavy Chain Sequencing Primers | |||
| M13 Forward primer | GTAAAACGACGGCCAGTACTCGAGGCTGAGCAAAGC | ||
| M13 Reverse primer | CAGGAAACAGCTATGACGGGAAGTGTCCTTGACCA | ||
| Light Chain Sequencing Primers | |||
| M13 Forward primer | TGTAAAACGACGGCCAGTCTGTCATAAAGTTGTCACGG | ||
| M13 Reverse primer | CAGGAAACAGCTATGACCCCTTGGTACCCTGTCCG | ||
| PCR SETTINGS | |||
| Step 1. | 94 °C for 3:00 min | ||
| Step 2. | 94 °C for 0:30 min | ||
| Step 3. | 55 °C for 0:30 min | ||
| Step 4. | 72 °C for 1:00 min | ||
| Return to Step 2 and repeat 24x | jump to step 2 | ||
| Step 5. | 72 °C for 7:00 min | ||
| Step 6. | 12 °C hold | ||
Table 2. PCR Master Mix and Settings.
| Interpretation of decontamination results | |||
| Plate | Expected carbenicillin titer | If not? | |
| Pre-infection control | 0 | Cells were pre-infected; nothing can be concluded from the experiment. | |
| Contamination control | lawn | Insufficient contamination to be able to assess efficacy of subsequent decontamination; consider submerging manifold in phage solution rather than aspirating it. | |
| Decontamination test | 0 | Decontamination not complete; increase soak time, bleach concentration or try SDS and EtOH washes instead. | |
Table 3. Evaluation of washer decontamination. To determine the efficacy of decontamination, it is necessary to show that washer was contaminated with phage, and that the decontamination protocol successfully eliminated that contamination. Using phage that confer carbenicillin resistance, the expected outcomes from such a test are listed, along with suggestions should the real outcome differ from the expected outcome.
Discussion
When conducting in vitro antibody selections, the two primary determinants of selection success are 1) the isolation of well-folded antigenic targets for selecting upon and 2) the availability of a high functional diversity antibody library. In many cases, the availability of sufficient quantities of well-folded, full-length protein can be limiting. One approach to overcoming this limitation is to use domains identified from bioinformatic analysis22 and synthesized for insertion into a bacterial expression vector with an appropriate affinity tag.
There are a variety of tags that are used in biotech applications to increase solubility, facilitate purification and to stabilize unstable domains.11 The ability to synthesize virtually any desired insert sequence enables a highly versatile antigen generation platform. Although the GST-tagged format is common, successful results have been previously obtained using hexahistidine, Fc, Halo, maltose binding protein and site-specific biotinylation tags and it is believed that myriad other affinity tags may be optimized for use in a similar platform. However, affinity tags can have both beneficial and unintended negative consequences in downstream applications11 and should be thoroughly investigated before establishing a pipeline based on a particular format.
Often new users will pose the question of how to assess the quality of an antibody library and though there is no straightforward answer, one should try to assess several key features (whether natural or synthetic) including overall diversity, relative display levels, the nature of and extent of randomization (i.e., library design versus library content determined by sequencing) and the physicochemical properties of the antibody versus intended application. Although beyond the scope of this protocol, a more detailed treatment of these features and how they can affect the selection of antibodies can be found here2,23. Knowledge of library diversity is, in particular, important to ensure adequate coverage of the diversity during selections. Typically, a phage concentration that provides 100–1,000 copies of each phage clone in the library is desirable and should ensure that any true positive binders present in the library can be recovered. However, non-specific binding of phage to microplate surfaces increases dramatically above 1013 phage/ml, and for very diverse libraries, more than a single well per antigen may be needed to ensure sufficient coverage. On the other hand, if the input library is too dilute, the absolute concentration of true positive binders will be so far below the KD (typically nM) that they will not be recovered. Given these constraints, phage libraries should generally be resuspended to 1012-13 phage/ml for the first round of selections. At this concentration, a 100 L aliquot of phage represents a 1,000–100 fold coverage of a 109-1010 diversity library and can routinely yield binding clones to diverse antigens. At lesser concentrations, or increased diversities, it may be necessary to increase the number of wells of antigen selected against in the first round (see Section 2.9). Subsequent to round 1, however, most of the non-binding diversity of the library has been removed; excess coverage of the round 1 output diversity is most often easily achievable using a single well. In cases of parallel selections in 96 well plates, this can reduce the number of antigen selection plates required to just one, subsequent to the first round.
During actual selections, phage titration can offer objective measures to characterize and monitor progress, while helping to identify areas of concern particularly in the early rounds of selection where enrichment is not sufficient to be detected. This can be particularly useful during scaling of selections to determine protocol performance. In general, input phage titres (i.e., naïve library or post-amplification of eluted phage) should remain relatively constant (1012–1013 phage/ml) between rounds, with poor cell growth / low phage titres indicating potential contamination and/or poor output from the previous round. Although output phage titres are expected to increase during enrichment in later rounds of selection, a range of 103–105 cfu/ml in the first two rounds is expected. Deviation from this range can indicate potential issues with the selection such as contamination or excess wash stringency. In later rounds of selection (i.e., rounds 3 and 4), the enrichment of clones that specifically bind the target antigen can be assessed via pooled ELISAs that measure binding of the phage pool against both target antigen and negative control protein (See Section 4).
After 3-4 rounds of selection (or once enrichment on target protein has reached a target antigen:negative control protein signal ratio of 2:1 or higher in comparison to background protein), infected bacterial cells are plated to isolate single phagemid colonies for sequencing, initial characterization and cloning if necessary. At this time, it is highly advisable to convert Fab-phage to free Fab before additional characterization. Although Fab-phage can be used for initial characterization by diluting 10 µl of phage supernatant (described in Section 5.1) in 20 µl PBT buffer and detecting with anti-M13-HRP antibodies (described in Steps 4.4 – 4.5), in our experience, binding properties can change substantially in anywhere from 5-20% of clones upon liberation from the phage particle and early conversion can obviate issues with false positives. The specific methodology used for conversion will depend on the unique architecture of the phagemid and thus will not be dealt with explicitly here. However, in the most commonly used vectors, this can generally be accomplished by 1) transformation of purified phagemid (or phagemid pool) in to a competent non-suppressor E. coli strain such as HB2151 or 55244 if an amber stop (TAG) codon intervenes the Fab and pIII protein, 2) insertion of an amber stop codon between the Fab and pIII proteins to allow expression of soluble antibody fragments in a non-suppressor strain or 3) by cloning immunoglobulin genes (from individual phagemid or phagemid pool) in to a suitable expression vector. To further streamline selection and characterization, pooled cloning methods may offer effective means for converting binding clones en masse in to expression constructs, both eliminating the potential for false-positive binding of Fab-phage particles and where expression lysates are used for early characterization, even the cost and effort of purification can be initially avoided.
For clones isolated directly from Library F (described in24), antibody variable domains can be sequenced using 1-2 μl of phage supernatant as template to PCR amplify complementarity determining regions (CDRs) (See Table 2 of Materials for Library F-directed primers). Using these primers, the heavy and light chain variable regions will yield products of ~700 and ~500 bp, respectively covering all three CDRs. Primers include annealing sites for M13 primers such that products can be sequenced after cleanup (as described in Section 5.4.5) using standard primers available in most sequencing core facilities. Alternately, for Fab-phage pools that have been cloned in to an expression vector and transformed directly in to E. coli for expression, single colonies can be isolated and resuspended in 15 µl of 2YT and 1-2 µl of the cell suspension used directly as template for single colony PCR using appropriate primers.
Automating selections will require optimization and validation of several key robotic functions. In general, pipette-based liquid handling must be accurate and consistent across all 96 channels, plate washing must remove unbound phage efficiently without stripping adsorbed antigen or bound Fab-phage from the selection plate. Effective decontamination of the plate washer is also necessary between rounds to allow enrichment and effective counter-selection, and between subsequent selections to prevent cross-contamination. To help optimize the high-throughput selection procedure, simple approaches for evaluating these functions have been used previously and are described below and detailed within the protocol.
To evaluate the consistency of volumes delivered by all channels of a liquid dispensing / aspiration head, a colored solution is pipetted to a flat-bottom 96 well plate and the absorbance in each well measured on a plate reader. A chromophore such as bromophenol blue is helpful for dyeing solutions similar to those used for real selections to evaluate the effects of different surface tension and cohesion properties on volumes delivered. After confirming that all channels are dispensing consistent volumes, accuracy of the total delivered volume can be determined by measuring the mass of liquid transferred to each plate.
Automation of plate washing to remove unbound clones primarily requires optimization of the rate at which wash buffer is to be dispensed and the number of washes to be employed. A reasonable method of evaluating this parameter employs positive controls (specific Fab-phage binding clones) to assess the effects of varying the dispense rate and/or number of washes to determine whether 1) adsorbed antigen or bound phage is being removed by overly stringent washing or 2) non-specific binding to non-cognate proteins is excessive due to wash conditions that are not sufficiently stringent. Residual/bound phage can be detected and quantified either by infection into bacteria and plating for single colony counts, or by ELISA using secondary antibodies specific for antibody affinity tags or phage coat proteins. Negative controls are used to assess residual levels of non-binding Fab-phage to target proteins or specific binding Fab-phage to non-cognate/background proteins, which may be high if washing is not sufficiently stringent. In this case, we used immobilized BSA as a control for non-specific binding and an anti-FLAG antibody that recognizes a FLAG epitope tag on the Fab-phage as a positive control. We compared eight and sixteen wash cycles using the robot versus washing by hand, at a medium flow rate, to show that these techniques were equivalent. Alternately, measuring input and output phage titres (See Section 7) during ongoing selections can also help to make fine adjustments in the wash parameters.
Finally, decontamination of plate washers used for selections must be effective in order to eliminate carry-over of phage from prior selections. In our experience, freshly diluted 0.5% sodium hypochlorite is fast, effective and cheap. Detergents such as SDS are also effective but require much more extensive washing to remove from the system. Check reagent compatibility for the system prior to any tests. To assess decontamination, we test for the presence of residual phage in solutions handled by the fluidics system and interpret results according to Table 3.
In summary, this protocol provides a scalable method for isolating Fabs from combinatorial libraries by parallel in vitro selections against protein domains and offers techniques for validating an automated phage selection system. Cost and infrastructure barriers that extensive animal facilities can pose are mitigated since this method does not rely upon the immunization of animals. Further, by integrating antigen generation with antibody-phage selection, quality factors that affect selection success are more easily monitored and adjusted. When the time frame from antigen expression to selecting and initial characterization is on the order of 6-8 weeks, a more responsive system of production may be realized. Antibodies isolated from in vitro selections are also both easily modified (due to genotype-phenotype linkage) and renewable, requiring only stored DNA of the expression construct to repeatedly and reproducibly re-generate antibody. Finally by showing that this protocol can be conducted using either a quasi-manual or fully automated approach that employs a custom-designed, robotic selection system, throughput can be scalable and accessible to small academic and industrial labs alike.
Using the high-throughput methods described above and the Library F synthetic antibody library24, high affinity (low nM) and specificity Fab antibody fragments can be routinely obtained and are functional in a variety of immuno-applications including Western blotting, immunoprecipitation and immunofluorescence of both in vitro– and in situ-expressed proteins. Although success rates for any given selection will depend both on the quality (purity, foldedness, mono-dispersity etc.) of the antigen targets as well as the quality and diversity of the antibody library, it has been observed that anywhere from 25-80% of antigens will yield binders for a high throughput selection. Importantly, it is worth noting that clones with the ability to modulate target function can also often be selected. This library is constructed using a fully human framework, which makes these antibodies amenable to potential therapeutic application25, thus underscoring the importance of this pipeline as an alternate approach for the generation of affinity reagents with broad-based value.
In summary, the robustness and versatility of the approach presented in this protocol will continue to aid in the optimization, industrialization and ultimate widespread use of this technology as a viable means of achieving the long-term goal of generating affinity reagents to virtually all members of the human proteome.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We would like to acknowledge the NIH Common Fund – Protein Capture Reagents Program for funding the development of the Recombinant Antibody Network antibody selection and characterization pipeline and the Canadian Foundation for Innovation for funding purchase of the robotic selection platform.
Materials
| MATERIALS | |||
| Name | Company | Catalog Number | |
| New Brunswick Innova44 stackable incubator shaker | Eppendorf | M1282-0004 | |
| Liquidator 96 channel manual benchtop pipetting system | Anachem Ltd | LIQ-96-200 | |
| Microplate shaker | VWR | 12620-926 | |
| ThermoScientific Sorvall ST-40 benchtop centrifuge | Thermo Product | 75004525 | |
| ELx405 Select Deep Well Microplate Washer | Biotek | ELX405USD | |
| Custom High Throughput Automated Phage Selection Robot | S&P Robotics | ||
| Plastic conical Falcon tube: 50 mL | VWR | 21008-178 | |
| Plastic conical Falcon tube: 15 mL | VWR | 89039-668 | |
| Axygen 1.7 mL microcentrifuge tubes | Corning | MCT-175-L-C | |
| 96-well/384-well Maxisorp plate | Sigma | M9410-1CS | |
| Corning 96-well V-bottom deep-well block | Corning | 3960 | |
| Axygen Mini-tube 96-well sterile microplate blue box | Corning | AXY-MTS-06-C-R-S | |
| Breathable adhesive plate sealing film | VWR | 60941-084 | |
| REAGENTS | |||
| Name | Company | Catalog Number | |
| polyethylene glycol | Bioshop | PEG800.1 | |
| yeast extract | Bioshop | YEX401.1 | |
| bio-tryptone | Bioshop | TRP402.1 | |
| N-Z amine | Sigma | C7290 | |
| glucose | Sigma | G8270-1KG | |
| lactose | Bioshop | LAC234 | |
| glycerol | Bioshop | GLY001.500 | |
| carbenicillin | Bioshop | CAR544.10 | |
| kanamycin | Bioshop | KAN201.25 | |
| tetracycline | Bioshop | TET701.25 | |
| Tween 20 | Bioshop | TWN510.500 | |
| monobasic potassium phophate | Bioshop | PPM302.1 | |
| dibasic sodium phosphate | Anachemia | 84486-440 | |
| sodium chloride | Bioshop | SOD001.10 | |
| potassium chloride | Bioshop | POC308.1 | |
| calcium chloride | Bioshop | CCL302.500 | |
| magnesium sulphate | EMD | MX0070-1 | |
| magnesium chloride | Bioshop | MAG510.500 | |
| NH4Cl | Amresco | 0621-1KG | |
| Na2SO4 | Bioshop | SOS513.500 | |
| agar | Bioshop | AGR001.500 | |
| SybrSafe DNA gel stain | Invitrogen | S33102 | |
| phosphoric acid | Acros Organics | 201140010 | |
| lysozyme | Bioshop | LYS702.25 | |
| benzonase | Novagen | 71205 | |
| Triton X-100 | Bioshop | TRX506.500 | |
| protease inhibitor cocktail tablets | Roche | 11 836 170 001 | |
| Ni-NTA resin | Qiagen | 1018240 | |
| 1000x helper phage (M13K07) stock (1013 phage per mL) | NEB | N0315S | |
| HRP conjugated M13-specific antibody (anti-M13-HRP). | GE Healthcare | 27-9241-01 | |
| TMB substrate: mix equal volumes of TMB and H2O2 peroxidase substrate | KPL | 50-76-00 | |
| dNTPs | Biobasic | DD0056 | |
| Taq polymerase | Genscript | E00007 | |
| Exonuclease | GE Healthcare | EZ0073X-EZ | |
| Shrimp alkaline phosphatase | GE Healthcare | E70092Z-EZ | |
References
- Winter, G., Milstein, C. Man-made antibodies. Nature. 349, 293-299 (1991).
- Miersch, S., Sidhu, S. S. Synthetic antibodies: concepts, potential and practical considerations. Methods. 57, 486-498 (2012).
- Schofield, D. J., et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 8, R254 (2007).
- Hust, M., et al. A human scFv antibody generation pipeline for proteome research. J. Biotechnol. 152, 159-170 (2011).
- Colwill, K., Graslund, S. A roadmap to generate renewable protein binders to the human proteome. Nat. Methods. 8, 551-558 (2011).
- Mersmann, M., et al. Towards proteome scale antibody selections using phage display. Nat. Biotechnol. 27, 118-128 (2010).
- Pershad, K., et al. Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display. Protein Eng. Des. Sel. 23, 279-288 (2010).
- Turunen, L., Takkinen, K., Soderlund, H., Pulli, T. Automated panning and screening procedure on microplates for antibody generation from phage display libraries. J Biomol. Screen. 14, 282-293 (2009).
- Hughes, R. A., Miklos, A. E., Ellington, A. D. Gene synthesis: methods and applications. Methods Enzymol. 498, 277-309 (2011).
- Boersma, Y. L., Pluckthun, A. DARPins and other repeat protein scaffolds: advances in engineering and applications. 22, 849-857 (2011).
- Terpe, K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 60, 523-533 (2003).
- Lichty, J. J., Malecki, J. L., Agnew, H. D., Michelson-Horowitz, D. J., Tan, S. Comparison of affinity tags for protein purification. Protein Expr. Purif. 41, 98-105 (2005).
- Fellouse, F. A., et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 373, 924-940 (2007).
- Paduch, M., et al. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods. 60, 3-14 (2013).
- Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207-234 (2005).
- Huang, H., Sidhu, S. S. Studying binding specificities of peptide recognition modules by high-throughput phage display selections. Methods Mol. Biol. 781, 87-97 (2011).
- McLaughlin, M. E., Sidhu, S. S. Engineering and analysis of peptide-recognition domain specificities by phage display and deep Sequencing. Methods Enzymol. 523, 327-349 (2013).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248-254 (1976).
- den Engelsman, J., et al. Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharm. Res. 28, 920-933 (2011).
- Lavinder, J. J., Hari, S. B., Sullivan, B. J., Magliery, T. J. High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J. Am. Chem. Soc. 131, 3794-3795 (2009).
- Fellouse, F. A., Sidhu, S. S., Howard, G. C., MR, K. a. s. e. r. . Making antibodies in bacteria. in Making and Using Antibodies: A Practical Handbook. , 151-172 (2013).
- Acton, T. B., et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods in Enzymology. 483, 23-47 (2011).
- Mahon, C. M., et al. Comprehensive interrogation of a minimalist synthetic CDR-H3 library and its ability to generate antibodies with therapeutic potential. J. Mol. Biol. 425, 1712-1730 (2013).
- Persson, H., et al. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. J. Mol. Biol. 425, 803-811 (2013).
- Bradbury, A. R., Sidhu, S., Dubel, S., McCafferty, J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 29, 245-254 (2011).

