In Vitro Differentiation of Human Pluripotent Stem Cells into Trophoblastic Cells
Summary
Here, we present a protocol to efficiently generate human trophoblastic cells from human pluripotent stem cells using bone morphogenic protein 4 and inhibitors of the Activin/Nodal pathways. This method is suitable for the efficient differentiation of human pluripotent stem cells and can generate large quantities of cells for genetic manipulation.
Abstract
The placenta is the first organ to develop during embryogenesis and is required for the survival of the developing embryo. The placenta is comprised of various trophoblastic cells that differentiate from the extra-embryonic trophectoderm cells of the preimplantation blastocyst. As such, our understanding of the early differentiation events of the human placenta is limited because of ethical and legal restrictions on the isolation and manipulation of human embryogenesis. Human pluripotent stem cells (hPSCs) are a robust model system for investigating human development and can also be differentiated in vitro into trophoblastic cells that express markers of the various trophoblast cell types. Here, we present a detailed protocol for differentiating hPSCs into trophoblastic cells using bone morphogenic protein 4 and inhibitors of the Activin/Nodal signaling pathways. This protocol generates various trophoblast cell types that can be transfected with siRNAs for investigating loss-of-function phenotypes or can be infected with pathogens. Additionally, hPSCs can be genetically modified and then differentiated into trophoblast progenitors for gain-of-function analyses. This in vitro differentiation method for generating human trophoblasts starting from hPSCs overcomes the ethical and legal restrictions of working with early human embryos, and this system can be used for a variety of applications, including drug discovery and stem cell research.
Introduction
The placenta is required for the growth and survival of the fetus during pregnancy and facilitates the exchange of gases, nutrients, waste products, and hormones between maternal and fetal circulation. The first organ formed during mammalian embryogenesis is the placenta, which begins developing 6-7 days post-conception in humans and 3.5-4.5 days in mice1,2,3,4. Trophoblastic cells are the most important cells of the placenta, and these cells represent one of the earliest lineage differentiation events of the mammalian embryo. They arise from the outer extra-embryonic trophectoderm cells of the preimplantation blastocyst. Our knowledge of the early stages of placental development is limited by ethical and logistical restrictions on modeling early human development.
During embryonic implantation, trophoblasts invade the maternal epithelium and differentiate into specialized progenitor cells5. Cytotrophoblasts (CTBs) are mononucleated, undifferentiated progenitors that fuse and differentiate into syncytiotrophoblasts (SYNs) and extravillous invasive trophoblasts (EVTs), which anchor the placenta to the uterus. SYNs are multinucleated, terminally differentiated cells that synthesize hormones necessary for sustaining pregnancy. The early differentiation events that generate EVTs and SYNs are essential for placental formation, as impairments in trophoblastic cells result in miscarriage, pre-eclampsia, and intrauterine growth restriction1. The types of human trophoblast cell lines that have been developed include immortalized CTBs and choriocarcinomas, which produce placental hormones and display invasive properties6. Primary trophoblastic cells from human first-trimester placentas can be isolated, but the cells quickly differentiate and stop proliferating in vitro. Importantly, transformed and primary cell lines have different gene expression profiles, indicating that tumorigenic and immortalized trophoblast cell lines may not accurately represent primary trophoblasts7. Additionally, these lines are unlikely to resemble placental trophoblast stem cell progenitors because they are derived from later-stage first through third trimesters.
There is a need for a robust in vitro culture system of early-stage human trophoblasts in order to study the early events of placental formation and function. Human embryonic stem cells (hESCs), which share properties with the inner cell mass of the preimplantation embryo, are frequently used to model early human development, including the formation of the early placenta. Both human induced pluripotent stem cells (hiPSCs) and hESCs can be differentiated into trophoblasts in vitro using Bone Morphogenic Protein 4 (BMP4)8,9,10,11,12,13,14,15. This conversion of pluripotent cells to trophoblastic cells using BMP4 is specific for human cells and is widely used to study the development of the early human placenta because it does not require access to early human embryos9,16. Recently, it was discovered that the addition of the inhibitors A83-01 (A) and PD173074 (P), which block the SMAD2/3 and MEK1/2 signaling pathways, increases the efficiency of hPSC differentiation into trophectoderm-like progenitors, mainly SYNs and EVTs, without the extensive generation of mesoderm, endoderm, or ectoderm cells9,17. Using these medium conditions, hESCs differentiated for 12 days have similar gene expression profiles as trophectoderm cells isolated from human blastocyst-stage embryos and secrete various placental-specific growth hormones, supporting the validity of this in vitro model system9,11. Here, we present a detailed protocol for the in vitro differentiation of hPSCs into human trophoblast progenitors using BMP4/A/P culture medium. These conditions produce abundant numbers of cells for a wide variety of applications, including RNA sequencing, gene disruption using siRNAs, pathogen infections, and genetic modification using lipofection-mediated transfection.
Protocol
NOTE: For the differentiation of either hESCs or hiPSCs into trophoblast progenitors, hPSCs grown on mouse embryonic fibroblasts (MEFs) are transitioned to feeder-free conditions for two passages before initiating differentiation with BMP4/A/P. This process eliminates the MEF contamination of differentiated cells. Here, we present a protocol for hESC differentiation, and the same protocol can be applied to hiPSCs.
1. Culture and Recovery of hESCs on Irradiated Mouse Embryonic Fibroblasts (MEFs) (Preparations)
- Gamma-irradiation of MEFs
- Make 500 ml of medium for culturing the MEFs: DMEM with 10% FBS, 2 mM L-glutamine, 1x penicillin/streptomycin (Pen/Strep), and 0.1 mM non-essential amino acids (NEAA).
- Thaw one vial of primary MEFs and use MEF medium for culturing the cells. Expand the primary MEFs to 30 plates using MEF culture medium.
NOTE: Oxygen concentrations are not critical for this step, so either physiological (4%) or ambient (20%) conditions within the incubator can be used. - Harvest the MEFS. Use 0.25% trypsin to remove the MEFs from the plates and transfer the cells to a conical tube. Place the MEFs into an irradiation instrument and expose the cells to 3,500 grays.
NOTE: The amount of time will depend on the activity of the source inside the irradiator unit. - Resuspend the irradiated MEFs with MEF culture medium and count the number of cells using a hemocytometer.
- Pellet the MEFs using a centrifuge and add 50% MEF culture medium and 50% cell freezing medium (80% FBS and 20% MEF culture medium). Aliquot 1 x 106 cells per vial.
- Place the irradiated MEF vials in a -80 °C freezer overnight, and then transfer them to liquid nitrogen for long-term storage.
- Thawing frozen MEFs and hESCs for culture
- Make 500 ml of hESC medium: DMEM/F-12 with 20% Serum Replacement, 0.1 mM NEAA, 2 mM L-Glutamine, 10 ng/ml bFGF, 0.1 mM ß-mercaptoethanol, and 1x Pen/Strep.
- Thaw one vial of MEFs one day before thawing the hESCs. Coat a 6-well plate with 0.1% gelatin, using 1 ml for each well. Incubate for at least 20 min at room temperature, and then remove the gelatin.
- Remove a vial of MEFs from storage in liquid nitrogen and immerse the vial in a 37 °C water bath. Watch the vial intermittently until only small ice crystals remain. Quickly transfer the contents of the vial to 9 ml of MEF medium inside a 15 ml conical tube.
- Pellet the cells by centrifuging at 200 x g for 5 min. Aspirate the supernatant and resuspend the cell pellet in MEF medium. Aliquot the MEFs evenly onto a 6-well plate. Put the plate into a 37 °C low-oxygen (4%) incubator overnight.
- Routine culture of hESCs on MEFs
- Remove an hESC vial from liquid nitrogen and quickly thaw the vial using a 37 °C water bath. Gently pipette the hESCs into a 15 ml conical tube containing 9 ml of hESC medium and spin down at 200 x g for 5 min. Remove the medium and resuspend the cells with 1 ml of hESC medium.
- Aspirate the MEF medium from the plate prepared in step 1.2.4 and add 1 ml of hESC medium. Add Rock inhibitor Y-27632 (10 µM) to the hESC medium. Transfer the hESCs onto MEFs.
- Place the cells into a 37 °C low-oxygen (4%) incubator overnight. Replace the hESC medium daily. Scrape off differentiated cells with a Pasteur pipette, visualized under an upright microscope at 4X.
- Passage the hESCs every 4-6 days, depending on the cell confluency and the size of the hESC colonies. Prepare a plate of MEFs the day before passaging. When the cells are ready to passage, remove the hESC medium and wash the well with 1 ml of PBS.
- Add 1 mg/ml collagenase (pre-warmed to 37 °C), incubate at 37 °C for 5 min, and remove the collagenase. Wash the cells with PBS, add 1 ml of hESC medium per well, and manually scrape the cells into small clumps using the tip of a 5 ml pipette. Transfer the suspended cell clumps into a new MEF-coated well(s).
2. Transition of hESCs from MEFs to Feeder-free Conditions on Extracellular Matrix-coated Plates
- Prepare MEF Conditioned Medium (CM)
- Plate the irradiated MEFs in a T75 flask at 90-100% confluency; it is important that the MEFs are densely plated. Observe the cells using a microscope to determine whether they have attached to the bottom of the flask (MEFs attach approximately 6 hr after plating or the next day).
NOTE: Unattached cells float in the medium when observed using a microscope. The next day, remove MEF medium and add 25 ml of hESC medium lacking B-FGF. - Collect the conditioned medium after 24 hr of incubation. Replace with fresh medium daily for a maximum of 12 days.
- Filter the collected CM using a 0.22 µM filter. Prior to use, add fresh B-FGF to the CM.
NOTE: CM can be frozen at -80 °C and stored for up to 1 year. Alternatively, store the CM at 4 °C for 2 weeks.
- Plate the irradiated MEFs in a T75 flask at 90-100% confluency; it is important that the MEFs are densely plated. Observe the cells using a microscope to determine whether they have attached to the bottom of the flask (MEFs attach approximately 6 hr after plating or the next day).
- Transition the hESCs from culture on the MEFs to feeder-free extracellular matrix-coated plates.
- Preparation of extracellular matrix for coating tissue culture plates
- Thaw an aliquot of extracellular matrix on ice (about 3-4 hr). Using ice-cold tips, one cold, 50 ml conical tube, and DMEM/F12 medium, transfer 2 mg of extracellular matrix into 24 ml of ice-cold DMEM/F-12 (the dilution depends on the extracellular matrix concentration from the supplier).
- Immediately transfer the 50 ml conical tube to ice and store it at 4 °C. For coating tissue culture plates, add the appropriate amount of diluted extracellular matrix to the well (e.g., 1 ml per well in a 6-well plate). Swirl to coat the plate and incubate at room temperature for at least 1 hr.
- Passage hESCs from the MEFs (as in step 1.3) using CM containing B-FGF (10 ng/ml).
- Aspirate to remove the extracellular matrix from the new plate and add CM containing B-FGF. Transfer the scraped cellular clumps from the MEF plate using a pipette and aliquot it into the extracellular matrix-coated plate. Incubate in the 37 °C low-oxygen incubator overnight.
- Replace the CM with B-FGF medium daily; the amount of medium will depend on the size of the culture flask. Use 2 ml of medium for one 6-well well. Scrape off differentiated cells with a Pasteur pipette.
- Passage hESCs every 6-7 days, when colonies are bright when viewed under the microscope.
NOTE: hESC colonies grown on extracellular matrix-coated plates grow larger than MEF-cultured cells. - When the feeder-free cells are ready to passage, remove the medium, wash by adding 1 ml of PBS using a pipette, aspirate to remove the PBS, add 0.5 mg/ml dispase (pre-warmed to 37 °C) with a pipette, and incubate at 37 °C for 5 min.
- Wash the cells with PBS by adding 2 ml of PBS per well and aspirating afterwards. Add 1 ml of CM per well and manually scrape the cells into small clumps using the tip of a 5 ml pipette.
- Plate the suspended cell clumps into new extracellular matrix-coated plates; the cells should adhere after 24 hr.
- Preparation of extracellular matrix for coating tissue culture plates
3. Differentiation of hESCs Using BMP4/A/P
- Prepare the differentiation medium. Use CM (lacking B-FGF) and add (fresh) BMP4 (50 ng/ml), A83-01 (1 µM), and PD173074 (0.1 µM). Add these inhibitors to the CM prior to use.
- Use feeder-free hESCs growing on extracellular matrix-coated plates for 1 passage, cultured inside an incubator set at 4% oxygen. After the first passage onto extracellular matrix-coated plates, let the cells attach for 24 hr. The next day, initiate differentiation. Remove the CM (containing B-FGF) and replace it with CM containing BMP4/A/P (2 ml per well in a 6-well plate). Continue culturing the cells using 4% oxygen levels.
- Replace the CM containing BMP4/A/P (2 ml/well for a 6-well plate) every 2 days. Aspirate to remove the old medium and add new CM using a pipette. On the second day after adding BMP4 and removing B-FGF (considered differentiation day 2), the cell morphology will change, appearing larger when observed using a microscope.
NOTE: Undifferentiated cells, which exhibit bright round edges, will not be present. - Collect cells at desired time-points. Differentiated cells will stop dividing after around 2 weeks. Transfect cells during this time period (see the next section).
4. Transfection of Trophoblastic Cells with siRNAs or Plasmid DNA
- Prepare trophoblastic cells for transfection
- Culture hESCs for two passages on extracellular matrix after transferring them from the MEFs (see step 2.2). Use hESC medium containing B-FGF.
- Split the hESCs onto a new extracellular matrix plate one day before differentiation. High cellular confluency is important, so split the cells 1:1 onto a new plate/well, which will ensure at least 50% confluency the next day. Incubate the cells overnight in the low-oxygen incubator.
NOTE: hESCs do not need to be counted during this step. - The next day, replace the medium with the differentiation medium (CM containing BMP4/A/P and lacking B-FGF, using 2 ml/well for a 6-well plate). Remove the old medium by aspiration and transfer the new medium (2 ml) using a pipette. Place the cells in a low-oxygen incubator (4% oxygen) overnight.
- Transfections using siRNAs for gene disruption or using plasmid DNA
- Differentiate the cells until the desired time-point (between days 1 and 14). The day before transfection, trypsinize the cells using 0.05% trypsin for 5 min. Plate them to the desired confluency (depending on the transfection reagent protocol) onto a gelatin-coated plate (add 0.1% gelatin to a tissue culture plate for 20 min and aspirate to remove). Add CM containing BMP4/A/P (lacking B-FGF) and incubate overnight.
- The next day, add fresh differentiation medium to the cells. Transfect siRNAs using an siRNA transfection reagent. For plasmid DNA, use an appropriate lipofectamine reagent.
- Follow the product protocol as described for each transfection reagent. Transfer the siRNA-lipofectamine complexes to each well/plate of cells, mix gently, and culture the cells overnight in a low-oxygen incubator.
NOTE: Some transfection reagents are inhibited by antibiotics, which require CM lacking Pen/Strep. - The next day, replace with fresh differentiation media.
- Harvest the cells at the desired time-points (e.g., 24 hr, 48 hr, and 72 hr) to check the gene disruption efficiency using quantitative RT-PCR. For harvesting cells, use 0.05% trypsin, adding 1 ml per well and incubating for 5 min at 37 °C. Add 5 ml of MEF medium per well to neutralize the trypsin. Spin down the cells at 200 x g for 5 min and use the cell pellet for RNA isolation.
5. Pathogen Infection of Trophoblastic Cells: Sendai Viral Infection
- Prepare hESCs for differentiation
- Culture hESCs for two passages on extracellular matrix after the transfer from the MEFs (see step 2.2). Use hESC medium containing B-FGF.
- Split the hESCs onto a new extracellular matrix plate one day before differentiation. High cellular confluency is important, so split cells 1:1 onto a new plate/well, which will ensure at least 50% confluency the next day. Incubate the cells overnight in a low-oxygen incubator (4% oxygen).
NOTE: hESCs do not need to be counted during this step. - The next day, replace the medium with the differentiation medium (CM containing BMP4/A/P and lacking B-FGF) and culture overnight in a low-oxygen incubator (4% oxygen).
- Sendai viral infection of trophoblastic cells
- One day before the desired differentiation day, trypsinize the differentiating cells (to single cells) using 0.05% trypsin for 5 min and transfer them to a gelatin-coated plate. Add differentiation medium and incubate overnight in a low-oxygen incubator.
- The next day, add medium for infection (this will vary depending on the virus). Use CM lacking Pen/Strep containing BMP4/A/P, using 0.5 ml for one well of a 6-well plate. Add Sendai virus for MOI = 1. Incubate for 8 hr in a low-oxygen incubator.
- If additional time-points are necessary, remove the virus-containing medium after 4 hr and replace it with BMP4/A/P CM. Collect cells for RNA isolation using a cell scraper.
- Determine the efficiency of viral infection by performing qRT-PCR for genes involved in the viral response18.
Representative Results
Overview of In Vitro Differentiation of hPSCs
This in vitro differentiation protocol begins with undifferentiated hESCs grown on MEFs that are transitioned to feeder-free conditions for one passage (Figure 1A). While we described the differentiation of hESCs in this protocol, we used this protocol to successfully differentiate hiPSCs into trophoblastic cells. The transition to extracellular matrix removes the majority of the irradiated MEFs, which are undesirable for analyses that require pure populations of human trophoblastic cells. Undifferentiated hPSCs are grown on extracellular matrix and cultured with CM containing B-FGF until the colonies are ready for passaging (approximately one week). This maintains the pluripotent state prior to inducing BMP4/A/P-mediated differentiation. The colonies are disrupted into clumps of ~50-100 cells using dispase and passaged a second time onto an extracellular matrix-coated plate. The day after passaging, the adherent cells are ready to induce differentiation, and the medium is switched to contain BMP4/A/P lacking B-FGF. The differentiated cells proliferate for approximately 2 weeks, during which time the cells can be collected for RNA isolation or used for transfection experiments. Morphological changes appear 1-2 days after differentiation has been initiated, and results from one representative experiment are shown in Figure 1B. Notice that cells on differentiation days 1-2 are larger in size than the cells from day 0 (Figure 1B). The differentiated cell colony grows rapidly, and this change is noticeable every day. As differentiation proceeds, the cells divide and expand out from the center of the colony, growing in an outward direction (days 3-5; Figure 1B). The outer portion of the colony contains larger cells compared to the cells at the center of the colony. The differentiating cells become darker and flatter than pluripotent stem cells cultured on extracellular matrix; the changes in cell brightness are more apparent when viewing the cells with an upright microscope used for routine cell culture. The individual colonies merge together by differentiation day 5 (Figure 1B), with cells growing on top of each other. After differentiation day 4-5, cells containing multiple nuclei are present (not shown) (Figure 1B).
BMP4/A/P Induces the Differentiation and Expression of Trophoblast Markers
We examined gene expression changes during the first three days of BMP4/A/P differentiation using quantitative RT-PCR (qRT-PCR). RNA was isolated at differentiation days 1, 2, and 3 and converted to cDNA. The disappearance of pluripotent stem cells can be assessed both by morphology, visually using a microscope, and by qRT-PCR, using primers for pluripotency genes. The relative expression levels of NANOG, a marker of pluripotent stem cells, is reduced by ~75% after one day of differentiation when cells are cultured with BMP4, and levels are reduced by ~90% in the presence of BMP4/A/P (Figure 2). By differentiation day 2, the levels of NANOG are very low for cells cultured in BMP4 alone or in BMP4/A/P compared to those in hESC CM medium (containing B-FGF). This is an expected result, because we do not observe undifferentiated cells after two days of differentiation (Figure 1B), which are brighter and smaller in size compared to differentiated cells. The efficiency of BMP4-mediated differentiation to trophoblastic cells can be directly assessed by qRT-PCR using primers specific for two trophoblast markers: KRT7 and CDX2. Both of these genes are not expressed in human pluripotent stem cells, and their expression increases after 2-3 days of BMP4 treatment (Figure 2). Using the conditions described here, KRT7 transcript levels increase on differentiation day 1 in BMP4/A/P medium and remain constant for the first 3 days of differentiation (Figure 2). KRT7 expression when using BMP4 alone has a delayed increase by day 2, in agreement with previous observations that Activin/Nodal inhibitors increase the differentiation rate of hESCs9. Another way to assess the efficiency of using these inhibitors is to determine the steady-state abundance of CDX2 and KRT7 transcripts in cells treated with BMP4 alone. CDX2 levels are similar for differentiation days 1-3 when using either BMP4 alone or BMP4 together with Activin/Nodal inhibitors. However, KRT7 transcripts are more abundant after one day of differentiation in the presence of these inhibitors, which suggests a more rapid differentiation (Figure 2). CDX2 expression typically peaks at differentiation day 2 for both conditions and decreases after day 3 (Figure 2). Undifferentiated hESCs do not express either KRT7 or CDX2 (Figure 2), as expected. In conclusion, BMP4/A/P conditions rapidly differentiate hESCs, and placental marker expression can be detected as early as differentiation day 3.
Transfections of siRNAs and Plasmid DNA and Viral Infections Using In Vitro-derived Trophoblastic Cells
In vitro derived human trophoblastic cells can be transfected with siRNAs to disrupt the gene expression of either coding or noncoding RNAs. We initiated the differentiation of hESCs using BMP4/A/P, performed two rounds of siRNA transfections (using 75 pmol of siRNAs) on differentiation days 1 and 2, and collected the cells for RNA isolation. Quantitative RT-PCR is an effective method to determine the knockdown efficiency for either coding or noncoding genes. Using two siRNAs complementary to different regions of the long, noncoding RNA lncRHOXF119, we obtained 80% knockdown of lncRHOXF1 transcripts (Figure 3A). Next, we transfected one siRNA (25 pmol) for the protein-coding gene hnRNP U, also on differentiation days 1 and 2. We observed an 80% reduction in hnRNP U transcripts following two consecutive transfections. In vitro differentiated trophoblast progenitor cells can also be used to investigate innate immune responses. We performed infections of differentiation day 2 cells using Sendai virus at an MOI of 1 and collected the cells after 8 hr of viral incubation. Using qRT-PCR, we detected abundant viral protein transcripts (Sev-NP) in infected cells (Figure 3B), indicative of active viral propagation in trophoblastic cells. Importantly, infected cells (using MOI = 1) do not change in appearance and are viable (data not shown). In vitro derived trophoblastic cells can also be transfected with plasmid DNA. We performed transfections using a GFP plasmid and introduced this construct into BMP4/A/P-treated cells on differentiation day 1 and day 3 and imaged for GFP the following day (Figure 4). We typically obtained 30-50% transfection efficiency at 24 hr post-transfection. In conclusion, in vitro derived trophoblastic cells can be utilized for gain- and loss-of-function experiments.
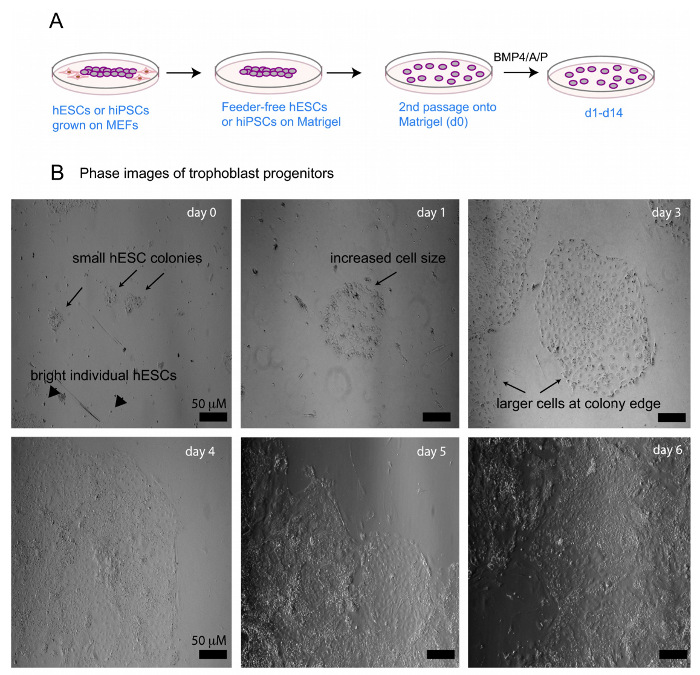
Figure 1: In vitro differentiation of human pluripotent stem cells to early trophoblastic cells using BMP4/A/P. (A) Schematic of BMP4 differentiation of hPSCs to trophoblastic cells using BMP4/A/P. (B) Representative bright-field images of HUES9 cells during d0, d2, and d6 of BMP4/A/P differentiation. Scale bar = 50 μM. The arrowheads denote single-cell hESCs that are not differentiated. The arrows highlight morphological features during differentiation. Please click here to view a larger version of this figure.
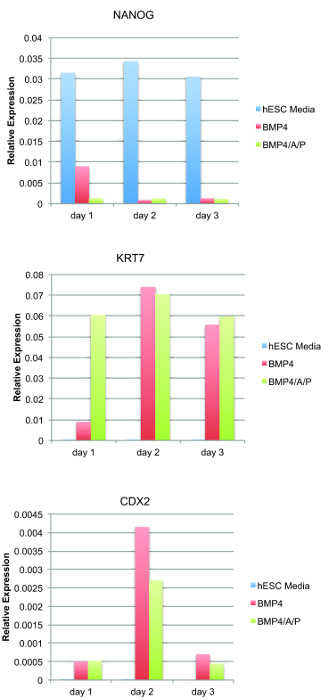
Figure 2: BMP4/A/P differentiation quickly downregulates the pluripotency marker NANOG and upregulates trophoblast genes CDX2 and KRT7. qRT-PCR analysis for NANOG (pluripotency marker), CDX2, and KRT7 (trophoblast markers). The values are averages from triplicate measures. Please click here to view a larger version of this figure.
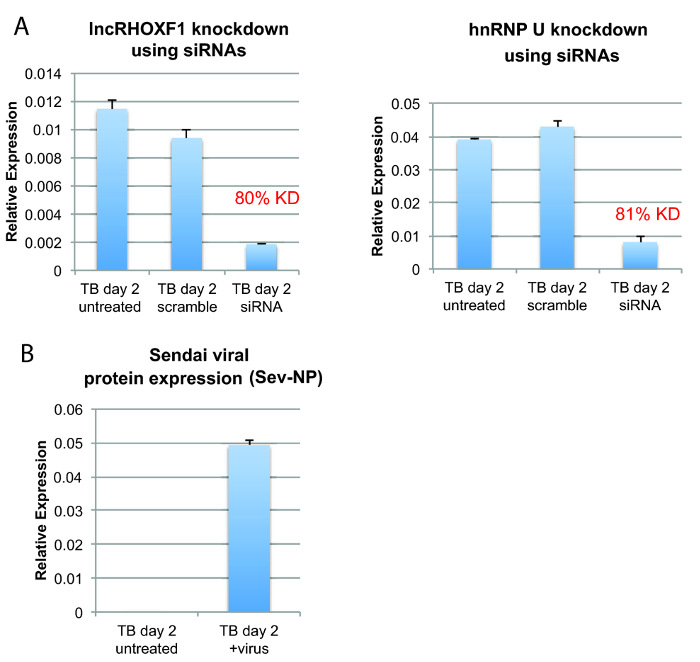
Figure 3: Gene disruption and viral infection using in vitro differentiated human trophoblastic cells. (A) qRT-PCR analysis of hESCs (HUES9) differentiated and then transfected with siRNAs specific to the long, noncoding RNA lncRHOXF1 (left) or the protein-coding gene hnRNP U (right) for two consecutive days. The knockdown efficiency (KD) is typically 70-80%, and the results from one transfection experiment are shown. (B) qRT-PCR analysis of the Sendai viral protein transcript of differentiation day 2 trophoblastic cells infected with Sendai virus (MOI = 1) for 8 hr. The error bars denote the SEM. Please click here to view a larger version of this figure.
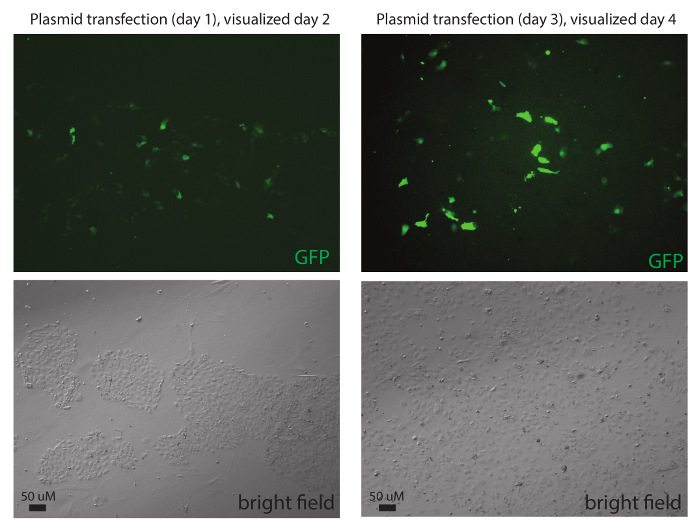
Figure 4: Introduction of plasmid DNA into in vitro differentiated human trophoblastic cells. Transfection of GFP plasmid DNA into differentiation day 1 and day 3 trophectoderm progenitor cells, visualized the following day. Scale bar = 50 μM. Please click here to view a larger version of this figure.
Discussion
We presented the basic steps for differentiating hESCs into trophoblast progenitors. This protocol has recently been optimized to rapidly differentiate hESCs with the addition of Activin/Nodal signaling inhibitors, increasing the differentiation to trophoblastic cells and avoiding the generation of mesoderm progenitors, which are typically observed with BMP4 treatment alone. The BMP4 model system allows for the examination of the earliest stages of human trophoblast lineage specification and expansion. In addition, this BMP4 model system is also useful for investigating the differentiation of trophoblastic cells to specific sub-lineages, which is not possible using choriocarcinoma cell lines and extravillous trophoblast cell lines. In vitro derived trophoblastic cells can also be used for loss-of-function experiments using siRNAs19. Additionally, hESCs or hiPSCs can be genetically modified for the inducible expression of transgenes19 or for the insertion of reporter genes at endogenous loci20, and these cells can be differentiated into trophoblastic cells using the conditions described here.
Critical Steps within the Protocol
It is imperative to omit B-FGF (FGF2) from the CM differentiation medium to obtain trophoblastic progenitor cells from hESCs. The presence of B-FGF together with BMP4 in the culture medium enhances the expression of mesoderm and endoderm progenitors16. The inclusion of B-FGF in trophoblast differentiation medium is believed to explain why a recent study found that BMP4-mediated differentiation generates mesoderm and endoderm cells instead of trophoblasts21. It is well-established that B-FGF together with Activin A and BMP4 promotes endoderm and mesoderm differentiation in a dose-dependent fashion22,23. In the mouse, the development of the epiblast, trophectoderm, and primitive endoderm lineages are dependent on FGF signaling pathways24. In differentiating hESCs, B-FGF-induced signaling is required for mesoderm formation when BMP4 is used to induce differentiation21,25. In contrast, the inhibition of both FGF and the Activin/Nodal signaling pathways, together with BMP4, favors SYN formation in hESC-derived trophoblast progenitors and prevents the formation of mesoderm and primitive endoderm progenitors26.
Limitations of the Technique
The most typical problem with this protocol of hPSC differentiation to trophoblast progenitors is associated with starting with a predominantly undifferentiated population of hESCs or hiPSCs on MEFs. Because hPSCs accumulate differentiated cells at both the inner and outer edges of the colonies, it is important to monitor the amount of differentiation daily and to remove differentiated colonies (or portions of colonies). The presence of differentiated cells can complicate the differentiation process, particularly when the cells are transferred to the extracellular matrix. The differentiated cells will quickly proliferate on the extracellular matrix in the presence of CM, and these cells will rapidly take over the culture and out-compete the hPSCs. Therefore, it is ideal to begin with healthy, undifferentiated colonies of hPSCs grown on MEFs before transferring the cells to the extracellular matrix.
Significance of the Technique with Respect to Existing/Alternative Methods
When using this differentiation protocol, the resulting trophoblast progenitors are a heterogenous mixture of cell types. Trophoblasts are comprised of villous cytotrophoblasts, syncytiotrophoblasts (SYNs), and extravillous cytotrophoblasts (EVTs)27. Villous cytotrophoblasts are the progenitors that generate EVTs and SYNs. BMP4-differentiated hPSCs will generate a mixture of villous cytotrophoblasts, SYNs, and EVTs9,15, which have been defined by the gene or protein expression of placental-specific markers. The distinct differentiation pathways can be distinguished by the secretion of HLA-G (characteristic of EVTs) or human chorionic gonadotropin (from SYNs)28,29. Recently, it has been shown that the inhibition of Activin/Nodal signaling favors EVT differentiation from hESCs, and the removal of this inhibition favors differentiation to SYN17. Indeed, an earlier publication found that using BMP4 alone to differentiate hESCs generates mostly SYN cells26. The data using BMP4/A/P and conditioned medium (lacking B-FGF) indicate that there are both EVT and SYN cells present by differentiation day 5. If pure EVT or SYN populations are desired, additional purification using cell surface markers, such as HLA-G conjugated to beads for the isolation of EVTs, is possible29. In conclusion, the culture method described here is an efficient way to generate significant amounts of trophoblastic cells from hPSCs that can be used for a variety of applications. Because this protocol generates both SYNs and EVTs, it is a suitable model system for examining the early human placenta. These cells can be used for a variety of applications, including drug discovery and genetic engineering.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a Pennsylvania Health Research Formula Fund.
Materials
| DMEM/F12 | Invitrogen | 11330-057 | |
| Knock Out Serum Replacement | Invitrogen | 10828-028 | This is referred to as "serum replacement" in this protocol. |
| NEAA | Invitrogen | 11140-050 | |
| FBS | Invitrogen | 16000-044 | |
| L-Glutamine | Invitrogen | 10828-028 | |
| Penicillin/Streptomycin | Invitrogen | 15140-155 | |
| 2-Mercaptoethanol | Sigma | M-7522 | |
| B-FGF | Millipore | GF-003 | |
| DMEM | Invitrogen | 11965-118 | |
| Dispase | Invitrogen | 17105-041 | |
| Collagenase Type IV | Invitrogen | 17104-019 | |
| Rock inhibitor Y27632 | Calbiochem | 688000 | |
| Irradiated CF1 MEFs | GlobalStem | 6001G | MEFs can be generated from embryonic day 13.5 embyos and irradiated. |
| 0.22 um syringe filter | Millipore | SLGS033SS | |
| Heracell 150i low oxygen incubator | Heracell/VWR | 89187-192 | Any tissue culture incubator with capacity to regulate oxygen concentrations is sufficient. |
| BMP4 | R&D Systems | 314-BP-01M | |
| A 83-01 | R&D Systems | 2939/10 | |
| PD173074 | R&D Systems | 3044/10 | |
| RNAiMax | Invitrogen | 13778150 | |
| Trizol | ThermoFisher | 15596026 | Trizol is used to isolate total RNA. |
| X-tremeGENE 9 | Roche | 6365779001 | |
| Matrigel | Corning | 356231 | This is referred to as "extracellular matrix" in this protocol. |
References
- Rugg-Gunn, P. J. Epigenetic features of the mouse trophoblast. Reproductive biomedicine online. 25 (1), 21-30 (2012).
- Rossant, J., Cross, J. C. Placental development: lessons from mouse mutants. Nature reviews. Genetics. 2 (7), 538-548 (2001).
- Hertig, A. T., Rock, J., Adams, E. C., Menkin, M. C. Thirty-four fertilized human ova, good, bad and indifferent, recovered from 210 women of known fertility; a study of biologic wastage in early human pregnancy. Pediatrics. 23 (1 Part 2), 202-211 (1959).
- Steptoe, P. C., Edwards, R. G., Purdy, J. M. Human blastocysts grown in culture. Nature. 229 (5280), 132-133 (1971).
- Delorme-Axford, E., Sadovsky, Y., Coyne, C. B. The placenta as a barrier to viral infections. Annual Review of Virology. 1, 133-146 (2014).
- Ji, L., et al. Placental trophoblast cell differentiation: physiological regulation and pathological relevance to preeclampsia. Molecular aspects of medicine. 34 (5), 981-1023 (2013).
- Bilban, M., et al. Identification of novel trophoblast invasion-related genes: heme oxygenase-1 controls motility via peroxisome proliferator-activated receptor gamma. Endocrinology. 150 (2), 1000-1013 (2009).
- Xu, R. H., et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nature biotechnology. 20 (12), 1261-1264 (2002).
- Amita, M., et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proceedings of the National Academy of Sciences of the United States of America. 110 (13), E1212-E1221 (2013).
- Genbacev, O., et al. Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells. 29 (9), 1427-1436 (2011).
- Marchand, M., et al. Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biology of reproduction. 84 (6), 1258-1271 (2011).
- Hyslop, L., et al. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem cells. 23 (8), 1035-1043 (2005).
- Harun, R., et al. Cytotrophoblast stem cell lines derived from human embryonic stem cells and their capacity to mimic invasive implantation events. Human reproduction. 21 (6), 1349-1358 (2006).
- Lichtner, B., Knaus, P., Lehrach, H., Adjaye, J. BMP10 as a potent inducer of trophoblast differentiation in human embryonic and induced pluripotent stem cells. Biomaterials. 34 (38), 9789-9802 (2013).
- Chen, Y., Wang, K., Chandramouli, G. V., Knott, J. G., Leach, R. Trophoblast lineage cells derived from human induced pluripotent stem cells. Biochemical and biophysical research communications. , (2013).
- Roberts, R. M., et al. Differentiation of trophoblast cells from human embryonic stem cells: to be or not to be?. Reproduction. 147 (5), D1-D12 (2014).
- Sarkar, P., et al. Activin/nodal signaling switches the terminal fate of human embryonic stem cell-derived trophoblasts. The Journal of biological chemistry. 290 (14), 8834-8848 (2015).
- Penkala, I., et al. lncRHOXF1, a Long Noncoding RNA from the X Chromosome That Suppresses Viral Response Genes during Development of the Early Human Placenta. Mol Cell Biol. 36 (12), 1764-1775 (2016).
- Penkala, I., et al. lncRHOXF1, a Long Noncoding RNA from the X Chromosome That Suppresses Viral Response Genes during Development of the Early Human Placenta. Molecular and cellular biology. 36 (12), 1764-1775 (2016).
- Hockemeyer, D., et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature biotechnology. 29 (8), 731-734 (2011).
- Bernardo, A. S., et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell stem cell. 9 (2), 144-155 (2011).
- Zhang, P., et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 111 (4), 1933-1941 (2008).
- Vallier, L., et al. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PloS one. 4 (6), e6082 (2009).
- Arman, E., Haffner-Krausz, R., Chen, Y., Heath, J. K., Lonai, P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proceedings of the National Academy of Sciences of the United States of America. 95 (9), 5082-5087 (1998).
- Yu, P., Pan, G., Yu, J., Thomson, J. A. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell stem cell. 8 (3), 326-334 (2011).
- Sudheer, S., Bhushan, R., Fauler, B., Lehrach, H., Adjaye, J. FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast. Stem cells and development. 21 (16), 2987-3000 (2012).
- Bischof, P., Irminger-Finger, I. The human cytotrophoblastic cell, a mononuclear chameleon. The international journal of biochemistry & cell biology. 37 (1), 1-16 (2005).
- Cole, L. A. Hyperglycosylated hCG, a review. Placenta. 31 (8), 653-664 (2010).
- Apps, R., et al. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology. 127 (1), 26-39 (2009).

