Kinetic Measurement and Real Time Visualization of Somatic Reprogramming
Summary
The protocol presented in this study describes methods for the real-time monitoring of reprogramming progression via the kinetic measurement of positive and negative pluripotent stem cell markers using flow cytometry analysis. The protocol also includes the imaging-based assessment of morphology, and marker or reporter expression during iPSC generation.
Abstract
Somatic reprogramming has enabled the conversion of adult cells to induced pluripotent stem cells (iPSC) from diverse genetic backgrounds and disease phenotypes. Recent advances have identified more efficient and safe methods for introduction of reprogramming factors. However, there are few tools to monitor and track the progression of reprogramming. Current methods for monitoring reprogramming rely on the qualitative inspection of morphology or staining with stem cell-specific dyes and antibodies. Tools to dissect the progression of iPSC generation can help better understand the process under different conditions from diverse cell sources.
This study presents key approaches for kinetic measurement of reprogramming progression using flow cytometry as well as real-time monitoring via imaging. To measure the kinetics of reprogramming, flow analysis was performed at discrete time points using antibodies against positive and negative pluripotent stem cell markers. The combination of real-time visualization and flow analysis enables the quantitative study of reprogramming at different stages and provides a more accurate comparison of different systems and methods. Real-time, image-based analysis was used for the continuous monitoring of fibroblasts as they are reprogrammed in a feeder-free medium system. The kinetics of colony formation was measured based on confluence in the phase contrast or fluorescence channels after staining with live alkaline phosphatase dye or antibodies against SSEA4 or TRA-1-60. The results indicated that measurement of confluence provides semi-quantitative metrics to monitor the progression of reprogramming.
Introduction
Patient-derived induced pluripotent stem cells (iPSCs) are promising tools for cell therapy and drug screening. They provide an autologous source of cells for therapy. In addition, they encompass a very broad set of genetic backgrounds, enabling a detailed in vitro analysis of genetic diseases beyond what current embryonic stem cell (ESC) lines would allow. Recent advances have led to the development of several methods for generating iPSCs, including reprogramming with Sendai virus, episomal plasmids or mRNAs 1,2. Notably, different reprogramming methods are associated with varying levels of efficiency and safety, and are likely to differ in other ways that influence their appropriateness for various applications. With the availability of a variety of reprogramming technologies, it has become important to develop methods for assessing the reprogramming process. Most existing methods rely on the qualitative inspection of morphology or staining with stem cell-specific dyes and antibodies. One recently developed method makes use of lentiviral fluorescence reporters that are sensitive to PSC-specific miRNAs or differentiated cell-specific mRNAs 3. Such monitoring methods facilitate the selection and optimization of reprogramming techniques for different situations. For example, CDy1 has been used as a fluorescent probe for early iPSCs in order to screen for reprogramming modulators 4. The ability to observe and compare different reprogramming experiments is also critical for gaining a better understanding of the process itself. For instance, it is now known that some somatic cell types are easier to reprogram than others 5, and that cells go through intermediate states during reprogramming 6-8. Unfortunately, the mechanisms underlying the reprogramming process are still not completely understood and consequently, the exact differences between reprogramming methods also remain to be defined. Thus, methods for monitoring, assessing, and comparing reprogramming events continue to be critical for the stem cell field.
The methods described in this protocol permit the monitoring and assessing of the reprogramming process and illustrate how these techniques can be used to compare different sets of reprogramming reagents. The first approach involves flow cytometry analyses using combinations of antibodies against positive and negative pluripotent stem cell (PSC) markers. The second approach couples real-time imaging and the measurement of total confluence (the percent surface area covered by the cells) and confluence of marker signals (the percent surface area covered by the fluorescent signals).
Protocol
1.Solution and Medium Preparation
- Basement Membrane Matrix (Purified from Engelbreth-Holm-Swarm Tumor)
- Slowly thaw the basement membrane matrix (5 ml) on ice at 4 °C overnight.
- Dilute the stock solution 1:1 with 5 ml of ice cold, sterile DMEM/F-12 medium in a sterile, pre-chilled 15 ml conical tube. Dispense aliquots into pre-chilled, 1.5 ml sterile micro centrifuge tubes and immediately store at -20 °C.
- Prior to usage, thaw the frozen 1:1 basement membrane matrix aliquot overnight at 4 °C. At the time of use, further dilute the 1:1 aliquot another 50 fold with ice cold, sterile DMEM/F-12 medium to a final dilution of 1:100.
- Add the appropriate amount of the 1:100 diluted basement membrane matrix solution to the appropriate tissue culture (TC) vessels and incubate at 37 °C for 1 hr. Typically, use 3 ml of the diluted basement membrane matrix solution to coat a TC dish of 20 cm2 surface area, coat all other types of TC dishes/plates at a ratio of 0.15 ml of diluted matrix per cm2 of surface area. Aspirate the remaining solution prior to addition of any culture medium and cells.
- Fibroblast Medium
- Thaw a 100 ml bottle of ES Qualified Fetal Bovine Serum (ES-FBS) at 4 °C overnight.
- Add 50 ml of the thawed ES-FBS and 5 ml of MEM Non-Essential Amino Acid Solution (1x) to 445 ml of DMEM medium. Sterilize the combined components through a 0.22 µm filter and store the medium at 4 °C for up to 4 weeks.
- iMEF Medium
- Thaw a 100 ml bottle of ES-FBS at 4 °C overnight.
- Add 50 ml of the thawed FBS, 5 ml of MEM Non-Essential Amino Acid Solution (1x), and 500 µl of 2-mercaptoethanol to 445 ml of DMEM medium.
- Sterilize the combined components through a 0.22 µm filter and store the medium at 4 °C for up to 4 weeks.
- Basic Fibroblast Growth Factor solution (b-FGF)
- Add 10 µl of Serum Replacement to 990 µl of D-PBS and thoroughly mix by pipetting up and down.
- Add the 1 ml of the 1% (v/v) Serum Replacement solution to one vial of 10 µg lyophilized b-FGF. Thoroughly mix by gently pipetting up and down.
- Aliquot the 10 µg/ ml b-FGF solution into 200 µl aliquots in micro-centrifuge tubes and store at -20 °C until needed for up to 4 weeks.
- Human Pluripotent Stem Cell (hPSC) Medium
- Thaw a 100 ml bottle of Serum Replacement at 4 °C overnight.
- Add 100 ml of the thawed Serum Replacement, 5 ml of MEM Non-Essential Amino Acid Solution (1x), and 500 µl of 2-mercaptoethanol to 395 ml of DMEM/F-12 medium.
- Sterilize the combined components through a 0.22 µm filter and store the medium at 4 °C for up to 4 weeks.
- At the time of usage, pre-warm the hPSC medium to 37 °C and supplement with 4 ng/ml of bFGF.
- iMEF Conditioned Medium (CM)
- Add 18 ml of 0.1% gelatin to a T-175 Culture flask and incubate at 37 °C for 1 hr.
- Remove the 0.1% gelatin solution after one hour of incubation. Add 9.1 x 106 mitotically-inactivated mouse embryonic fibroblasts (iMEF) in 45 ml of iMEF medium and incubate in the 37 °C incubator overnight.
- Following overnight incubation, remove the old iMEF medium and wash once with 25 ml of D-PBS for 5 min at room temperature.
- Aspirate off the D-PBS wash and add 90 ml of pre-warmed hPSC medium, supplemented with 4 ng/ml of bFGF. Incubate in the 37 °C incubator for exactly 24 hr.
- After 24 hr of incubation, aseptically remove the hPSC medium from the iMEF flask and sterilize through a 0.22 µm filter. Aliquot into 50 ml conical tubes and freeze at -20 °C until needed. iMEF CM can be stored for up to 6 months at -20 °C.
- Add another 90 ml of pre-warmed hPSC medium supplemented with 4 ng/ml of bFGF to the iMEF flask. Incubate in the 37 °C incubator for exactly 24 hr. Repeat the harvesting of CM for up to 7 days.
- At the time of usage, pre-warm the iMEF-CM to 37 °C and supplement with 4 ng/ml of bFGF.
- E8 Medium
- Thaw a 10 ml E8 supplement at 4 °C overnight.
- Remove 10 ml of the E8 basal medium and aseptically add the contents of the thawed E8 supplement to the remaining basal medium. Mix thoroughly and sterilize through a 0.22 µm filter. Store the complete medium at 4 °C for up to 2 weeks.
- At the time of usage, pre-warm the aliquot of the medium to be used to room temperature. It is not recommended to pre-warm E8 Medium to 37 °C.
- Cell Sorting Buffer
- Prepare fresh sorting buffer right before cell sorting by adding EDTA solution (1 mM final concentration), HEPES buffer (25 mM final concentration) Penicillin/Streptomycin solution (100 units/ml final concentration) and Bovine Serum Albumin (1% final concentration) into Dulbecco's Phosphate Buffered Saline (Calcium and Magnesium free).
- Sterilize the buffer through a 0.22 µm filter prior to using.
- Cell Sorting Recovery Medium
- Modify 50 ml of Fibroblast Medium by adding Penicillin/Streptomycin (100 units/ml final concentration) and Rock Inhibitor Y-27632 (10 µM final concentration). Pre-warm this medium to 37 °C prior to seeding the sorted cells.
2. Sendai Mediated Reprogramming of Human Fibroblasts
- 24 hr prior to transduction with the Sendai viruses, seed the fibroblasts at a density of 2 × 105 cells per well into the desired wells of a 6-well TC plate.
- Perform the transduction on Day 0 as follows.
- Pre-warm 2 ml of Fibroblast Medium in a 37 °C water bath. Remove one set of Sendai virus tubes from -80 °C storage and thaw each tube one at a time by first immersing the bottom of the tube in a 37 °C water bath for 5 – 10 sec, and then remove the tube from the water bath and allow it to thaw at room temperature. Once thawed, briefly centrifuge the tube and place it immediately on ice.
- Use commercially second generation Sendai viruses by adding the appropriate volumes (as per the COA) for each of the tubes in order to achieve the following multiplicity of infection (MOI) to 1 ml of pre-warmed Fibroblast Medium per well to be transduced as follow: KOS = 5 x 106 CIU, hc-Myc = 5 x 106 CIU, hKlf4 = 3 x 106 CIU .
- Thoroughly mix the viral solution by pipetting the mixture gently up and down. Complete the next step within 5 min.
- Aspirate the Fibroblast Medium from the well of the fibroblasts to be reprogrammed, and add 1 ml of the viral solution to each well. Place the cells in a 37 °C, 5% CO2 incubator and incubate overnight.
- Following overnight incubation, aspirate off the transduction medium and dispose of the old medium properly (10% bleach solution for 30 min). Replace with 2 ml of pre-warmed Fibroblast Medium and continue to culture the transduced cells.
- Culture the cells for 6 more days, replacing the old medium with fresh Fibroblast Medium every other day.
Note: Do not passage the reprogrammed cells until 7 days post transduction. - Day 6: Prepare Dishes for iPSC Generation
- For feeder-dependent iPSC generation, add 1.5 ml of 0.1% gelatin solution to each well of a 6-well tissue culture (TC) dish and incubate at 37 °C for 1 hr.
- Remove the 0.1% gelatin solution after 1 hr of incubation. Add 3 x 105 mitotically-inactivated mouse embryonic fibroblasts (iMEF) in 2 ml of iMEF medium per well and incubate in the 37 °C incubator overnight.
- For feeder-independent iPSC generation, add 1.5 ml of diluted basement membrane matrix (1:100 dilution) to each well of a 6-well tissue culture (TC) dish and incubate at 37 °C for 1 hr.
- Seven days after transduction, harvest the transduced fibroblasts and re-plate them onto the pre-made culture dishes for iPSC colony generation.
- Aspirate off the normal culture medium from the fibroblasts, and wash the cells once with 2 ml of D-PBS.
- Aspirate off the D-PBS wash and add 0.5 ml of pre-warmed 0.05% trypsin/EDTA. Incubate the cells for 3 – 5 min at 37 °C.
- When the cells have rounded up, add 2 ml of pre-warmed Fibroblast Medium into each well to stop trypsinization. Dislodge the cells from the well by gently pipetting the medium across the surface of the dish. Collect the cell suspension in a 15-ml conical centrifuge tube.
- Centrifuge the cells at 200 x g for 2 min. Aspirate the supernatant, and re-suspend the cells in 2 ml fresh, pre-warmed Fibroblast Medium.
- Count the cells using the desired method (hemocytometer or automated cell counter) and seed the pre-prepared culture dishes with 5 x 104 cells for feeder-dependent conditions and 1 x 105 cells for feeder-independent conditions, per well of a 6-well dish and incubate at 37 °C, 5% CO2 incubator overnight.
- Following the overnight incubation, change the medium to hPSC medium, E8 medium or iMEF-conditioned media, depending on the application. Replace the old medium with fresh medium everyday thereafter.
3. Sendai Reprogramming of BGO1v hOct4-GFP Reporter Human Embryonic Stem Cells (hESC) Derived Secondary Fibroblasts
- Derive secondary human fibroblasts from the BG01v hOct4-GFP reporter hESC line as per the procedure previously described 9.
- Two days before transduction, plate BGO1v/hOG derived fibroblasts into two wells of a 6-well plate at 2 x 105 cells per well, using Fibroblast Medium.
- On the day of transduction, pre-warm 2 ml of Fibroblast Medium to 37 °C.
- Remove one set of Sendai tubes from the -80 °C storage. Remove the vials from the pouch and thaw each tube one at a time by first immersing the bottom of the tube in a 37 °C water bath for 5 – 10 sec, after which, allow the tubes to thaw at room temperature. Once thawed, briefly centrifuge the tube and place it immediately on ice.
- Add the indicated volumes of each of the three Sendai tubes (see the CoA for the appropriate volume) to 2 ml of Fibroblast Medium, pre-warmed to 37 °C and thoroughly mix the viral solution by pipetting up and down. Complete the next step within 5 min.
Note: For this experiment, a MOI of 5:5:6 was utilized. - Aspirate the Fibroblast Medium from the wells of fibroblasts to be transduced. Add 1 ml of the viral suspension to each of two wells to be transduced. Place the cells in a 37 °C, 5% CO2 incubator and incubate overnight.
- Following overnight incubation, remove the transduction medium and dispose of it properly (10% bleach solution for 30 min). Replace each well with 2 ml of pre-warmed Fibroblast Medium and continue to culture the transduced cells.
- Culture the cells for 6 more days, changing the old medium with fresh Fibroblast Medium every other day.
- Seven days post transduction, harvest the transduced fibroblasts and re-plate them onto the pre-made culture dishes for iPSC colony generation.
- Aspirate off the normal culture medium from the fibroblasts, and wash the cells once with 2 ml of D-PBS.
- Aspirate off the D-PBS wash and add 0.5 ml of pre-warmed 0.05% trypsin/EDTA. Incubate the cells for 3 – 5 min at 37 °C.
- When the cells have rounded up, add 2 ml of pre-warmed Fibroblast Medium into each well to stop trypsinization. Dislodge the cells from the well by gently pipetting the medium across the surface of the dish. Collect the cell suspension in a 15-ml conical centrifuge tube.
- Centrifuge the cells at 200 x g for 2 min. Aspirate the supernatant, and re-suspend the cells in an appropriate amount of fresh, pre-warmed Fibroblast Medium.
- Count the cells using the desired method (hemocytometer or automated cell counter).
- Seed the transduced cells at a density of 1 x 105 cells per well of a basement membrane matrix -coated 6-well plate and incubate in a 37 °C, 5% CO2 incubator overnight.
- Following the overnight incubation, change the medium to iMEF Conditioned Medium, supplemented with 10 ng/ml of b-FGF. Replace the old medium with fresh iMEF CM everyday thereafter.
- Three weeks post transduction, mechanically dissect and transfer the desired colonies for clonal expansion or use for analysis.
4. Live Cell Imaging of Fibroblast Reprogramming
- Real-Time Monitoring
- 7 days post transduction with the Sendai reprogramming kit, seed the desired amount of cells onto 6-well dishes with the desired medium and matrix systems as previously described. Place the seeded dishes inside the imager, located in a humidified incubator set at 37 °C, and 5% CO2.
- Set the imaging software to capture whole well images of each individual well at a set interval (every 6 – 8 hr) of continuous image capturing for the next 14 days according to manufacturer's protocol.
Note: Phase contrast image settings are selected for normal reprogramming to evaluate colony progression. In the case of BG01v/hOG derived fibroblast reprogramming, it is best to capture images in phase contrast and in the green fluorescent channel as the re-activation of the endogenous OCT4-GFP reporter can be tracked throughout the reprogramming process. - Change the medium daily, it can be hPSC medium, E8 medium, or iMEF-CM, depending on the experiment, as previously described from Day 8 to 21 post-transduction. The wells can be further analyzed for colony formation using indirect immunofluorescence.
- At the end of the experiment on Day 19 to 21, remove the culture medium from each well to be probed with antibodies for fibroblast and stem cell markers of choice.
- Wash each well once with 2 ml of pre-warmed DMEM/F-12 medium.
- Prepare aliquots of each positive/negative primary antibody pairs in 1 ml of DMEM/F-12 medium as follows: anti-mouse SSEA4 (1:200) and anti-rat CD44 (1:50), anti-mouse TRA-1-60 (1:200) and anti-rat CD44 (1:50), and Alkaline Phosphatase Live Stain (1:50) and anti-rat CD44 (1:50). Add the 1 ml primary antibody solutions to each corresponding well. Incubate at 37 °C for 45 min.
- Aspirate off the primary antibody solution and wash twice with pre-warmed DMEM/F-12 medium.
- Prepare secondary antibody solutions (1 ml for each well) using goat-anti-mouse Alexa Fluor 488 conjugated secondary (1:500) for green fluorescence and goat-anti-rat Alexa Fluor 594 conjugated secondary (1:500) for red fluorescence in DMEM/F-12 medium. Add to each well after the final wash. Incubate the secondary antibodies for 30 min at 37 °C.
Note: Due to Alkaline Phosphatase Live Stain's (APLS) transient signal, the anti-CD44 primary incubation should be first performed by itself and the APLS should be added at the time of incubation with the secondary antibodies - Following the secondary antibody incubation, aspirate off the solution and wash twice with DMEM/F-12 medium. Add fresh pre-warmed DMEM/F-12 medium to each well prior to final image capture.
- Set the imager software to acquire whole well images of each well using all three scanning parameters: phase contrast, green fluorescence and red fluorescence. Create images at various magnifications and create time-lapse movies with the software to monitor growth and marker expression. Use manufacturer's protocol.
- Utilizing the analysis software, evaluate confluence of the well over time in phase contrast and in green fluorescent units for the BG01v/hOG reprogramming.
- Monitoring Reprogramming Intermediates using Cell Surface Markers
- Monitor reprogramming events at different time points by probing the reprogramming dishes at various time points with antibodies against different stem cell positive/negative markers in order to monitor the reprogramming process. Probe cultures every 2 to 3 days depending on availability of replicates. Probe the reprogramming dishes at day 19 to 21 using the dual staining strategy to identify fully reprogrammed iPSC colonies and partially reprogrammed colonies based on the staining patterns.
- At the desired time point, aspirate off the normal growth medium from each well to be probed with antibodies for the markers of choice.
- Wash each well once with 2 ml of pre-warmed DMEM/F-12 medium.
- Prepare aliquots of each positive/negative primary antibody pairs in 1 ml of DMEM/F-12 medium as follows: SSEA4 conjugated to the green fluorescent fluorophore (1:200), TRA-1-60 conjugated to the green fluorescent fluorophore(1:50), Alkaline Live Stain (1:500), CD24 conjugated to a green fluorescent fluorophore (1:50), B2M conjugated to a green fluorescent fluorophore (1:50), EpCAM conjugated to a green fluorescent fluorophore(1:50), mouse CD73 (1:100) which was probed with anti-mouse secondary antibody conjugated to a green fluorescent fluorophore(1:500), and rat CD44 (1:50) which is probed with anti-rat secondary antibody conjugated to the red fluorescent fluorophore(1:500). Add the 1 ml primary antibody solutions to each corresponding well. Incubate at 37 °C for 45 min.
- Aspirate off the primary antibody solution and wash twice with pre-warmed DMEM/F12 medium.
- For unconjugated antibodies, proceed to using a 2-step method by preparing the appropriate secondary antibody solutions and incubate for 30 min at 37 °C. Confirm that secondary antibodies are to the primary hosts and do not cross react. Perform additional wash steps as previously described.
- After all appropriate washes, add fresh pre-warmed DMEM/F-12 medium to each well prior to final image capture.
- Capture the fluorescent images under the appropriate filter at 100X magnification using a fluorescent microscope and analyze the images using available analysis software.
5. Measurement of Reprogramming Kinetics Using Flow Cytometry
- Quantitative Kinetic Measurement of Reprogramming using Cell Surface Antibodies
- Prior to reprogramming, set up the appropriate number of reprogramming experiments for each time point. For measuring kinetics of reprogramming for the first 7 days of reprogramming, individual transductions are performed for each desired time point. For time points after Day 7, seed sufficient feeder-independent time points to continue to measure the reprogramming events. A single well of a 6-well plate will yield sufficient amounts of cells for performing flow cytometry analysis.
- Measure all desired reprogramming time points from individual wells, as this is an end point assay. Aspirate off the Fibroblast Medium from the desired well. Wash once with D-PBS.
- Aspirate off the D-PBS wash. Add 1 ml of 0.05% trypsin-EDTA solution to the well to be tested. Incubate at room temperature for 5 min.
- Following the incubation, dissociate the cells into single cells by flushing the 1 ml cell suspension against the surface of the well and transfer the suspension into a 15 ml conical tube. Use 3 ml of D-PBS to wash off any remaining cells from the well and add them to the 15 ml conical. Use 10 ml of additional D-PBS to further dilute the cell solution in the conical tube.
- Centrifuge the cells at 200 x g for 2 min. Aspirate the supernatant and re-suspend the pellet into 1 ml of DMEM/F-12 medium. Perform a cell count as needed to create a standard cell suspension that is less than 1 x 106 cells/ml.
- Prepare each aliquot of positive/negative directly-conjugated antibodies of choice in 1 ml of DMEM/F-12 medium for less 1 x 106 cells/ml using the following combinations: SSEA4 conjugated to a green fluorescent fluorophore (1:200) with CD44 conjugated to PE-Cy5, or CD44 conjugated to a green fluorescent fluorophore and SSEA4 conjugated to a far- red fluorescing fluorophore. Incubate the antibodies with the cell suspension for 45 min at room temperature.
- Aspirate the primary antibody solution and wash twice with pre-warmed DMEM/F-12 medium.
- Aspirate the final wash and re-suspend the cell pellet in 1 ml of D-PBS. Pipet the content of each suspension in individual FACS tubes.
- Using the appropriate negative controls (unstained and isotype controls), confirm the proper forward and side scatter profile and voltages on the cytometer. Set the voltages for the corresponding fluorophores according to initial single stain parameters, such that the green fluorophores are activated by the Blue Laser and signals acquired in the BL1 channel, while far red fluorescing fluorophore and PE-Cy5 fluorophore are activated by the Red Laser and signals acquired in the RL1 channel. Acquire the dual-stained population for each time point.
- Analyze the FCS files for each time point using the appropriate analysis software and use the editor to arrange data to observe the population expression shift over time.
- Follow steps 5.1.1 to 5.1.6, if cell sorting is desired at different time points. Use the appropriate fluorophore combinations according to the laser configuration of the cell sorter to be used.
- Following the final wash, re-suspend the cell pellet in 1 ml of the cell sorting buffer. Set up the cell sorter to the proper forward and side scatter settings using an unstained control. Use the single stained controls to set up the proper settings for each fluorophore.
- Use a small sample of the double-stained cell suspension; select the 2 populations to be sorted and assign the respective charge to ensure proper collection of the sorted cells.
- Add 200 µl of the cell recovery medium into the collection tubes to ensure that the sorted cell suspension is cushioned and protected at the time of sorting. Harvest the desire number of cells and transfer onto new iMEF dishes containing freshly prepared recovery medium.
- Change the medium following overnight incubation with the recovery medium. The cultures must be subsequently fed and routinely passaged using hPSC medium containing Penicillin/Streptomycin (100 units/ml final concentration).
6. Endpoint Analysis
- Terminal Alkaline Phosphatase (AP) Staining
- Use terminal chromogenic AP staining to correlate observed kinetic and morphological events with final reprogramming efficiencies.
- Following 21 days of reprogramming, remove the culture medium from the well to be stained with terminal AP stain and wash once with 200 mM Tris-HCl (pH 8.0).
- Prepare the appropriate amount of staining solution as directed by the manufacturer's manual in 200 mM Tris-HCl.
- Aspirate the wash and add 2 ml of the prepared staining solution to each well. Incubate for 20 to 30 min at room temperature and away from the light.
- Aspirate the staining solution and wash once with 200 mM Tris HCl. Remove the wash and add distilled water prior to visualization.
- Capture the colorimetric signal using a normal picture camera. Count the number of positively stained colonies by hand. Visualize the stain using fluorescent analysis in the Trit-C channel, as the stain is also fluorescent in nature and perform whole-well imaging and analyzed using the whole-well imager. Use manufacturer's protocol.
Representative Results
Monitoring Reprogramming Kinetics Using Flow Cytometry
CD44 is a fibroblast marker while SSEA4 is a PSC marker 6,10. As expected from this expression pattern, flow cytometry of BJ fibroblasts shows an SSEA4– CD44+ population that facilitates the creation of quadrant gates in combination with the unstained sample. During reprogramming of DF1 fibroblasts with the Sendai viruses, CD44 is gradually lost while SSEA4 is slowly expressed. At Day 3 after transduction with the Sendai Viruses, there is a large population of SSEA4– CD44+ cells and a small population of SSEA4+ CD44+ cells that becomes more distinct at Day 7 (Figure 1). At Day 13, the double-positive population transitions towards the SSEA4+ CD44– state. By Day 17, a large percentage of the cells in culture are already SSEA4+ CD44–. This time course shows that flow cytometry with CD44 and SSEA4 can be used to monitor the progression and rate of reprogramming.
A quantitative comparison confirms that reprogramming BJ fibroblasts with Sendai reprogramming viruses generates SSEA4+ CD44– cells at a different rate than reprogramming of BJ Fibroblasts (Figure 2A). The data demonstrates that an early comparison of the percentage of PSC-like SSEA4+ CD44– cells tracks the progression of reprogramming. Interestingly, at Day 8 of DF1 fibroblast reprogramming, sorting and separately culturing the population of cells that are in the process of upregulating SSEA4 and downregulating CD44 generates AP+ colonies 11 (Figure 2B). In contrast, the original SSEA4– CD44+ population generates a much smaller number of AP+ colonies at Day 21 of reprogramming, which is the typical day of final colony analysis. This suggests that it is possible to quantify and compare the transitioning population in order to predict differences in reprogramming kinetics even before the SSEA4+ CD44– population is formed. An important point to note here is that reprogramming is a variable process dependent on several factors such as starting somatic cell population, transduction efficiency, medium, etc. However the general pattern and progression of the above described surface markers is consistent between different reprogramming experiments and starting fibroblasts. The analysis of reprogramming kinetics can also be done with different markers, such as the newly identified negative PSC markers CD73 and B2M 6, as well as the established PSC markers CD24 12,13 and EPCAM 14,15. Similar to CD44, CD73 and B2M are expressed in parental BJ fibroblasts, but are downregulated during the course of reprogramming, becoming undetectable in fully reprogrammed iPSCs (Figure 3A). A flow cytometry time course using CD44 and CD73 or CD44 and B2M shows that CD44 and CD73 are downregulated at a similar rate, whereas B2M is downregulated faster than CD44 (Figure 3B). In both cases, the rate of reprogramming can be observed by measuring the accumulation of the double negative population. In contrast to these negative PSC markers, CD24 and EPCAM are absent in fibroblasts and are expressed in iPSCs (Figure 3A). In a flow cytometry time course, CD24– CD44+ and EPCAM– CD44+ fibroblasts eventually form double-negative populations that later transition into CD24+ CD44– and EPCAM+ CD44– PSC-like cells, respectively (Figure 3B). Thus, during reprogramming, both positive PSC markers are expressed after CD44 is downregulated. Similar to what was observed with SSEA4 and CD44, the formation and accumulation of the different cell populations indicate the progression of reprogramming.
Tracking Reprogramming Using Real-time Imaging Analysis
Real-time imaging of reprogramming cultures after reseeding shows the gradual formation of colonies (Figure 4A), which corresponds to a logarithmic increase in confluence under phase contrast (Figure 4B). Confluence under phase contrast thus provides another metric for monitoring reprogramming quantitatively, but it encompasses both, the colonies that are positive for PSC markers and the continued proliferation of unreprogrammed or partially reprogrammed cells (Figure 4A, last panel). Quantifying the areas that have been stained for pluripotency markers TRA-1-60, SSEA4 and AP 6,10 at Day 21 indicates that iPSCs only account for less than half of the total confluence (Figure 4B). To measure the progress of reprogramming more stringently, it is possible to use a reporter line for reprogramming. Here, secondary fibroblasts were generated from the BG01v/hOG ESC line, which has been engineered with an OCT4-GFP reporter 16. GFP is expressed as the cells are reprogrammed and as colonies are formed (Figure 5A). Measuring confluence under phase contrast and the green channels demonstrates that the GFP confluence increases more slowly than the total confluence, and the GFP confluence at Day 19 is less than half of the total confluence (Figure 5B), reminiscent of TRA-1-60, SSEA4 and AP staining at Day 21.
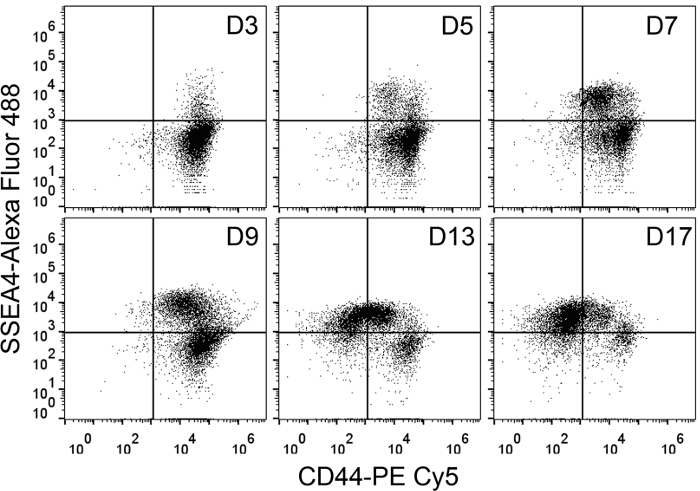
Figure 1. Monitoring the Progress of Reprogramming Using Flow Cytometry. Flow Cytometry Dot Plots for DF1 Fibroblasts Reprogrammed with Sendai Viruses under Feeder-free Conditions. Cells were harvested and stained with SSEA4 and CD44 antibodies at different intervals from Day 3 (D3) to Day 19 (D19) of the reprogramming process. Cultures at Day 9 (D9) onwards were analyzed after the cells were reseeded at Day 7 (D7). Dot plots show 8,000 singlets. Please click here to view a larger version of this figure.
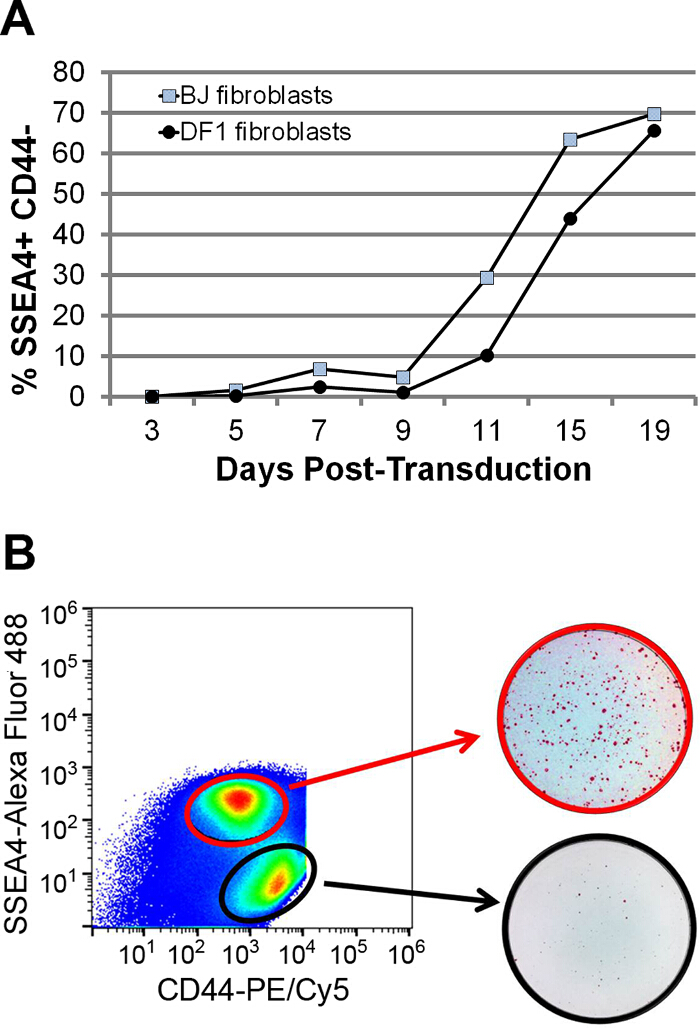
Figure 2. Evaluating Reprogramming Kinetics and Efficiency under Feeder-free Conditions. (A) Graph showing changes in the percentage of SSEA4+ CD44– reprogrammed cells after transducing DF1 fibroblasts and BJ fibroblasts with the Sendai viruses. Cells were analyzed by flow cytometry at Day 3 to 19 post-transduction, with cultures at Day 9 onwards undergoing reseeding at Day 7. (B) Pseudo-color flow cytometry plots for DF1 cells transduced Sendai viruses and stained with SSEA4 and CD44 antibodies. SSEA4– CD44+ cells (black circle) and SSEA4+ cells (red circle) were sorted at Day 8 and allowed to grow until Day 19 before staining for alkaline phosphatase (red). Please click here to view a larger version of this figure.
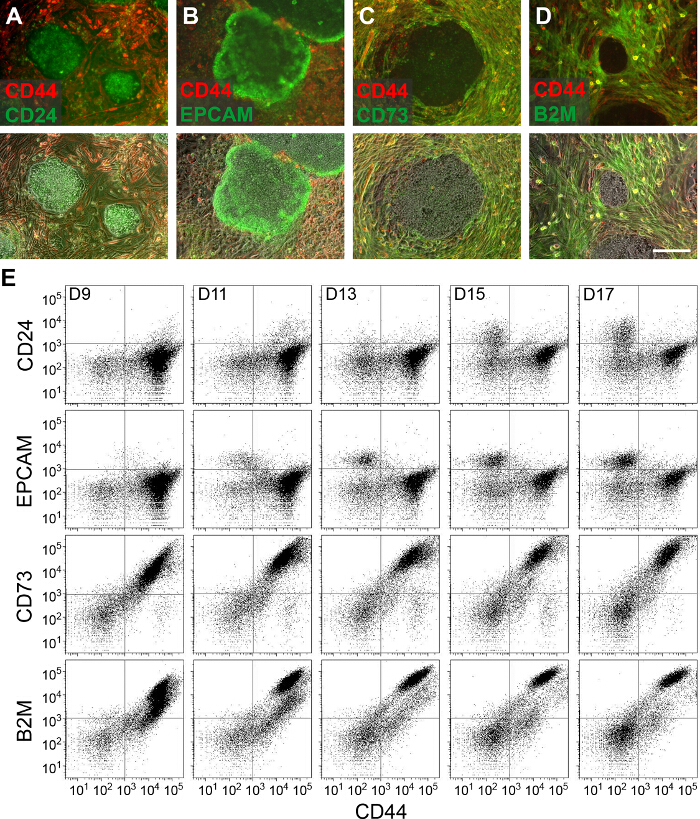
Figure 3. Observing Reprogramming Kinetics of Feeder-free Cultures Using Different Positive and Negative PSC Markers. (A-D) Live staining of Day-18 DF1-derived reprogramming cultures using antibodies for the positive PSC markers CD24 and EPCAM and negative PSC markers CD44, CD73, and B2M. The top panels merge the green and red fluorescence channels while the bottom panels also include the phase contrast image. The scale bar represents 200 µm. (E) Flow cytometry dot plots for DF1-derived reprogramming cultures harvested and stained using the same markers from Day 9 to 17 (D9 to D17) post-transduction. Prior to analysis, cultures were reseeded at Day 7. Dot plots show 8,000 singlets. Please click here to view a larger version of this figure.
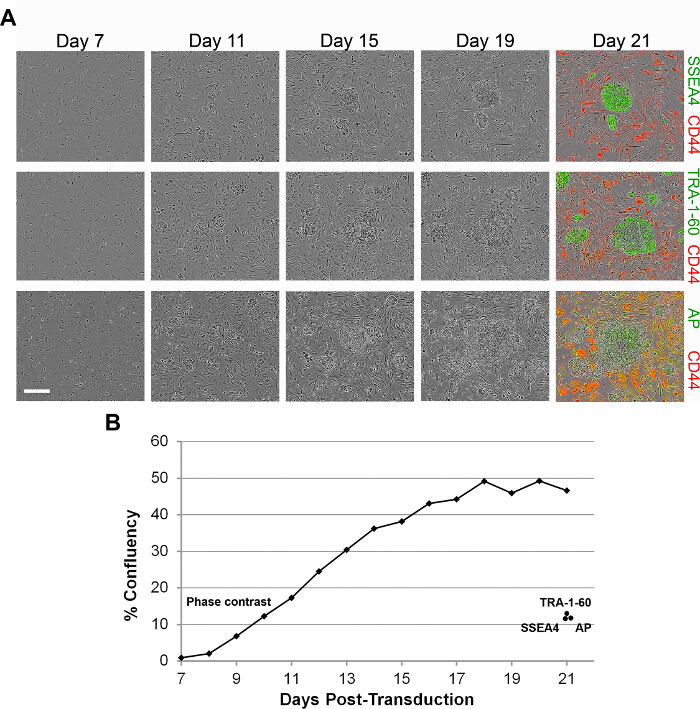
Figure 4. Tracking Reprogramming Kinetics by Measuring Total Confluence. (A) Real-time imaging of BJ fibroblasts reprogrammed with Sendai viruses under feeder-free conditions. Phase contrast images of the same colonies were taken at Day 7 to 19, then at Day 21 with additional staining for CD44 and SSEA4, TRA-1-60 or AP. Scale bar corresponds to 400 µm. (B) Quantification of total confluence (phase contrast) as reprogramming progresses from reseeding at Day 7 to Day 21. Total confluence at Day 21 is compared to SSEA4, TRA-1-60 and AP signal confluence (green channel). Please click here to view a larger version of this figure.
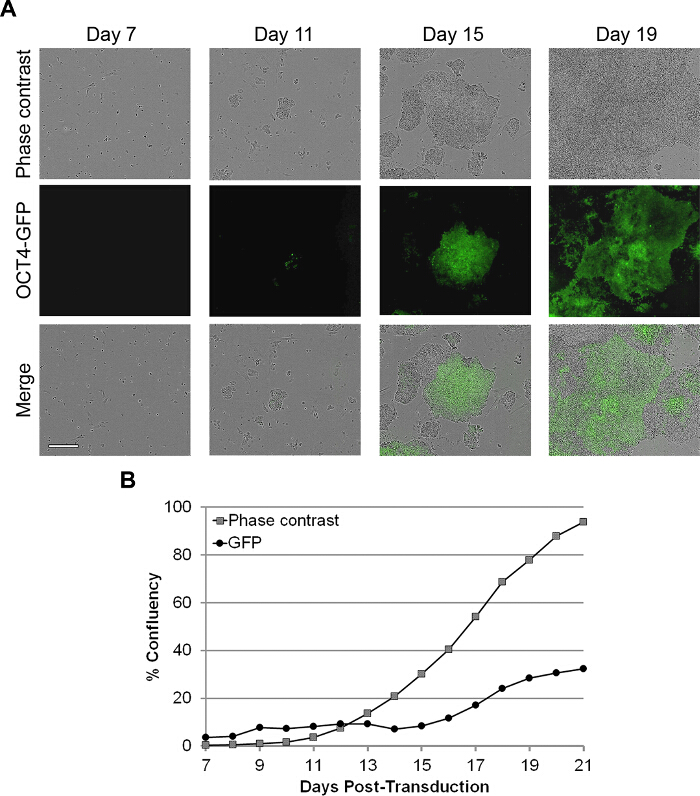
Figure 5. Assessing Reprogramming Kinetics by Measuring OCT4-GFP Confluence. (A) Real-time imaging of BG01v/hOG hESC-derived fibroblasts reprogrammed with Sendai viruses under feeder-free conditions. Phase contrast, green fluorescence and merged images of the same colonies were generated at Day 7 to 19. Scale bar corresponds to 400 µm. (B) Quantification of total confluence (phase contrast) and OCT4-GFP confluence (green channel) as reprogramming progresses from re-seeding at Day 7 to Day 21. Please click here to view a larger version of this figure.<!–
Discussion
This study provides strategies for monitoring and tracking of the reprogramming process using flow cytometry and real-time imaging-based analysis. The critical steps in the protocol are initiating reprogramming, measuring reprogramming progression based on marker expression and real-time monitoring of reprogramming. Any reprogramming method of choice can be used but here we focus on Sendai based reprogramming of human fibroblasts. The advantage of this method is the ease of use and consistent high efficiency of reprogramming.
To assess reprogramming progression, cells are harvested at discrete stages of reprogramming and subjected to an end-point flow cytometry analysis. Double staining of reprogramming cultures with the negative PSC marker CD44 and the positive PSC marker SSEA4 allows the assessment of reprogramming progression6. More specifically, the distribution of cells expressing either, neither or both markers at discrete stages of reprogramming enables the quantitative comparison of reprogramming kinetics under different conditions. This approach can demonstrate the differences in the kinetics and efficiency between different reprogramming methods and the media and matrices used during the reprogramming processes. The results obtained using different starting somatic cell samples confirms the importance of these factors in reprogramming efficiency and progression. They also demonstrate how this approach can be used to optimize reprogramming methods. The feasibility of monitoring reprogramming through flow cytometry using other combinations of pluripotent and non-pluripotent markers such as EPCAM, CD24, CD73, and B2M can be used in conjunction with the fore mentioned method14. Interestingly, these marker combinations show the formation of multiple cell populations through the course of reprogramming. Further studies will be required to fully characterize those populations and to determine their relevance in the reprogramming process. Regardless, the applicability of the flow cytometry approach with this diverse set of markers indicates that it can be applied more broadly, making use of different markers-of-interest and uncovering many other reprogramming intermediates. The limitation with this approach is that cells will need to be harvested for end point analysis. This limitation can be overcome with the use of live monitoring methods.
Monitoring of cells undergoing reprogramming in real time is done using the Live Cell Imaging system. Confluence, as measured under phase contrast, increased rapidly as colonies began to form. As such, measuring total confluence under phase contrast allows us to track the progression of reprogramming easily in real time, but because this measurement encompasses unreprogrammed and partially reprogrammed cells that are negative for AP, SSEA4 and TRA-1-60, this approach is only semi-quantitative. Linking the real-time phase contrast images to end-point positive marker staining with either dyes such as Live Alkaline Phosphatase or pluripotent markers such as SSEA4 or TRA-1-60 provides better insight into the kinetics of the reprogramming process11. However, this study shows that a more accurate and quantitative imaging-based method for tracking reprogramming would involve measuring fluorescence signal confluence while using an OCT4 reporter line16. Each of the methods presented here have their own advantages and disadvantages that make them appropriate for different situations. The flow cytometry approach is laborious and requires a lot of cells, but it provides the most accurate quantitative data. Measuring fluorescence confluence while using an OCT4 reporter line is easier and still quantitative, but it is not always feasible to obtain and use OCT4 reporter-containing parental cells. Finally, measuring total confluence under phase contrast is easy and applicable to any parental cell, but this approach is only semi-quantitative. Ultimately, the choice of method should depend on the available resources and the requirements of the actual experiments involved.
The methods outlined here provide a platform for comparing new reprogramming technologies, optimizing current work flows and extending the use of reprogramming to novel primary cell types.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Chad MacArthur for helpful discussions.
Materials
| DMEM, high glucose, GlutaMAXSupplement, pyruvate | Thermo Fisher Scientific | 10569-010 | |
| Fetal Bovine Serum, embryonic stem cell-qualified, US origin | Thermo Fisher Scientific | 16141-061 | |
| MEM Non-Essential Amino Acids Solution (100X) | Thermo Fisher Scientific | 11140-050 | |
| Trypsin-EDTA (0.05%), phenol red | Thermo Fisher Scientific | 25300-054 | |
| Mouse (ICR) Inactivated Embryonic Fibroblasts | Thermo Fisher Scientific | A24903 | |
| Attachment Factor Protein (1X) | Thermo Fisher Scientific | S-006-100 | |
| DMEM/F-12, GlutaMAX supplement | Thermo Fisher Scientific | 10565-018 | |
| KnockOut Serum Replacement | Thermo Fisher Scientific | 10828010 | |
| 2-Mercaptoethanol (55 mM) | Thermo Fisher Scientific | 21985-023 | |
| Collagenase, Type IV, powder | Thermo Fisher Scientific | 17104-019 | |
| TrypLE Select Enzyme (1X), no phenol red | Thermo Fisher Scientific | 12563-011 | |
| DPBS, no calcium, no magnesium | Thermo Fisher Scientific | 14190-144 | |
| Geltrex LDEV-Free, hESC-Qualified, Reduced Growth Factor Basement Membrane Matrix | Thermo Fisher Scientific | A1413302 | |
| Essential 8 Medium | Thermo Fisher Scientific | A1517001 | |
| FGF-Basic (AA 1-155) Recombinant Human Protein | Thermo Fisher Scientific | PHG0264 | |
| UltraPure 0.5M EDTA, pH 8.0 | Thermo Fisher Scientific | 15575-020 | |
| Bovine Albumin Fraction V (7.5% solution) | Thermo Fisher Scientific | 15260-037 | |
| HEPES (1 M) | Thermo Fisher Scientific | 15630-080 | |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | 15140-122 | |
| InSolution Y-27632 | EMD Millipore | 688001 | |
| CytoTune-iPS Sendai Reprogramming Kit | Thermo Fisher Scientific | A1378001 | |
| CytoTune-iPS 2.0 Sendai Reprogramming Kit | Thermo Fisher Scientific | A16517 | |
| Countess II Automated Cell Counter | Thermo Fisher Scientific | AMQAX1000 | |
| Countess Cell Counting Chamber Slides | Thermo Fisher Scientific | C10228 | |
| BJ ATCC Human Foreskin Fibroblasts, Neonatal | ATCC | CRL-2522 | |
| DF1 Adult Human Dermal Fibroblast | Thermo Fisher Scientific | N/A | |
| BG01V/hOG Cells Variant hESC hOct4-GFP Reporter Cells | Thermo Fisher Scientific | R7799-105 | |
| IncuCyte ZOOM | Essen BioScience | ||
| SSEA-4 Antibody, Alexa Fluor 647 conjugate (MC813-70) | Thermo Fisher Scientific | SSEA421 | |
| SSEA-4 Antibody, Alexa Fluor 488 conjugate (eBioMC-813-70 (MC-813-70)) | Thermo Fisher Scientific | A14810 | |
| SSEA-4 Antibody (MC813-70) | Thermo Fisher Scientific | 41-4000 | |
| TRA-1-60 Antibody (cl.A) | Thermo Fisher Scientific | 41-1000 | |
| CD44 Rat Anti-Human/Mouse mAb (clone IM7), PE-Cy5 conjugate | Thermo Fisher Scientific | A27094 | |
| CD44 Alexa Fluor 488 Conjugate Kit for Live Cell Imaging | Thermo Fisher Scientific | A25528 | |
| CD44 Rat Anti-Human/Mouse mAb (Clone IM7) | Thermo Fisher Scientific | RM-5700 (no longer available) | |
| Goat anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | A-11029 | |
| Goat anti-Rat IgG (H+L) Secondary Antibody, Alexa Fluor 594 conjugate | Thermo Fisher Scientific | A-11007 | |
| Alkaline Phosphatase Live Stain | Thermo Fisher Scientific | A14353 | |
| TRA-1-60 Alexa Fluor 488 Conjugate Kit for Live Cell Imaging | Thermo Fisher Scientific | A25618 | |
| CD24 Mouse Anti-Human mAb (clone SN3), FITC conjugate | Thermo Fisher Scientific | MHCD2401 | |
| beta-2 Microglobulin Antibody, FITC conjugate (B2M-01) | Thermo Fisher Scientific | A15737 | |
| EpCAM / CD326 Antibody, FITC conjugate (VU-1D9) | Thermo Fisher Scientific | A15755 | |
| CD73 / NT5E Antibody (7G2) | Thermo Fisher Scientific | 41-0200 | |
| VECTOR Red Alkaline Phosphatase (AP) Substrate Kit | Vector Laboratories | SK-5100 | |
| Zeiss Axio Observer.Z1 microscope | Carl Zeiss | 491912-0003-000 | |
| FlowJo Data Analysis Software | FLOJO, LLC | N/A | |
| Attune Accoustic Focusing Cytometer, Blue/Red Laser | Thermo Fisher Scientific | Use Attune NXT | |
| S3e Cell Sorter (488/561 nm) | BIO-RAD | 1451006 | |
| Falcon 12 x 75 mm Tube with Cell Strainer Cap | Corning | 352235 | |
| Falcon 15 mL, high-clarity, dome-seal screw cap | Corning | 352097 | |
| Falcon T-75 Flask | Corning | 353136 | |
| Falcon T-175 Flask | Corning | 353112 | |
| Falcon 6-well dish | Corning | 353046 | |
| HERAEUS HERACELL CO2 ROLLING INCUBATOR | Thermo Fisher Scientific | 51013669 | |
| Nonstick, RNase-free Microfuge Tubes, 1.5 mL | AM12450 | ||
| HulaMixer Sample Mixer | 15920D |
References
- Yamanaka, S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 10, 678-684 (2012).
- Robinton, D. A., Daley, G. Q. The promise of induced pluripotent stem cells in research and therapy. Nature. 481, 295-305 (2012).
- Kamata, M., Liang, M., Liu, S., Nagaoka, Y., Chen, I. S. Live cell monitoring of hiPSC generation and differentiation using differential expression of endogenous microRNAs. PLoS One. 5, e11834 (2010).
- Vendrell, M., Zhai, D., Er, J. C., Chang, Y. T. Combinatorial strategies in fluorescent probe development. Chem Rev. 112, 4391-4420 (2012).
- Aasen, T., et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 26, 1276-1284 (2008).
- Quintanilla, R. H., Asprer, J. S., Vaz, C., Tanavde, V., Lakshmipathy, U. CD44 is a negative cell surface marker for pluripotent stem cell identification during human fibroblast reprogramming. PLoS One. 9, e85419 (2014).
- Papp, B., Plath, K. Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res. 21, 486-501 (2011).
- Nefzger, C. M., Alaei, S., Knaupp, A. S., Holmes, M. L., Polo, J. M. Cell surface marker mediated purification of iPS cell intermediates from a reprogrammable mouse model. J Vis Exp. , e51728 (2014).
- Xu, C., et al. Immortalized fibroblast-like cells derived from human embryonic stem cells support undifferentiated cell growth. Stem Cells. 22, 972-980 (2004).
- International Stem Cell Initiative. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 25, 803-816 (2007).
- Singh, U., et al. Novel live alkaline phosphatase substrate for identification of pluripotent stem cells. Stem Cell Rev. 8, 1021-1029 (2012).
- Naujok, O., Lenzen, S. A critical re-evaluation of CD24-positivity of human embryonic stem cells differentiated into pancreatic progenitors. Stem Cell Rev. 8, 779-791 (2012).
- Ramirez, J. M., et al. Brief report: benchmarking human pluripotent stem cell markers during differentiation into the three germ layers unveils a striking heterogeneity: all markers are not equal. Stem Cells. 29, 1469-1474 (2011).
- Huang, H. P., et al. Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming. J Biol Chem. 286, 33520-33532 (2011).
- Kolle, G., et al. Identification of human embryonic stem cell surface markers by combined membrane-polysome translation state array analysis and immunotranscriptional profiling. Stem Cells. 27, 2446-2456 (2009).
- Thyagarajan, B., et al. Creation of engineered human embryonic stem cell lines using phiC31 integrase. Stem Cells. 26, 119-126 (2008).

