Measurement of Chitinase Activity in Biological Samples
Summary
Presented here is a simple method to measure chitinase activity in biological fluids such as bronchoalveolar lavage or serum.
Abstract
Chitinases are the enzymes that cleave chitin. Even in the absence of chitin, mammalians have significant amounts of chitinases present in the body including at baseline. The precise role of chitinase is not known, however it was believed to play important role in digestion and host defense against chitin-containing food and pathogens, respectively. Recent work, including ours, has shown an important role of chitinase and chitinase-like proteins in host immunity and allergic diseases. Importantly, chitinase activities serve as important biomarkers of disease severity in a wide-range of diseases including type 2 inflammatory diseases such as asthma and pulmonary fibrosis. Similarly, patients with genetic disorders like Gaucher disease have significantly elevated chitinase levels, which not only correlate with disease severity but also serve as a reliable biomarker for therapeutic effectiveness. The protocol outlined here describes a simple, quick, and straightforward way to measure chitinase activity in BAL or serum samples of mice and can be widely adapted to human subjects and other model organisms due to the highly conserved nature of the enzymes.
Introduction
Chitin is the second most abundant polysaccharide on the earth after cellulose, serving as a major structural component of a variety of organisms including the exoskeleton of insects, fungi, yeast, and algae; some vertebrates also have chitin1. Chitinases are a family of enzymes that are capable of breaking down chitin and are highly conserved throughout evolution in species ranging from bacteria to mammals2,3. In addition to chitinases, mammals also have chitinase-like proteins which are similar to chitinases in their ability to bind chitin but differ in that they lack enzymatic ability to cleave the chitin4.
Although chitin and chitinases have long been studied—with investigations dating back to the early 1900’s—the main focus has been on their role in insects and other invertebrates. In fact, it was not until the 1960s when vertebrates were found to have chitinases at all. Using a chitobiase-based assay, chitinases were shown to be found in the digestive tract of a number of vertebrates including lizards and blackbirds—an observation hypothesized to be due to the consumption of chitin-containing organisms such as insects5.
Mammals have two enzymatically active forms of chitinase: acidic mammalian chitinase (AMCase; also known as CHIT2 and CHIA) and chitotriosidase (CHIT1)4. Both of these proteins are capable of hydrolyzing chitin. However, CHIT1 is more enzymatically active in humans, where almost all of chitinase activity is derived from CHIT1.4 In mice, both Chit1 and AMCase almost equally contribute to the overall chitinase activity6. On the other hand, chitinase-like proteins lack chitinase activity.
Chit1 is mainly secreted by macrophages and is often considered to be an immune response to chitin-containing pathogens7. The enzyme has also been shown to be involved in the maturation of monocytes into both M1 and M2 macrophage subtypes, even without the presence of the substrate chitin8. Furthermore, it may also be involved in the maturation of other immune cells including T helper type 2 (Th2) cells and eosinophils—as was shown to be the case in cryptococcal lung infection9. These studies point to a complex role of chitinases in the immune system.
In recent years, Chit1 levels have been found to serve as an important biomarker of progression for over 40 different human diseases including lysosomal storage diseases, infectious diseases, respiratory diseases, endocrinological diseases, cardiovascular diseases, neurological diseases, and others (reviewed10). In many of these diseases, the levels of Chit1 are a strong predictor of disease severity and therapeutic effectiveness10.
As chitinases have gained a reputation as a biomarker for numerous medical conditions including Gaucher Disease, tools and assays have been developed to facilitate testing for chitinase presence. Older methods include Schales’ procedure, a protocol adapted from a blood glucose test, and the 3,5 dinitrosalicylic acid (DNS) method. However, these methods are often time sensitive and technically difficult11. The procedure for both of these tests requires the reduction of inorganic oxidants, ferricyanide in the case of the Schales’ procedure for example, producing a color change that can only be measured spectrophotometrically. Additionally, both tests involve a heating or boiling step that is both time consuming and necessary for the color to develop12,13.
Described here is a quick and simple fluorimetric assay to determine chitinase levels in mammalian samples14,15. Two of the samples used here include serum and bronchoalveolar lavage fluids (BAL); chitinase activity has also been measured in breast milk and urine samples, and the technique can be performed in those type of samples as well as in any other biological fluid16,17.
Protocol
All animal procedures were performed under an IACUC-approved protocol at Yale University School of Medicine.
1. Mouse Sample Collection
- Anesthetization
- Anesthetize mice using ketamine and xylazine (100 mg/kg of ketamine and 10 mg/kg of xylazine).
- Blood sample collection
- Confirm the depth of anesthesia by non-responsiveness to a toe pinch.
- Surgically open the chest cavity to expose the heart.
- Insert a 26.5 G needle into the left ventricle and collect blood into a syringe.
NOTE: Typically, about 0.5–1 mL of blood can be harvested from a healthy 20–25 g mouse. - Collect the blood in heparinized tubes, for plasma collection, or non-heparinized tubes for serum collection.
- Collection of bronchoalveolar lavage fluid (BAL)
- To collect the BAL, make a vertical cut on the neck to expose the trachea.
- Cannulate the trachea with a 22 G catheter and secure it firmly using a string in order to prevent leakage out of the nose of the mouse.
- Slowly inject two aliquots of sterile phosphate-buffered saline (PBS, 0.75 mL each) into the lungs via the trachea and retrieve back.
- Pool aliquots and keep on ice for further analysis.
- Processing the biological samples.
- Blood sample: Either fresh (plasma) or after allowing coagulation for 4 h (serum), centrifuge at 600 x g for 10 min. Take the supernatant and either freeze for storage or use for experiment.
- BAL: Centrifuge the sample at 350 x g for 5 min. Take the supernatant and either freeze for storage or use for experiment.
2. Chitinase Assay
NOTE: The science behind the assay is relatively simple. A non-fluorescent substrate is cleaved by enzymatically active chitinase to produce a fluorescent product, which is then measured as an indirect marker of chitinase activity. The chitinases within the samples break down the substrate 4-methylumbelliferyl-D-N, N’-diacetylchitobiose present in the 1x McIlvain buffer. A fluorescent molecule 4-methylumbelliferyl is released. By measuring the total fluorescence of each well, we obtain an accurate measurement of the active chitinase in each sample. Because the breakdown of chitin by chitinase is a hydrolytic reaction, the stop buffer, a mixture of 0.3 M glycine and NaOH (12.0 g/L at pH 10.6), ends the breakdown of chitin by creating an environment that is too basic for the enzyme to function.
- Prepare the substrates, standards, and solutions.
- Prepare McIlvain buffer. McIlvain Buffer is comprised of 0.1 M citrate and 0.2 M phosphate at a pH of 5.2. Dilute at a ratio of 1:1 for 1x McIlvain buffer.
- Dissolve both 4-methylumbelliferyl-D-N, N’-diacetylchitobiose and 4-methylumbelliferyl ß-D-N, N’, N”-triacetylchitotriose in 1x McIlvain Buffer to a concentration of 500 µM each.
NOTE: Stock solution can be stored at -20 °C for further uses; best stored in aliquots. - Dissolve standard, 4-methylumbelliferone, to a concentration of 0.1 mM in stop buffer (mixture of 0.3 M glycine and NaOH (12.0 g/L at pH 10.6)) to create the standard curve stock solution.
- Create and plate the working solution.
- Combine 1x McIlvain Buffer and chitin substrate 4-methylumbelliferyl-D-N, N’-diacetylchitobiose to create the working solution. For every 10 samples, mix 100 µL of substrate with 2.17 mL of 1x McIlvain buffer. See Table 1 for additional calculations based on sample size.
NOTE: Use 4-methylumbelliferyl ß-D-N, N’, N”-triacetylchitotriose for human samples. While both substrates can be cleaved by either human or mouse enzymes, they have been assigned for either human or mouse samples based on efficiency of cleavage6. - Pipette 95 µL of working solution into each well of a 96-well plate.
- Combine 1x McIlvain Buffer and chitin substrate 4-methylumbelliferyl-D-N, N’-diacetylchitobiose to create the working solution. For every 10 samples, mix 100 µL of substrate with 2.17 mL of 1x McIlvain buffer. See Table 1 for additional calculations based on sample size.
- Add biospecimen samples to the working solution.
- Next, add 5 µL of the test sample (blood/BAL/tissue lysate/other biological fluid) to each well.
- Mix the sample into the working solution for the most accurate and reliable results.
NOTE: Samples should be run in duplicates/triplicates with special attention paid to potential edge effects. All the samples should be pre-warmed to minimize the edge effect. As the assay involves a reaction between chitinases in the sample and substrate in the working solution, it is best to work relatively quickly to minimize the difference in time between when the first sample is plated and the last sample is plated. Multichannel pipette is recommended.
- Incubate at 37 °C.
- Cover the plate and shake briefly (5 s).
- Incubate the plate at 37 °C for 15 min. This allows the enzymatic reaction to take place.
- Prepare the standard curve.
- While the samples are incubating, prepare a serial dilution of 4-methylumbelliferone, a standard often used in the fluorimetric determination of enzyme activity.
- Dilute the stock of the standard solution in stop buffer to a concentration of 5 µM.
- Perform a series of dilutions as indicated in Table 2. Add the components in the amount indicated and mix well.
NOTE: The standard curve helps to ensure that measured samples fall in the linear range of the standard curve.
- Stop the reaction with stop buffer.
- Add 200 µL of stop buffer to each well to stop the reaction.
- Plate the standards.
- Add 300 µL of each standard per well in duplicates on the same plate.
- Read the plate using a fluorometric reader.
- Read the plate at an excitation of 360 nm, and an emission of 455 nm.
3. Data Analysis
- Subtract the blanks
- Average the values of the duplicate standard dilutions that have no 4-methylumbelliferone. Subtract this value from all of the other recorded values.
- Take average of technical replicates and plot the standards.
- Average the two readings for each concentration and graph the average reading by concentration.
NOTE: The resulting plot should look linear and have an R2 value close to 1. If it does not, it may be necessary to redo the standards and reread the plate to ensure quality of data analysis.
- Average the two readings for each concentration and graph the average reading by concentration.
- Standardize the read-out values.
- Create a best-fit line for the plotted standards data. Divide the read-outs of the samples by the slope of the best-fit line.
- Compare values from control group and variable group.
- Use a t-test or ANOVA, for more than two groups, to determine if the difference between groups is statistically significant. Values will take the units nmol/mL·h.
Representative Results
The results shown here are from a study measuring the chitinase activity in serum and BAL samples of wild type (C57BL/6) and Chit1 transgenic mice (on C57BL/6 background) that overexpress the Chit1 gene on a doxycycline promoter. Our data show that both serum and BAL samples have detectable chitinase activity at baseline 698.2 ± 189.9 nmol/mL·h and 485.7 ± 114 nmol/ml·h, respectively. Overexpression of the Chit1 gene using doxycycline in drinking water for 4 weeks resulted in the elevation of chitinase activity that was observed in both BAL (4,575 ± 673.8 nmol/mL·h, p = 0.003) and serum 1556 ± 251.2 nmol/mL·h, p = 0.0272, when compared to the baseline) samples.
Using the protocol described here, the assay confirms that overexpression of the Chit1 gene in mice results in increased chitinase activity. A t-test was used to compare the means of the two groups. Each group contained 5 mice.
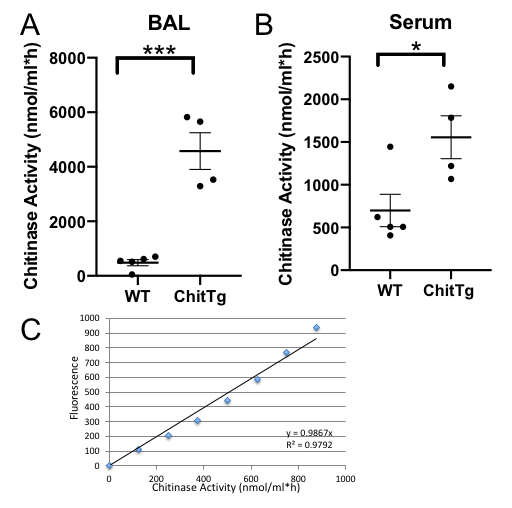
Figure 1: Chitinase activity. Chitinase activity was measured in the (A) serum and the (B) bronchoalveolar lavage fluid of wild type (WT) and chitotriosidase (ChitTg) over-expressing transgenic mice. Each dot represents an individual mouse sample. (C) Values were standardized using the linearity of the standard curve as described in the protocol. Fluorescence levels were measured using a plate reader at wavelength of 360 nm for excitation and 455 nm for emission and presented as AU. AU = arbitrary unit *p < 0.05, ***p < 0.001. Please click here to view a larger version of this figure.
| Number of Sample Wells | Substrate (µL) | McIlvain Buffer (mL) |
| 20 | 100 | 2.17 |
| 50 | 250 | 5.425 |
| 100 | 500 | 10.85 |
Table 1: Sample calculations for creating the working solution.
| (µL) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Conc. (nM) | 0 | 125 | 250 | 375 | 500 | 625 | 750 | 875 |
| Standard | 0 | 20 | 40 | 60 | 80 | 100 | 120 | 140 |
| 2x McIlvian | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 |
| Distilled H2O | 200 | 180 | 160 | 140 | 120 | 100 | 80 | 60 |
| Stop Buffer | 400 | 400 | 400 | 400 | 400 | 400 | 400 | 400 |
Table 2: Dilutions for standard curve measurements.
Discussion
Chitinase activity has emerged as an important biomarker for predicting disease severity, disease progression, therapeutic effectiveness and the presence of specific pathogens18. Although many of the long-postulated theories about the role of chitinases have not been experimentally proven19, new studies have provided important insights into the role of chitinases and chitinase like proteins in various diseases2,9,20.
The protocol described here is a simple, quick and effective way to measure chitinase activity in biological samples. Little to no modification is required to measure the chitinase activity in a wide variety of biological samples and species due to the highly conserved nature of chitinases. Listed in this protocol are two substrates, one of which as been designated for mouse samples and one for human samples; however, both substrates can be used on either species, as they have been designated as such based on previous papers’ demonstration of the efficiency with which the enzymes can cleave the substrate.
Compared to previously existing techniques, such as the Schales’ procedure and DNS method, the assay outlined here removes the need for time intensive steps such as boiling or heating that were necessary in older methods. Additionally, the assay is less technically difficult and requires fewer complicated steps than preceding measurement techniques. For best results both biological and technical replicates should be used.
If the chitinase activity is too high, dilutions of the biological sample are sufficient to reduce the chitinase activity for measurement, and the true values can be calculated based on the dilution. When performing this experiment on new biological samples, it may be helpful to perform repeat measurements on the same plate over time until saturation to ensure a proper incubation time for best results. Additionally, the protocol can be scaled up or down as long as consistency between the groups remains. The future applications of this protocol are far reaching as research is just picking up in the role of chitinase activity in a number of diseases.
One of the first category of diseases that chitinase activity was found to be relevant in was lysosomal storage diseases including Gaucher’s disease, Niemann-Pick A/B and C, GM-1 gangliosidosis, and many others21. Gaucher disease is a lysosomal storage disease caused by insufficient activity of glucocerebrosidase and is characterized by the buildup of glucosylceramide, the substrate of glucocerebrosidase, in the lysosome of macrophages15. It is a genetic disease, with clinical symptoms that include spleen and liver enlargement, low platelet count, anemia, fatigue, and bone problems22. Clinical research has demonstrated that a majority of people suffering from Gaucher disease have a median chitinase activity that is over 600 times the median value of normal control volunteers; additionally, when people with Gaucher disease were put on enzyme supplementation therapy—the current treatment for the disease—their chitinase activity levels dropped significantly lending chitinase activity to be an excellent marker of both disease progression and treatment monitoring15. No role has yet been found for Chit1 in the disease however. Similar research has led to Chit1 activity monitoring for numerous other lysosomal storage diseases10.
Additionally, further research has indicated a potential role for Chit1 activity during various pathogen infection including fungal infection, malaria, tuberculosis and K. pneumoniae to name a few2,3,10. A 2012 study found that chitinase activity was elevated in patients with active tuberculosis (TB) infection even when the sputum smear was negative, indicating that chitinase activity could be used as an effective biomarker in TB diagnosis, especially during the long wait time that often is necessary to accurately diagnosis the disease23. Similar findings were seen during Plasmodium falciparum infection, with chitotriosidase serum levels significantly increased in those with acute infection24. While many of these diseases take days to diagnose, quick measurements of chitinase activity levels in the blood can be provide beneficial insights in the diagnosis. Additionally, as malaria parasites and tuberculosis bacterium become increasingly resistant to treatments, measuring chitinase activity levels in the serum may help with treatment monitoring for its effectiveness in real time.
More and more research has begun to focus on the role of chitinase and chitinase-like proteins in the immune response, particularly its role in inflammatory pathways. As this research continues, this protocol will allow for quick and accurate measurement of this chitinase activity to further research. The protocol is easily adjusted for any number of species samples including both mouse and human, making it widely applicable.
In addition to the research benefit, better methods for detecting and measuring chitinase activity in samples will be beneficial in the clinical realm. Recent research has demonstrated the role of chitinase as a biomarker in the pathology of numerous chronic diseases including Gaucher disease, diabetes mellitus, sarcoidosis, atherosclerosis, inflammatory bowel disease and cancers3,21,25,26.
Because of this role as a biomarker, quick and accurate measurement of chitinase activity as shown here is incredibly relevant to the field of medicine, allowing healthcare providers to have more information for a proper diagnosis and treatment plan.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by American Lung Association and American Thoracic Society awards to LS. Authors sincerely want to thank Dr. Jack Elias and Dr. Chun Geun Lee for providing transgenic mouse strains.
Materials
| 4-methylumbelliferone | Sigma | M1381 | Standard: commonly used in flourimetric assays for determination of enzyme activity |
| 4MU-GlcNAc2 | Sigma | M9763 | fluorescent chitinase substrate for use in mouse samples |
| 4MU-GlcNAc3 | Sigma | M5639 | fluorescent chitinase substrate for use in human samples |
| Citric Acid-monohydrate | for use in McIlvain Buffer | ||
| Glycine | for use in Stop Buffer at a concentration of 0.3 M | ||
| Na2HPO4xH2O | for use in McIlvain Buffer | ||
| NaOH | for use in Stop Buffer at a concentration of 12 g/L | ||
| Vision Plate: Non-sterile, untreated black 96 well plate | 4titude | 4ti-0224 |
References
- Hamid, R., Khan, M. A., Ahmad, M. Chitinases: An update. Journal of Pharmacy and Bioallied Sciences. 5 (1), 21-29 (2013).
- Sharma, L., et al. Regulation and Role of Chitotriosidase during Lung Infection with Klebsiella pneumoniae. Journal of Immunology. 201 (2), 615-626 (2018).
- Kanneganti, M., Kamba, A., Mizoguchi, E. Role of chitotriosidase (chitinase 1) under normal and disease conditions. Journal of Epithelial Biology & Pharmacology. 5, 1-9 (2012).
- Lee, C. G., et al. Role of Chitin and Chitinase/Chitinase-Like Proteins in Inflammation, Tissue Remodeling, and Injury. Annual Review of Physiology. (73), 479-501 (2011).
- Jeuniaux, C. Chitinase: An Addition to the List of Hydrolases in the Digestive Tract of Vertebrates. Nature. 192 (4798), 135-136 (1961).
- Boot, R. G., et al. Identification of a Novel Acidic Mammalian Chitinase Distinct from Chitotriosidase. Journal of Biological Chemistry. 276 (9), 6770-6778 (2000).
- Arndt, S., Hobbs, A., Sinclaire, I., Lane, A. B. Chitotriosidase Deficiency: A Mutation Update in an African Population. JIMD Reports. 10, 11-16 (2013).
- Di Rosa, M., Malaguarnera, G., De Gregorio, C., Drago, F., Malaguarnera, L. Evaluation of CHI3L-1 and CHIT-1 Expression in Differentiated and Polarized Macrophages. Inflammation. 36 (2), 482-492 (2013).
- Wiesner, D. L., et al. Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection. PLOS Pathogens. 11 (3), e1004701 (2015).
- Elmonem, M. A., vanden Heuvel, L. P., Levtchenko, E. N. Immunomodulatory Effects of Chitotriosidase Enzyme. Enzyme Research. 2016, 1-9 (2016).
- Ferrari, A. R., Gaber, Y., Fraaije, M. W. A fast, sensitive and easy colorimetric assay for chitinase and cellulase activity detection. Biotechnol Biofuels. 7 (37), 1-8 (2014).
- Schales, O., Schales, S. S. Simple method for the determination of glucose in blood. Proc Am Fed Clin Res. 2, 78 (1945).
- Zarei, M., et al. Characterization of Chitinase with Antifungal Activity from a Native Serratia Marcescens B4A. Brazilian Journal of Microbiology. 42 (297), 1017-1029 (2011).
- Zhu, Z., et al. Acidic Mammalian Chitinase in Asthmatic Th2 Inflammation and IL-13 Pathway Activation. Science. 304 (5677), 1678-1682 (2004).
- Hollak, C. E., van Weely, S., van Oers, M. H., Aerts, J. M. Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. Journal of Clinical Investigation. 93 (3), 1288-1292 (1994).
- Musumeci, M., Malaguarnera, L., Simpore, J., Barone, R., Whalen, M., Musumeci, S. Chitotriosidase activity in colostrum from African and Caucasian women. Clinical Chemistry and Laboratory. 43 (2), 198-201 (2005).
- Maddens, B., et al. Chitinase-like Proteins are Candidate Biomarkers for Sepsis-induced Acute Kidney Injury. Mol Cell Proteomics. 11, 1-13 (2012).
- Hector, A., et al. Chitinase activation in patients with fungus-associated cystic fibrosis lung disease. Journal of Allergy and Clinical Immunology. 138 (4), 1183-1189 (2016).
- Hall, A. J., Morroll, S., Tighe, P., Götz, F., Falcone, F. H. Human chitotriosidase is expressed in the eye and lacrimal gland and has an antimicrobial spectrum different from lysozyme. Microbes and Infection. 10 (1), 69-78 (2008).
- Kim, L. K., et al. AMCase is a crucial regulator of type 2 immune responses to inhaled house dust mites. Proceedings of the National Academy of Sciences of the United States of America. 112 (22), e2891-e2899 (2015).
- Olkhovych, N. V. Chitotriosidase activity as additional biomarker in the diagnosis of lysosomal storage diseases. Ukrainian Biochemical Journal. 88 (1), 69-78 (2016).
- Bouayadi, O., et al. Disease: An Underdiagnosed Pathology in the Eastern Moroccan Population. eJIFCC. 30 (1), 82-87 (2019).
- Tasci, C., et al. Efficacy of serum chitotriosidase activity in early treatment of patients with active tuberculosis and a negative sputum smear. Therapeutics and Clinical Risk Management. 8, 369-372 (2012).
- Barone, R., Simporé, J., Malaguarnera, L., Pignatelli, S., Musumeci, S. Plasma chitotriosidase activity in acute Plasmodium falciparum malaria. Clinica Chimica Acta. 331 (1-2), 79-85 (2003).
- Kitamoto, S., et al. Chitinase inhibition promotes atherosclerosis in hyperlipidemic mice. American Journal of Pathology. 183 (1), 313-325 (2013).
- Kzhyshkowska, J., Gratchev, A., Goerdt, S. Human Chitinases and Chitinase-Like Proteins as Indicators for Infl Ammation and Cancer. Biomark Insights. 2, 128-146 (2007).

