Probing The Structure And Dynamics Of Nucleosomes Using Atomic Force Microscopy Imaging
Summary
Here, we present a protocol to characterize nucleosome particles at the single-molecule level using static and time-lapse atomic force microscopy (AFM) imaging techniques. The surface functionalization method described allows for the capture of the structure and dynamics of nucleosomes in high-resolution at the nanoscale.
Abstract
Chromatin, which is a long chain of nucleosome subunits, is a dynamic system that allows for such critical processes as DNA replication and transcription to take place in eukaryotic cells. The dynamics of nucleosomes provides access to the DNA by replication and transcription machineries, and critically contributes to the molecular mechanisms underlying chromatin functions. Single-molecule studies such as atomic force microscopy (AFM) imaging have contributed significantly to our current understanding of the role of nucleosome structure and dynamics. The current protocol describes the steps enabling high-resolution AFM imaging techniques to study the structural and dynamic properties of nucleosomes. The protocol is illustrated by AFM data obtained for the centromere nucleosomes in which H3 histone is replaced with its counterpart centromere protein A (CENP-A). The protocol starts with the assembly of mono-nucleosomes using a continuous dilution method. The preparation of the mica substrate functionalized with aminopropyl silatrane (APS-mica) that is used for the nucleosome imaging is critical for the AFM visualization of nucleosomes described and the procedure to prepare the substrate is provided. Nucleosomes deposited on the APS-mica surface are first imaged using static AFM, which captures a snapshot of the nucleosome population. From analyses of these images, such parameters as the size of DNA wrapped around the nucleosomes can be measured and this process is also detailed. The time-lapse AFM imaging procedure in the liquid is described for the high-speed time-lapse AFM that can capture several frames of nucleosome dynamics per second. Finally, the analysis of nucleosome dynamics enabling the quantitative characterization of the dynamic processes is described and illustrated.
Introduction
In eukaryotic cells, DNA is highly condensed and organized into chromosomes.1 The first level of DNA organization within a chromosome is the assembly of nucleosomes in which 147 bp of DNA is tightly wrapped around a histone octamer core.2,3 Nucleosome particles assemble on a long DNA molecule forming a chromatin array which is then organized until a highly compact chromosome unit is formed.4 The disassembly of chromatin provides the access to free DNA required by critical cellular processes such as gene transcription and genome replication, suggesting that chromatin is a highly dynamic system.5,6,7 Understanding the dynamic properties of DNA at various chromatin levels is critically important for elucidating genetic processes at the molecular level where mistakes can lead to cell death or the development of diseases such as cancer.8 A chromatin property of great importance is the dynamics of nucleosomes.9,10,11,12 The high stability of these particles has allowed for the structural characterization by crystallographic techniques.2 What these studies lack are the dynamic details of nucleosomes such as the mechanism of DNA unwrapping from the histone core; the dynamic pathway of which is required for transcription and replication processes.7,9,13,14,15,16 Furthermore, special proteins termed remodeling factors have been shown to facilitate the disassembly of nucleosomal particles17; however, the intrinsic dynamics of nucleosomes is the critical factor in this process that contributes to the entire disassembly process.14,16,18,19
Single-molecule techniques such as single-molecule fluorescence19,20,21, optical trapping (tweezers)13,18,22,23 and AFM10,14,15,16,24,25,26 have been instrumental in understanding the dynamics of nucleosomes. Among these methods, AFM benefits from several unique and attractive features. AFM allows one to visualize and characterize individual nucleosomes as well as the longer arrays27. From AFM images, important characteristics of nucleosome structure such as the length of DNA wrapped around the histone core can be measured 10,14,26,28; a parameter that is central to the characterization of nucleosome unwrapping dynamics. Past AFM studies have revealed nucleosomes to be highly dynamic systems and that DNA can spontaneously unwrap from the histone core14. The spontaneous unwrapping of DNA from nucleosomes was directly visualized by AFM operating in the time-lapse mode when the imaging is done in aqueous solutions 14,26,29.
The advent of the high-speed time-lapse AFM (HS-AFM) instrumentation made it possible to visualize the nucleosome unwrapping process at the millisecond time-scale 14,15,24. Recent HS-AFM 16,30 studies of centromere specific nucleosomes revealed several novel features of the nucleosomes compared with the canonical type. Centromere nucleosomes constitute of a centromere, a small part of the chromosome critically important for chromosome segregation31. Unlike canonical nucleosomes in bulk chromatin, the histone core of centromere nucleosomes contains CENP-A histone instead of histone H332,33. As a result of this histone substitution, DNA wrapping in centromere nucleosomes is ~120 bp instead of the ~147 bp for canonical nucleosomes; a difference that can lead to distinct morphologies of the centromere and canonical nucleosomes arrays34, suggesting that centromere chromatin undergoes higher dynamics compared with the bulk one. The novel dynamics displayed by centromere nucleosomes in HS-AFM16,30 studies exemplify the unique opportunity provided by this single-molecule technique to directly visualize the structural and dynamic properties of nucleosomes. Examples of these features will be briefly discussed and illustrated at the end of the paper. This progress was made due to the development of novel protocols for AFM imaging of nucleosomes as well as the modifications of existing methods. The goal of the protocol described here is to make these exciting advances in single-molecule AFM nucleosome studies accessible to anyone who would like to utilize these techniques in their chromatin investigations. Many of the techniques described are applicable to problems beyond the study of nucleosomes and can be used for investigations of other protein and DNA systems of interest. A few examples of such applications can be found in publications35,36,37,38,39,40,41,42,43,44,45,46,47,48,49 and prospects of AFM studies of various biomolecular systems are given in reviews29,50,51,53,54.
Protocol
1. Continuous Dilution Assembly of Mono-nucleosomes
- Generate and purify an approximately 400 bp DNA substrate that contains an off-centered Widom 601 nucleosome positioning sequence.55
NOTE: To limit the unwanted formation of di-nucleosomes, each ‘arm’ flanking the positioning sequence should not exceed ~150 bp.- Use plasmid pGEM3Z-601 along with the designed primers and amplify the substrate DNA using PCR. For the 423 bp substrate with 122 and 154 bp arm lengths used here, use forward (5’-CAGTGAATTGTAATACGACTC-3’) and reverse (5’-ACAGCTATGACCATGATTAC-3’) primers.
- Add tubes containing the reaction mixture to a thermal cycler preheated to 95 °C. Run the following program for 33 cycles after an initial denaturation for 5 min at 95 °C: 30 s denaturation at 95 °C, 30 s annealing at 49 °C, 35 s extension at 72 °C. Set a final extension at 72 °C for 10 min following the 33 cycles.
- Purify the DNA from the PCR mixture using a commercially available PCR Purification Kit. When eluting the DNA from the PCR cleanup column, use 10 mM Tris buffer (pH 7.5) in place of the kit provided elution buffer.
NOTE: Take extra care not to transfer buffers between the purification steps. A contaminated eluent can cause issues downstream when measuring DNA concentration and/or can alter the starting salt concentration of the nucleosome assembly mixture.
- Determine the DNA concentration by measuring the absorbance of purified DNA at 260 nm.
- Collect a blank on the UV VIS Spectrophotometer using only the 10 mM Tris pH 7.5 elution buffer. Collect a measurement of the purified DNA.
- With the concentration determined, aliquot 25 pmol of the purified DNA into a 0.6 mL microfuge tube and place it in a vacuum centrifuge until the solution is barely visible; this is typically 30 min to 1 h.
NOTE: The DNA substrate is now ready for nucleosome assembly. Otherwise, the protocol can be paused here and the DNA stored at -20 °C until use. - Place the microfuge tube containing the 25 pmol DNA on the ice and add the nucleosome assembly components in Table 1, in the order listed. When all components have been added, remove the mixture from the ice and incubate at room temperature (RT) for 30 min.
NOTE: It is critical that the salt content of the stock histone buffer is considered when calculating the NaCl needed to achieve the 2 M final concentration.15,56 - Assemble the nucleosomes by reducing the 2 M salt concentration of the mixture to 200 mM using a continuous rate dilution.57
- Fill a syringe with 100 µL of dilution buffer containing 10 mM Tris pH 7.5 and place it on a syringe pump.
- Direct the needle of the syringe through a pre-punctured hole in the cap of the microfuge tube, ensuring contact is made with the assembly mixture (Figure 1).
- Run the syringe pump at a rate of 0.75 µL/min for 120 min.
NOTE: The resulting 100 µL solution contains 250 nM nucleosomes and 200 mM NaCl. - Transfer the mixture to a 10 K MWCO dialysis button and dialyze against 200 mL of a pre-chilled (4 °C) low salt buffer containing 10 mM Tris pH 7.5, 0.25 mM EDTA and 2.5 mM NaCl, for 1 hr at 4 °C.
- To assess the histone content of the nucleosome assembly, prepare a discontinuous SDS-PAGE gel with a 15% separating and 6% stacking as previously described30.
- Aliquot 10 – 20 µL of the nucleosome stock to a microfuge tube and add 4x Laemmli Sample Buffer to a working concentration of 1 – 2x.58
- As a control, repeat this preparation in a separate microfuge tube for 1 – 2 µg of the histone stock. Heat the samples at 95 °C for ~ 5 min.
- Load the samples in adjacent lanes to one another on the gel. Add sample buffer to the unused lanes to promote even band migration.
- Run the gel at 65 V until the dye front moves through the stacking gel. When the separating gel is reached, increase to 150 V and run until the dye front has migrated completely out of the gel.
- Dismantle the electrophoresis unit and gently transfer the gel to a staining container filled with dd H2O. Let the gel sit for 5 min with gentle agitation. Repeat this process twice more with fresh dd H2O used each time.
NOTE: The Coomassie stain used in this protocol does not require the typical fixing steps needed for Coomassie stains (see Table of Materials). If another Coomassie preparation is being used, adjust the fixing steps as needed. - Remove the water from the final rinse and add just enough stain to cover the gel. Let the gel sit with gentle agitation for at least 1 h.
NOTE: For the agitation, the staining container can be placed on any apparatus that promotes movement of the stain over the gel while also keeping the gel covered in liquid. - Remove the stain from the container and rinse the gel with dd H2O. Replace the dd H2O and soak the gel for 30 min with gentle agitation. (The gel should appear like that shown in Figure 2, with clear separation of the histone bands.)
- Store the nucleosomes at 4 °C until use.
NOTE: When stored in these conditions, nucleosomes remain stable for several months. The protocol can be paused here.
2. Functionalization of Mica Surface for Static AFM Imaging of Nucleosomes
- Prepare a 50 mM 1-(3-Aminopropyl) silatrane APS stock solution in deionized water as described.30 Store 1 mL aliquots of this solution at 4 °C until use.
NOTE: The aliquots can be stored for more than a year at 4 °C.59 - Prepare a working APS 1:300 solution for mica modification by dissolving 50 µL of the 50 mM APS stock in 15 mL dd H2O.
NOTE: This working solution can be stored at room temperature for several days. - Cut 1 x 3 cm strips of mica from high quality mica sheets (see Table of Materials for mica used here).
- Check that the piece fits when placed diagonally in a cuvette. Use the tip of sharp tweezers, a razor blade or scotch tape, to cleave layers of the mica until both sides are freshly cleaved and the piece is as thin as ~0.1 mm (Figure 3A). Immediately place the mica piece into the APS filled cuvette and incubate for 30 min (Figure 3B).
- Transfer the mica piece to a cuvette filled with dd H2O and soak for 30 s (Figure 3C). Completely dry both sides of the APS-mica strip under an argon flow.
NOTE: A non-woven cellulose and polyester cleanroom wipe (recommended wipe detailed in materials) can be used to aid in wicking water from the edge of the mica when drying. - Use the dry mica strip is now for the sample preparation. Otherwise, store the piece in a clean, dry cuvette (Figure 3D).
NOTE: Additional storage in a vacuum for 1-2 h is recommended when the environment is humid. The protocol can be paused here.
3. Preparation of Nucleosome Samples on APS-Mica for Static AFM Imaging
- Apply double-faced adhesive tape to several magnetic pucks and place them to the side.
- Cut the APS-mica substrate to the desired size (1 x 1 cm squares for the MM AFM instrument used here). Place these pieces in a clean petri dish and keep covered.
- Prepare three dilutions of the assembled nucleosomes (final nucleosome concentrations of 0.5, 1.0 and 2.0 nM) using a 0.22 µm filtered buffer containing 10 mM HEPES pH 7.5 and 4 mM MgCl2.
NOTE: To limit the loss of nucleosomes at the low final concentration, the dilutions should be done one at a time, immediately prior to deposition on the APS-mica. - Deposit 5-10 µL of the diluted nucleosome sample at the center of the APS-mica piece, and let incubate for two minutes. Gently rinse the sample with 2-3 mL of dd H2O to remove all buffer components. After each ~0.5 mL of dd H2O used, gently shake the mica to remove the excess rinse water.
NOTE: A disposable syringe is recommended for this rinsing step. - Dry the deposited sample under a light flow of clean argon gas.
NOTE: The sample is now ready to be imaged or can be stored in a vacuum cabinet or desiccator filled with argon. Samples prepared and stored as described have been imaged one year following preparation with no quality loss. The protocol can be paused here.
4. Static AFM Imaging of Nucleosomes
- Mount an AFM tip on the tip holder. Use a tip that has a spring constant of ~40 N/m and a resonance frequency between 300 and 340 kHz (see the Table of Materials for the cantilevers used here).
- Mount the sample prepared in section 3 on the AFM stage being careful not to contact the sample surface.
- Position the laser over the cantilever until the sum is at the maximum and adjust the vertical and lateral deflection values to near zero.
- Tune the AFM probe to find its resonance frequency and adjust the drive amplitude and set the image size to 100 x 100 nm. Click the engage button to begin the approach.
- Once approached, gradually optimize the Amplitude Setpoint until the surface of the sample is clearly seen. Increase the scan size to 1 x 1 µm and the resolution to 512 x 512 pixels. Click the capture button followed by the engage button to begin image acquisition.
NOTE: The images in Figure 4 show the smooth background that can be expected when imaging these samples. - To analyze the nucleosome sample, open the captured images using the AFM instruments analysis software.
- Flatten the image using a polynomial line subtraction or similar feature.
- Set the color table to reflect the lowest value for the minimum and the highest value for maximum. Keep a record of these values for each image as they will be needed in a later step.
NOTE: If these values are not used, height data will be incorrect in the later analysis as cross-sections of nucleosome cores will appear as plateaus of equal, rather than varying height. - Export the image as a .tiff image at the original size. De-select any options to save the image with a border or scale bars.
- Open the image in analysis software capable of contour length measurements.
- Set the x, y and z scales to match the image.
- As an internal length calibration, measure and record the contour length of free DNA from one end to the other. For this calibration factor, use the measurements to generate a histogram and fit it with a normal (Gaussian) distribution. Divide the peak center (xc) by the substrate length in base pairs.
NOTE: This value is the image specific conversion factor from nanometers to base pairs of DNA. - Measure and record the contour length of both arms for each nucleosome from the free end of the arm to the center of the core, for consistency (Figure 5A).
- For each nucleosome core, collect two full width at half maximum (FWHM) values from a perpendicular pair of core cross section measurements (Figure 5B Average the two FWHM measurements and subtract one-half of the resulting value from each nucleosome arm to correct for the measured length from the exit/entry DNA to the center of the core that is not part of the arm length.
- Divide each arm length by the calculated calibration factor (step 4.5.6) to obtain arm lengths in DNA base pairs.
- Calculate the extent of DNA wrapping by subtracting the nucleosome arm sum from the total base pair length of the unwrapped substrate. Plot these values as a histogram and fit the peak(s) with a normal (Gaussian) distribution to obtain the mean wrapped base pairs of DNA for the nucleosome population.
- Calculate the average nucleosome height from the measured cross sections. Plot this as a histogram and fit the peak(s) with a normal (Gaussian) distribution to obtain the height of the nucleosome population.
5. Time-Lapse AFM Imaging of Nucleosome Dynamics
- Mount the AFM tip on the tip holder. A probe with a spring constant of approximately 0.1 N/m and a resonance frequency of 7-10 kHz can be used (see the materials list for the cantilevers used here).
- Attach a 1 x 1 cm piece of APS-mica to a magnetic puck using double-stick tape and mount the puck on the AFM instrument.
- Dilute the nucleosomes to a 1 nM concentration in a 0.22 µm filtered imaging buffer containing 10 mM HEPES pH 7.5 and 4 mM MgCl2.
- Deposit 5 – 10 µL of the diluted nucleosome at the center of the mica piece for 2 min. Rinse the deposited sample with 20µL of the imaging buffer two times. After the second rinse, keep a droplet of imaging buffer on the surface.
- Use the top-view camera to find the tip and approach to the surface manually until the tip is ~100-500 µm from the surface.
- Add additional imaging buffer to fill the gap between the tip and the surface. In this example, approximately 50 µL of imaging buffer is sufficient to fill the gap. Find a resonance peak for the tip.
- Begin the computer-controlled approach to the surface. When approached, begin imaging with a 1-2 µm area to select an ~500 nm area of interest. With this area of interest selected, adjust the data acquisition density to 512 x 512 pixels.
- Adjust the set point voltage and drive amplitude parameters to improve image quality. A free amplitude of 10 nm or less and a scan rate of ~2 Hz can be used to capture quality images. An example of nucleosome dynamics captured using time-lapse AFM is shown in Figure 6.
6. High-Speed Time-Lapse AFM Imaging of Nucleosome Dynamics
NOTE: The protocol below is provided for the HS-AFM instrument developed by the Ando group (Kanazawa University, Kanazawa, Japan).60
- Prepare APS-mica for liquid imaging.
- Attach the glass rod to the AFM scanner stage using the glass rod – scanner glue (see Table of Materials). Let this dry for a minimum of 10 min.
- Make ~0.1 mm thick circular pieces of mica with a 1.5 mm diameter by punching them from a larger mica sheet. Use the HS-AFM mica-glass rod glue (see Table of Materials) to attach this mica piece to the glass rod on the HS-AFM and dry, untouched for a minimum of 10 minutes. Cleave layers using a pressure-sensitive tape until a well cleaved layer is seen on the tape.
- Dilute 1 µL of 50 mM APS stock in 99 µL of dd H2O to make a 500 µM APS solution. Deposit 2.5 µL of this solution on the freshly cleaved mica surface and let functionalize for 30 min.
NOTE: To prevent drying of the surface while functionalizing, the cap of a 50 mL conical centrifuge tube can be fit with a damp piece of filter paper and placed over the scanner. The APS stock is diluted 3 times less for liquid imaging than for static imaging to control the dynamics to a rate that can be observed with AFM. - Rinse the mica with 20 µL of dd H2O by applying several ~3 µL rinses. Remove water completely following each rinse by placing a non-woven wipe at the edge of the mica. After the final rinse, place ~3 µL of dd H2O on the surface and let it sit for a minimum of 5 min to remove any nonspecifically bound APS.
- Place the probe in the HS-AFM holder and position the holder on the AFM stage with the tip facing up. Rinse the holder using ~100 µL of dd H2O followed by two ~100 µL rinses of 0.22 µm filtered nucleosomes imaging buffer which contains 10 mM HEPES pH 7.5 and 4 mM MgCl2.
- With the rinses done, fill the chamber with ~100 µL of nucleosome imaging buffer, submerging the tip. Adjust the cantilever position until it is hit with the laser. Rinse the APS-mica with 20 µL of filtered nucleosome imaging buffer, using ~4 µL per rinse.
- Dilute 1 µL of the nucleosome assembly stock into 250 µL of filtered nucleosome imaging buffer for a final nucleosome concentration of 1 nM. Deposit 2.5 µL of this dilution on the surface and let it sit for 2 min. Rinse the surface with ~ 4 µL of nucleosome imaging buffer two times. After the final rinse, leave the surface covered in imaging buffer.
NOTE: If the surface is not rinsed after depositing the nucleosome sample, the surface will rapidly become overcrowded. - Set the scanner and sample on top of the tip holder so that the sample is face down. To begin the approach, use the auto-approach function with a set point amplitude, As close to the free oscillation amplitude A0.
NOTE: Ideally, As = 0.95 A0, however, operating at 82% of A0 will work as well if careful. - Adjust the set point until the surface is being well tracked.
NOTE: To minimize the transfer of energy from the AFM tip to the nucleosome sample, the amplitude of the cantilever should be kept small, with amplitudes as low as 1 nm optimal. - Set the image area around 150 x 150 nm to 200 x 200 nm with data acquisition rate of ~300 ms per imaging frame.
NOTE: This image size is typically sufficient to capture the dynamics of several nucleosomes simultaneously. A less populated surface may call for changes to these parameters. The suggested frame rate is sufficient to capture nucleosome dynamics such as looping, sliding and unwrapping, among others (see Representative Results section below).
7. Analysis of Nucleosome Dynamics Captured Using Time-Lapse AFM
- Convert the images from the HS AFM data type (.asd) to .tiff images.
- Flatten the images using the AFM system’s analysis software using either plane or line functions until the background has uniform contrast.
- Set the image contrast (color scale) to automatic.
NOTE: The contrast can be adjusted manually for presentation purposes but not for analysis. This causes height detail to be lost in the converted images. - Save the selected range of images as a .mov file. Deselect the scale bar and border options before saving.
NOTES: Do not do analysis on images that have scale bars in the frame. These alter the size of the image and will result in false measurements. - Convert the .mov file to tiff images using a suitable software (see Table of Materials). Use the same frame rate for conversion as was used when creating the .mov file.
- For arm length contour measurements, open the images in a measurement software capable of this measurement type (see Table of Materials).
- Set the image dimensions to match those at which it was captured.
- Measure the contour length of each nucleosome arm from the end of the arm to the center of the nucleosome core and record the measurements in a spreadsheet (Figure 4A).
- Measure the length of the unwrapped DNA substrate in the frames after a nucleosome unwraps. Use this for a movie specific calibration of the nm/bp ratio which is used for nucleosome wrapping calculations
- Collect two cross-section profiles for the nucleosome core and two for the bare DNA (Figure 4B).
- Import the cross-section profiles to a spreadsheet software and normalize the plots by subtracting the lowest z value from all points in the profile.
- Calculate the height and full width at half maximum (FWHM) for each cross section and average the values from the two profiles for each frame.
- Subtract half of the FWHM value from each of the nucleosome arms are plot as a scatter plot along with the calculated sum of the arms.
- Calculate the mean contour length from the DNA measurements made for the DNA in frames after the nucleosome unwrapped.
- Determine the nm/bp calibration factor by dividing the mean contour length of the free DNA by the base pair length of the DNA substrate. Use this value to calculate nucleosome wrapping in each frame, as described in section 5.12.
Representative Results
Mono-nucleosomes were first prepared for AFM imaging experiments using a continuous dilution assembly method (Figure 1). The prepared nucleosomes were then checked using discontinuous SDS-PAGE (Figure 2). A mica surface was next functionalized using APS, which captures nucleosomes at the surface while maintaining a smooth background for high-resolution imaging (Figure 3). Nucleosomes were deposited on APS-mica and were subsequently imaged using static AFM imaging. As a control for the assembly and deposition, H3 mono-nucleosomes were prepared and imaged using static AFM. An image of the H3 mono-nucleosomes (Figure 4A) provides a snapshot of the nucleosome population as it existed moments before deposition, confirming that nucleosomes were successfully assembled. The 2 nM nucleosome deposition provided a uniform distribution of nucleosome and DNA particles across the surface and very little to no crowding was observed.
With the H3 control assembly a success, the presented methods were next applied to the study of CENP-A nucleosomes. Static AFM imaging of this sample (Figure 4B) revealed that the assembly was a success. To demonstrate the influence of nucleosome concentration on the surface particle density, the CENP-A nucleosomes were deposited at 1 nM (Figure 4B), compared to the 2 nM used for H3 (Figure 4A). This resulted in a reduced surface particle density for the CENP-A sample to approximately half that of H3 sample. From the static AFM images, the height and turn number of mono-nucleosomes were characterized (Figure 5). Both the angle between the free DNA arms and the length of the free DNA arms was used to determine the number of DNA turns in the individual nucleosome.
Time-lapse AFM imaging of the nucleosomes in buffer was used to visualize the overall spontaneous unwrapping behavior of the nucleosomes (Figure 6). Measuring the angle between nucleosome arms and the contour length of the arms allowed for the turn number to be determined in each of the frames during this unwrapping process (Figure 6 B-C). As the turn number of the nucleosome decreases, a corresponding decrease in the nucleosome core volume is also observed (Figure 6C). High-speed time-lapse AFM was next used to probe the more intricate nucleosome dynamics that were missed using standard time-lapse imaging. The ability of this technique to capture the dynamics over a long period of time was essential to the visualization of a long-distance translocation of a CENP-A nucleosome core (Figure 7) which was captured over the course of ~1200 frames. This technique was also critical in capturing the rare transfer of a CENP-A nucleosome core from one DNA substrate to another (Figure 8). The fast image capture rate (~300 ms/frame) made visualization of this dynamic event possible, as it only took several frames to complete.
Table 1: Reagents needed for continuous salt gradient nucleosome assembly. Each of the components listed is added to the microfuge tube containing the purified DNA. This should be done in the order in which the reagents are listed in the table, with water and NaCl added first, followed by the H2A/H2B dimer and the histone tetramer added last. If pre-folded histone octamers are to be used, add at the same ratio as for the tetramer above. *Take note of the NaCl content in each of the histone stocks and adjust the 5M NaCl to add accordingly, the final [NaCl] should equal 2M. (See Table of Materials).

Figure 1: Schematic of the syringe pump used for microscale nucleosome assembly. The assembly mixture is positioned to be in contact with the end of the syringe needle. As the dilution buffer is delivered by the syringe pump to the assembly mixture the concentration of NaCl is decreased, promoting nucleosome assembly. This figure is adapted from Stumme-Diers et al.30 Please click here to view a larger version of this figure.
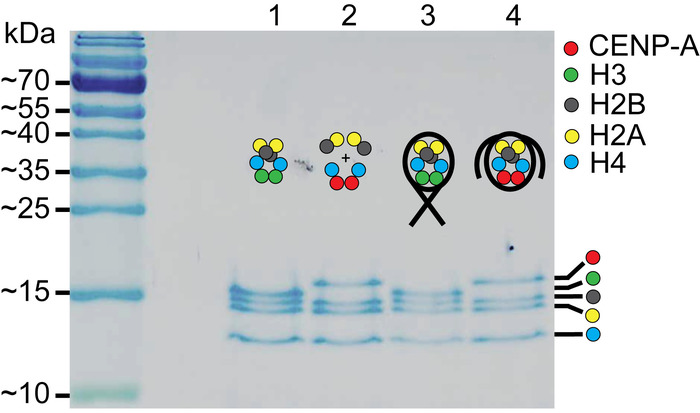
Figure 2: SDS-PAGE of assembled nucleosomes. Lanes 1 and 2 contain the H3 octamer and the CENP-A assembly of histones, respectively. Lanes 3 and 4 contain the assembled H3 nucleosomes and the assembled CENP-A nucleosomes, respectively. Comparison of the assembled nucleosomes to the histone only controls in lanes, confirm that nucleosomes were properly assembled. The cartoon schematic above each lane indicates which histone components are present. This figure is adapted from Stumme-Diers et al.30 Please click here to view a larger version of this figure.
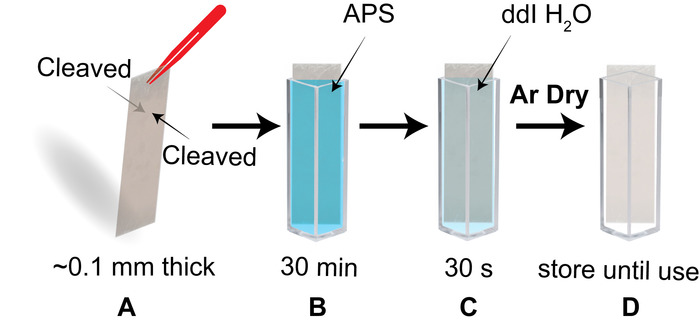
Figure 3: Schematic of the process to prepare APS functionalized mica for AFM imaging of nucleosomes. (A) a piece of mica ~0.1 mm in thickness has both sides freshly cleaved. (B) The cleaved mica piece is promptly placed diagonally in a cuvette containing the APS solution and is set to incubate for 30 min. (C) Following the APS functionalization step, the APS-mica piece is transferred to a cuvette filled with dd H2O for a 30 s rinse. (D) The APS-mica piece is stored in a cuvette until use. Please click here to view a larger version of this figure.
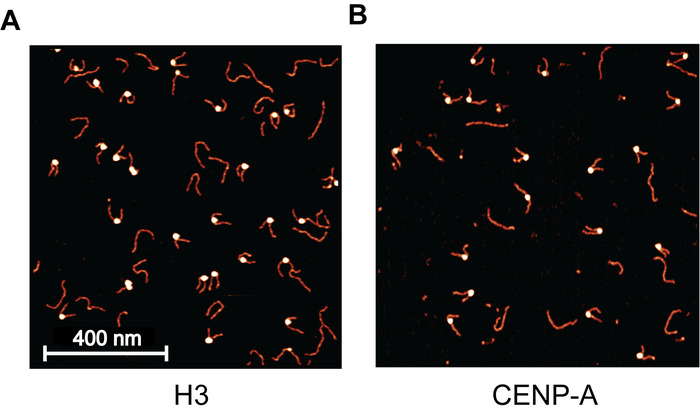
Figure 4: Example AFM images of H3 and CNEP-A nucleosomes. (A) Sample image of H3 mono-nucleosomes deposited on APS-mica, captured using static AFM. Each bright blob is a nucleosome core particle with the flanking DNA regions appearing as noodle-like arms. The long noodle-like features are free DNA particles that are not associated with a histone core. For this image, a 2 nM nucleosome concentration was used, providing a uniform distribution across the surface, with little to no crowding. (B) This nucleosome sample was deposited at 1 nM and is much less populated than the 2 nM used in (A). This demonstrates the direct effect that nucleosome dilution has on the surface density of nucleosomes. Please click here to view a larger version of this figure.
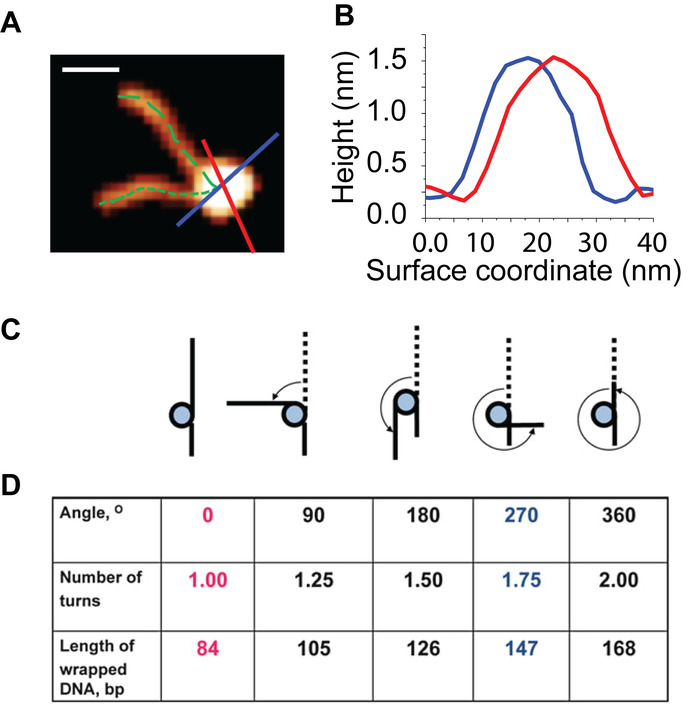
Figure 5: Visual depiction of the analysis used to characterize the wrapping and height of the nucleosome particles. (A) A representative nucleosome particle from images like those shown in Figure 3. The contour length of each nucleosome arm is measured from the end of the arm to the center of the core (dotted green lines). Plotting cross section profiles (red and blue lines) of a nucleosome produce the curves shown in (B) From these curves, height and width detail of particle can be determined. (C) Schematic of the various wrapped states of the nucleosomes. (D) Each wrapped state is characterized using the angle between the DNA arms, the number of DNA turns and the bp of wrapped DNA. Scale Bar = 20 nm. This figure is adapted from Lyubchenko et al. 24 Please click here to view a larger version of this figure.
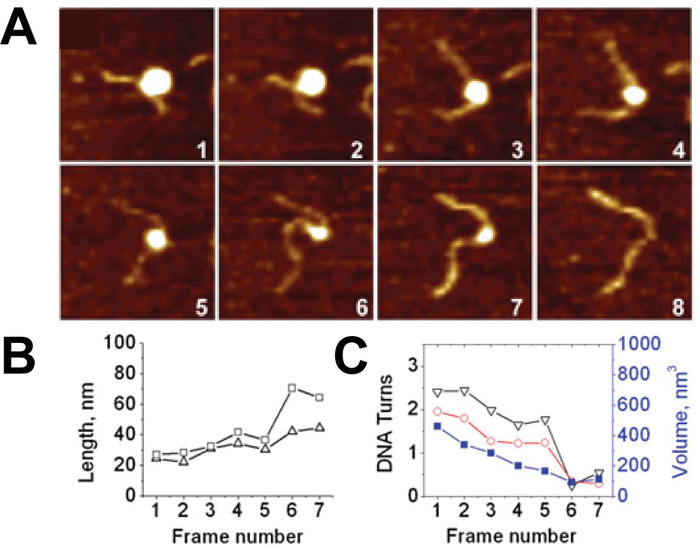
Figure 6. Example of time-lapse AFM images capturing the spontaneous unwrapping of nucleosomes. (A) A series of consecutive AFM images of the spontaneous unwrapping process of nucleosomes captured by continuous scanning in the buffer. The size of each frame is 200 nm and images were captured at a rate of ~170 s per frame. (B) As the unwrapping process progresses in each frame, the arm lengths of the nucleosome increase, (C) resulting in a decrease in DNA turns around the nucleosome. This turn number can be determined from either the measured arm lengths (black) or the angle between the nucleosome arms (red). As the turn number decreases, a reduction in nucleosome volume is also observed (blue curve, right axis). Each frame is 200 x 200 nm in size. This figure is adapted from Lyubchenko et al. 24 Please click here to view a larger version of this figure.
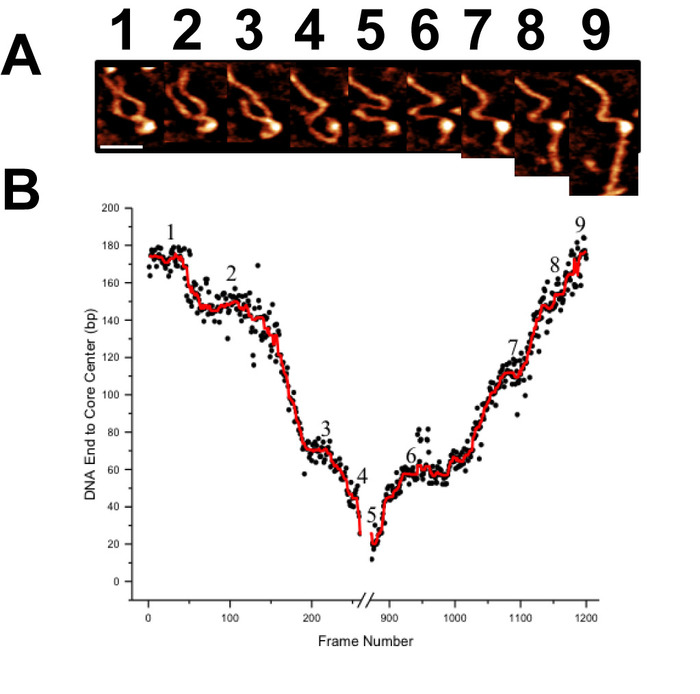
Figure 7. Demonstration of the high capacity of high-speed time-lapse AFM for long image acquisition times (A) A gallery of images selected from more than 1200 frames demonstrating the translocation behavior of a CENP-A nucleosomes core. Each image was captured at a rate of ~300 ms/frame. (B) Contour length measurements from one end of the DNA substrate to the CENP-A core are used to characterize this long translocation process. Scale Bar = 25 nm. This figure is adapted from Stumme-Diers et al. 16 Please click here to view a larger version of this figure.
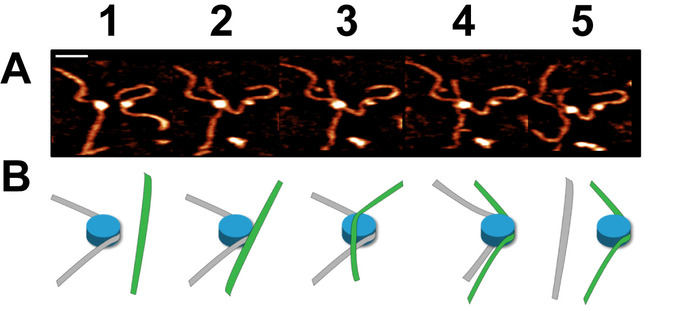
Figure 8. Example of a dynamic nucleosome core transfer captured using high-speed time-lapse AFM (figure adapted from Stumme-Diers et al.16 (A) Selected frames demonstrating the spontaneous transfer of a CENP-A nucleosome core from one DNA substrate to another. This process took place within several frames which were captured at a rate of ~300 ms/frame. (B) A schematic of the transfer process shown in (A). Scale Bar = 25 nm. This figure is adapted from Stumme-Diers et al. 16 Please click here to view a larger version of this figure.
Discussion
The protocol described above is rather straightforward and provide highly reproducible results, although a few important issues can be emphasized. Functionalized APS-mica is a key substrate for getting reliable and reproducible results. A high stability of APS-mica is one of the important features of this substrate that allows one to prepare the imaging substrate in advance for use that can be used at least two weeks after being prepared.59,61 However, the surface can be damaged by vapors of glue if it is used for mica mounting on the metal puck. Therefore, it is recommended to use double stick tape for the mica mounting as described in the protocol (section 2). In cases when the use of glue is necessary, for example, some users use glues for mounting of mica on metal disks for imaging in liquid, the following procedure modified from the one described in section 6.1.3 for the sample preparation for HS-AFM imaging is recommended. Mica is glued to the metal disk or other solid material and cleaved after the glue is solidified. Blow the mica with argon to remove potential vapors of the glue. Then the mica can be cleaved with a scotch tape and the APS mica working solution placed to cover the freshly cleaved mica strip. The amount of the solution depends on the mica strip size. Allow APS to react for 30 min. To minimize evaporation, run the reaction in a wet Petri dish. Use the following procedure: Soak lab wipe paper and place is at the bottom of the Petri dish. Place a 5 mm thick plastic disk on the wet wipes and put the metal disk with glued mica on it avoiding contact with water at the bottom of the Petri dish. After 30 min, remove the mica/disk assembly and thoroughly rinse the mica surface with dd water and pipette as described in section 6.1.3. The surface is ready for the sample deposition. The APS concentration needs to be adjusted in experiments with DNA, although working APS solution (1:300) described in section 2.2 should be a good starting point. In the protocol described in section 6.1.3 in which the procedure for imaging of nucleosomes with HS-AFM is described, the three-fold higher concentration of APS (1:100) was used.
The protocol described is based on mica functionalization with APS. This is the most robust, highly reproducible and simple procedure. The only disadvantage is that APS, aminopropyl silatrane is not commercially available. However, the procedure of the APS synthesis is straightforward and the protocol for its synthesis is described in detail. 59 If the synthesis procedure is problematic, commercially available reagent, aminopropyltriethoxy silane (APTES) can be used for mica functionalization (AP-mica). The same paper provides the protocol for the preparation of AP-mica using vapors of APTES. The procedure was tested and used for imaging of topologically different types of DNA 35,36,50,62,63 and various protein-DNA complexes including nucleosomes. 27,50,64 Similarly to APS-mica, AP-mica is stable over weeks and can be prepared in batches in advance; the AP-mica surface is as smooth as APS-mica and rather insensitive to the buffer composition, so the samples can be prepared in the buffer solutions with pH values up to pH10 and ionic strengths between 1 mM and 200 mM of NaCl. 59,65 The only drawback with the use of AP-mica is the possibility of aminopropyl silane to hydrolyze and assemble in aggregates on the surface during time-lapse imaging in water, although this process is rather slow with the aggregates that can be distinguished on the surface, so high-resolution images of DNA can be obtained.35 The hydrolysis of silatrane moiety is very slow, so no visible aggregates are seen during long-term imaging;26,35,56,66 , therefore, APS-mica is the preferable substrate when compared with AP-mica if imaging in water is employed. For reproducibility using APTES, the distillation of the reagent is highly recommended in the preparation of AP-mica. The protocol for the APTES distillation is described in paper 59.
AFM, as a single molecule technique, operates with nucleosome concentrations at the nanomolar range and that nucleosomes can spontaneously dissociate at such low concentrations. According to our data 25, the dissociation process occurs in dozens of minutes suggesting that the sample for AFM studies should be prepared just prior to the deposition. Some detergents such as 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) increase the nucleosome stability, so the nucleosomes increase their lifetime by several orders of magnitude 25, but the use of detergents should be done with caution. We showed in 25 that stabilization of nucleosomes is accompanied by the change of the sequence specificity, so the nucleosome even with the use of such sequence specific motif as Widom 601 template, assemble without a specificity of binding to the 601 sequence.
There are a number of important issues with the proposed method relative to other AFM sample preparation methods. First, APS-mica and AP-mica substrates are stable over weeks and can be prepared in batches in advance; the surfaces are smooth and rather insensitive to the buffer composition so the samples can be prepared in buffer solutions with pH values up to pH10 and ionic strengths between 1 mM and 200 mM of NaCl. 59,65 None of the existing methods match these properties. Second, AFM is a single molecule technique and requires a very little sample. The described protocol operates with nanoscale amounts of the preparation. Third, in time-lapse AFM studies, and the HS-AFM data acquisition, there is a significant amount of time required to generate and characterize the large data sets acquired. The methodology described here is well suited for analyzing hundreds of AFM images.
The sample preparation methodology is not limited to nucleosome type samples. Rather it is insensitive to the type of the protein-DNA complexes and has already been applied to a number proteins recognizing specific sequences on DNA, e.g. restrictions enzymes 67,68,69, single-stranded DNA binding proteins 44,45,56,70,71 along with complex protein systems involved in DNA replication 47,72 and recombination68,73. Importantly, HS-AFM has been applied to these systems to follow the dynamics of these systems. These studies make it possible to apply the developed methodology to the characterization of nucleosome arrays as proposed in and elucidate the role of such centromere specific proteins as centromere protein B (CENP-B) or C (CENP-C) in the centromere assembly and understanding their role in the development of many cancers33,34.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Author contributions: YLL and MSD designed the project; MSD assembled nucleosomes. MSD and ZS performed AFM experiments and data analyses. All authors wrote and edited the manuscript.
Materials
| Plasmid pGEM3Z-601 | Addgene, Cambridge, MA | 26656 | |
| PCR Primers | IDT, Coralville, IA | Custom Order | (FP) 5'- CAGTGAATTGTAATACGACTC-3' (RP) 5'-ACAGCTATGACCATGATTAC-3' |
| DreamTaq polymerase | ThermoFischer Scientific, Waltham, MA | EP0701 | Catalog number for 200 units |
| PCR purification kit | Qiagen, Hilden, Germany | 28104 | Catalog number for 50 units |
| Tris base | Sigma-Aldrich, St. Louis, MO | 10708976001 | Catalog number for 250 g |
| EDTA | ThermoFischer Scientific, Waltham, MA | 15576028 | Catalog number for 500 g |
| (CENP-A/H4)2, recombinant human | EpiCypher, Durham, NC | 16-0010 | Catalog number for 50 ug |
| H2A/H2B, recombinant human | EpiCypher, Durham, NC | 15-0311 | Catalog number for 50 ug |
| H3 Octamer, recombinant human | EpiCypher, Durham, NC | 16-0001 | Catalog number for 50 ug |
| Slide-A-Lyzer MINI Dialysis Device Kit, 10K MWCO, 0.1 mL | ThermoFischer Scientific, Waltham, MA | 69574 | Catalog number for 10 devices |
| Sodium Chloride | Sigma-Aldrich, St. Louis, MO | S9888-500G | Catalog number for 500 mg |
| Amicon Ultra-0.5 mL Centrifugal Filters | Millipore-sigma, Burlington, MO | UFC501008 | Catalog number for 8 devices |
| HCl | Sigma-Aldrich, St. Louis, MO | 258148-25ML | Catalog number for 25 mL |
| Tricine | Sigma-Aldrich, St. Louis, MO | T0377-25G | Catalog number for 25 g |
| SDS | Sigma-Aldrich, St. Louis, MO | 11667289001 | Catalog number for 1 kg |
| Ammonium Persulfate (AmmPS) | Bio-Rad, Hercules, CA | 1610700 | Catalog number for 10 g |
| 30% Acrylamide/Bis Solution, 37.5:1 | Bio-Rad, Hercules, CA | 1610158 | Catalog number for 500 mL |
| TEMED | Bio-Rad, Hercules, CA | 1610800 | Catalog number for 5 mL |
| 4x Laemmli protein sample buffer for SDS-PAGE | Bio-Rad, Hercules, CA | 1610747 | Catalog number for 10 mL |
| 2-ME | Sigma-Aldrich, St. Louis, MO | M6250-10ML | Catalog number for 10 mL |
| ageRuler Prestained Protein Ladder | ThermoFischer Scientific, Waltham, MA | 26616 | Catalog number for 500 uL |
| Bio-Safe™ Coomassie Stain | Bio-Rad, Hercules, CA | 1610786 | Catalog number for 1 L |
| Nonwoven cleanroom wipes: TX604 TechniCloth | TexWipe, Kernersvile, NC | TX604 | |
| Muscovite Block Mica | AshevilleMica, Newport News, VA | Grade-1 | |
| Aminopropyl silatrane (APS) | Synthesized as described in 22 | ||
| HEPES | Sigma-Aldrich, St. Louis, MO | H4034-25G | Catalog number for 25 g |
| Scotch Tape | Scotch-3M, St. Paul, MN | ||
| TESPA-V2 afm probe (for static imaging) | Bruker AFM Probes, Camarillo, CA | ||
| MSNL-10 afm probe (for standard time-lapse imaing) | Bruker AFM Probes, Camarillo, CA | ||
| Aron Alpha Industrial Krazy Glue | Toagosei America, West Jefferson, OH | AA480 | Catalog number for 2 g tube |
| MgCl2 | Sigma-Aldrich, St. Louis, MO | M8266-100G | Catalog number for 100 g |
| Millex-GP Filter, 0.22 µm | Sigma-Aldrich, St. Louis, MO | SLGP05010 | Catalog number for 10 devices |
| BL-AC10DS-A2 afm probe (for HS-AFM) | Olympus, Japan | ||
| Compound FG-3020C-20 | FluoroTechnology Co., Ltd., Kagiya, Kasugai, Aichi, Japan | ||
| Compound FS-1010S135-0.5 | FluoroTechnology Co., Ltd., Kagiya, Kasugai, Aichi, Japan | ||
| MultiMode Atomic Force Microscope | Bruker-Nano/Veeco, Santa Barbara, CA | ||
| High-Speed Time-Lapse Atomic Force Microsocopy | Toshio Ando, Nano-Life Science Institute, Kanazawa University, Kakuma-machi, Kanazawa, Japan |
References
- Kornberg, R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 184 (4139), 868-871 (1974).
- Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 389 (6648), 251-260 (1997).
- Clark, D. J. Nucleosome Positioning, Nucleosome Spacing and the Nucleosome Code. Journal of biomolecular structure. 27 (6), 781-793 (2010).
- Poirier, M. G., Oh, E., Tims, H. S., Widom, J. Dynamics and function of compact nucleosome arrays. Nature Structural & Molecular Biology. 16 (9), 938-944 (2009).
- Li, B., Carey, M., Workman, J. L. The Role of Chromatin during Transcription. Cell. 128 (4), 707-719 (2007).
- Venkatesh, S., Workman, J. L. Histone exchange, chromatin structure and the regulation of transcription. Nature Reviews Molecular Cell Biology. 16, 178 (2015).
- Lai, W. K. M., Pugh, B. F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nature Reviews in Molecular Cell Biology. 18 (9), 548-562 (2017).
- Adam, S., Polo, S. o. p. h. i. e. E., Almouzni, G. Transcription Recovery after DNA Damage Requires Chromatin Priming by the H3.3 Histone Chaperone HIRA. Cell. 155 (1), 94-106 (2013).
- Ahmad, K., Henikoff, S. Epigenetic Consequences of Nucleosome Dynamics. Cell. 111 (3), 281-284 (2002).
- Filenko, N. A., Palets, D. B., Lyubchenko, Y. L. Structure and dynamics of dinucleosomes assessed by atomic force microscopy. Journal of amino acids. 2012, 650840 (2012).
- Hihara, S., et al. Local nucleosome dynamics facilitate chromatin accessibility in living mammalian cells. Cell reports. 2 (6), 1645-1656 (2012).
- Jiang, C., Pugh, B. F. Nucleosome positioning and gene regulation: advances through genomics. Nature reviews. Genetics. 10 (3), 161-172 (2009).
- Brennan, L. D., Forties, R. A., Patel, S. S., Wang, M. D. DNA looping mediates nucleosome transfer. Nature Communications. 7, 13337 (2016).
- Lyubchenko, Y. L. Nanoscale nucleosome dynamics assessed with time-lapse AFM. Biophysical Reviews. 6 (2), 181-190 (2014).
- Miyagi, A., Ando, T., Lyubchenko, Y. L. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 50 (37), 7901-7908 (2011).
- Stumme-Diers, M. P., Banerjee, S., Hashemi, M., Sun, Z., Lyubchenko, Y. L. Nanoscale dynamics of centromere nucleosomes and the critical roles of CENP-A. Nucleic Acids Research. 46 (1), 94-103 (2018).
- Narlikar, G. e. e. t. a. J., Sundaramoorthy, R., Owen-Hughes, T. Mechanisms and Functions of ATP-Dependent Chromatin-Remodeling Enzymes. Cell. 154 (3), 490-503 (2013).
- Ngo, T. T., Zhang, Q., Zhou, R., Yodh, J. G., Ha, T. Asymmetric Unwrapping of Nucleosomes under Tension Directed by DNA Local Flexibility. Cell. 160 (6), 1135-1144 (2015).
- Ruth, B., Wietske, K., Kirsten, M., John van, N. spFRET reveals changes in nucleosome breathing by neighboring nucleosomes. Journal of Physics: Condensed Matter. 27 (6), 064103 (2015).
- Buning, R., van Noort, J. Single-pair FRET experiments on nucleosome conformational dynamics. Biochimie. 92 (12), 1729-1740 (2010).
- Koopmans, W. J. A., Brehm, A., Logie, C., Schmidt, T., van Noort, J. Single-Pair FRET Microscopy Reveals Mononucleosome Dynamics. Journal of Fluorescence. 17 (6), 785-795 (2007).
- Brower-Toland, B. D., et al. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proceedings of the National Academy of Sciences. 99 (4), 1960 (1960).
- Bennink, M. L., et al. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nature Structural Biology. 8 (7), 606-610 (2001).
- Lyubchenko, Y. L., Shlyakhtenko, L. S., Chellappan, S. P. . Chromatin Protocols. , 27-42 (2015).
- Menshikova, I., Menshikov, E., Filenko, N., Lyubchenko, Y. L. Nucleosomes structure and dynamics: effect of CHAPS. International Journal of Biochemistry and Molecular Biology. 2, 2129-2137 (2011).
- Shlyakhtenko, L. S., Lushnikov, A. Y., Lyubchenko, Y. L. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry. 48 (33), 7842-7848 (2009).
- Yodh, J. G., Lyubchenko, Y. L., Shlyakhtenko, L. S., Woodbury, N., Lohr, D. Evidence for nonrandom behavior in 208-12 subsaturated nucleosomal array populations analyzed by AFM. Biochemistry. 38 (48), 15756-15763 (1999).
- Filenko, N. A., et al. The role of histone H4 biotinylation in the structure of nucleosomes. PLoS One. 6 (1), e16299 (2011).
- Lyubchenko, Y. L., Meyers, R. . Encyclopedia of Analytical Chemistry. , 1-24 (2013).
- Stumme-Diers, M. P., Banerjee, S., Sun, Z., Lyubchenko, Y. L., Lyubchenko, Y. L. . Nanoscale Imaging: Methods and Protocols. , 225-242 (2018).
- Cleveland, D. W., Mao, Y., Sullivan, K. F. Centromeres and Kinetochores. Cell. 112 (4), 407-421 (2003).
- Rosin, L. F., Mellone, B. G. Centromeres Drive a Hard Bargain. Trends in Genetics. 33 (2), 101-117 (2017).
- McKinley, K. L., Cheeseman, I. M. The molecular basis for centromere identity and function. Nature Reviews Molecular Cell Biology. 17 (1), 16-29 (2016).
- Lyubchenko, Y. L. Centromere chromatin: a loose grip on the nucleosome. Nature Structural & Molecular Biology. 21 (1), 8 (2014).
- Lyubchenko, Y. L., Shlyakhtenko, L. S. Visualization of supercoiled DNA with atomic force microscopy in situ. Proceedings of the National Academy of Sciences. 94 (2), 496-501 (1997).
- Lyubchenko, Y. L., Shlyakhtenko, L. S., Aki, T., Adhya, S. Atomic force microscopic demonstration of DNA looping by GalR and HU. Nucleic Acids Research. 25 (4), 873-876 (1997).
- Herbert, A., et al. The Zalpha domain from human ADAR1 binds to the Z-DNA conformer of many different sequences. Nucleic acids research. 26 (15), 3486-3493 (1998).
- Oussatcheva, E. A., et al. Structure of branched DNA molecules: gel retardation and atomic force microscopy studies. Journal of Molecular Biology. 292 (1), 75-86 (1999).
- Gaillard, C., Shlyakhtenko, L. S., Lyubchenko, Y. L., Strauss, F. Structural analysis of hemicatenated DNA loops. BMC Struct Biol. 2 (1), 7 (2002).
- Potaman, V. N., et al. Unpaired structures in SCA10 (ATTCT)n.(AGAAT)n repeats. Journal of Molecular Biology. 326 (4), 1095-1111 (2003).
- Virnik, K., et al. “Antiparallel” DNA loop in gal repressosome visualized by atomic force microscopy. Journal of Molecular Biology. 334 (1), 53-63 (2003).
- Pavlicek, J. W., et al. Supercoiling-induced DNA bending. Biochemistry. 43 (33), 10664-10668 (2004).
- Karymov, M., Daniel, D., Sankey, O. F., Lyubchenko, Y. L. Holliday junction dynamics and branch migration: single-molecule analysis. Proceedings of the National Academy of Sciences. 102 (23), 8186-8191 (2005).
- Shlyakhtenko, L. S., et al. Nanoscale structure and dynamics of ABOBEC3G complexes with single-stranded DNA. Biochemistry. 51 (32), 6432-6440 (2012).
- Shlyakhtenko, L. S., Lushnikov, A. Y., Miyagi, A., Lyubchenko, Y. L. Specificity of binding of single-stranded DNA-binding protein to its target. Biochemistry. 51 (7), 1500-1509 (2012).
- Shlyakhtenko, L. S., et al. APOBEC3G Interacts with ssDNA by Two Modes: AFM Studies. Scientific Reports. 5, 15648 (2015).
- Sun, Z., Tan, H. Y., Bianco, P. R., Lyubchenko, Y. L. Remodeling of RecG Helicase at the DNA Replication Fork by SSB Protein. Scientific Reports. 5, 9625 (2015).
- Bianco, P. R., Lyubchenko, Y. L. SSB and the RecG DNA helicase: An intimate association to rescue a stalled replication fork. Protein Science. 26 (4), 638-649 (2017).
- Zhang, Y., et al. High-speed atomic force microscopy reveals structural dynamics of alpha-synuclein monomers and dimers. Journal of Chemical Physics. 148 (12), 123322 (2018).
- Lyubchenko, Y. L. DNA structure and dynamics: an atomic force microscopy study. Cell Biochem Biophys. 41 (1), 75-98 (2004).
- Lyubchenko, Y. L., Shlyakhtenko, L. S. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods. 47 (3), 206-213 (2009).
- Lyubchenko, Y. L., Shlyakhtenko, L. S., Gall, A. A. Atomic force microscopy imaging and probing of DNA, proteins, and protein DNA complexes: silatrane surface chemistry. Methods in Molecular Biology. 543, 337-351 (2009).
- Lyubchenko, Y. L. Nanoimaging methods for biomedicine. Methods. 60 (2), 111-112 (2013).
- Lyubchenko, Y. L., Shlyakhtenko, L. S. Imaging of DNA and Protein-DNA Complexes with Atomic Force Microscopy. Critical Reviews in Eukaryotic Gene Expression. 26 (1), 63-96 (2016).
- Lowary, P. T., Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of Molecular Biology. 276 (1), 19-42 (1998).
- Lyubchenko, Y. L., Shlyakhtenko, L. S., Ando, T. Imaging of nucleic acids with atomic force microscopy. Methods (San Diego, Calif). 54 (2), 274-283 (2011).
- Luger, K., Rechsteiner, T. J., Richmond, T. J. Preparation of nucleosome core particle from recombinant histones. Methods in enzymology. 304, 3-19 (1999).
- Gallagher, S. R. One-dimensional SDS gel electrophoresis of proteins. Current protocols in immunology. , (2006).
- Shlyakhtenko, L. S., Gall, A. A., Lyubchenko, Y. L., Taatjes, D. J., Roth, J. . Cell Imaging Techniques: Methods and Protocols. , 295-312 (2013).
- Uchihashi, T., Ando, T., Braga, P. C., Ricci, D. . Atomic Force Microscopy in Biomedical Research: Methods and Protocols. , 285-300 (2011).
- Lyubchenko, Y. L., Gall, A. A., Shlyakhtenko, L. S. Visualization of DNA and protein-DNA complexes with atomic force microscopy. Methods in molecular biology. 1117, 367-384 (2014).
- Lyubchenko, Y. L., Shlyakhtenko, L. S. . Proceeding of the Fourth International Workshop: STM-AFM-SNOM: New Nanotools for Molecular Biology. , 20-34 (1997).
- Kato, M., et al. Interarm interaction of DNA cruciform forming at a short inverted repeat sequence. Biophys J. 85 (1), 402-408 (2003).
- Yodh, J. G., Woodbury, N., Shlyakhtenko, L. S., Lyubchenko, Y. L., Lohr, D. Mapping nucleosome locations on the 208-12 by AFM provides clear evidence for cooperativity in array occupation. Biochemistry. 41 (11), 3565-3574 (2002).
- Lyubchenko, Y. L., Gall, A. A., Shlyakhtenko, L. S. Atomic force microscopy of DNA and protein-DNA complexes using functionalized mica substrates. DNA-Protein Interactions: Principles and Protocols. , 569-578 (2001).
- Lyubchenko, Y. L. Preparation of DNA and nucleoprotein samples for AFM imaging. Micron. 42 (2), 196-206 (2011).
- Gilmore, J. L., et al. Single-molecule dynamics of the DNA-EcoRII protein complexes revealed with high-speed atomic force microscopy. Biochemistry. 48 (44), 10492-10498 (2009).
- Shlyakhtenko, L. S., et al. Molecular mechanism underlying RAG1/RAG2 synaptic complex formation. J Biol Chem. 284 (31), 20956-20965 (2009).
- Suzuki, Y., et al. Visual Analysis of Concerted Cleavage by Type IIF Restriction Enzyme SfiI in Subsecond Time Region. Biophysical. 101 (12), 2992-2998 (2011).
- Shlyakhtenko, L. S., Lushnikov, A. J., Li, M., Harris, R. S., Lyubchenko, Y. L. Interaction of APOBEC3A with DNA assessed by atomic force microscopy. PloS one. 9 (6), e99354 (2014).
- Pan, Y., et al. Nanoscale Characterization of Interaction of APOBEC3G with RNA. Biochemistry. 56 (10), 1473-1481 (2017).
- Sun, Z., Hashemi, M., Warren, G., Bianco, P. R., Lyubchenko, Y. L. Dynamics of the Interaction of RecG Protein with Stalled Replication Forks. Biochemistry. 57 (13), 1967-1976 (2018).
- Pavlicek, J. W., Lyubchenko, Y. L., Chang, Y. Quantitative analyses of RAG-RSS interactions and conformations revealed by atomic force microscopy. Biochemistry. 47 (43), 11204-11211 (2008).

