Quantification of Self-renewal in Murine Mammosphere Cultures
Summary
Here, we describe the implementation and interpretation of the results of an in vitro mammosphere self-renewal quantitative assay.
Abstract
The mammary gland is characterized by extensive regeneration capacity, as it goes through massive hormonal changes throughout the life cycle of a female. The role of mammary stem cells (MaSCs) is widely studied both in the physiological/developmental context and with regards to breast carcinogenesis. In this aspect, ex vivo studies focused on MaSC properties are highly sought after. Mammosphere cultures represent a surrogate of organ formation and have become a valuable tool for both basic and translational research. Here, we present a detailed protocol for the generation of murine primary mammosphere cultures and the quantitation of MaSC growth properties. The protocol includes mammary gland collection and digestion, isolation of primary mammary epithelial cells (MECs), establishment of primary mammosphere cultures, serial passaging, quantitation of mammosphere growth parameters and interpretation of the results. As an example, we present the effect of low-level constitutive Myc expression on normal MECs leading to increased self-renewal and proliferation.
Introduction
Isolation and in vitro culture of mammary epithelial stem and progenitor cells have become essential for understanding their properties in mammary cell biology. Elegant lineage tracing and serial transplantation assays have enabled the study of stem cells (SCs) and other tissue subsets in the context of their in vivo niche. However, this approach is time consuming and requires the generation of reporter mouse models1,2,3,4,5. Therefore, in vitro culture and propagation of mammary stem cells (MaSCs) while sparing key stemness features, namely self-renewal and differentiation ability, is one of the biggest challenges in the field. In the last years, the mammosphere assay has been widely used to model both normal mammary tissue and breast cancer growth, to quantify normal or cancer SCs (CSCs) and assess their self-renewal ability as a surrogate reporter of their activity in their respective in vivo context6,7,8,9,10,11.
The mammosphere assay is an efficient and cost-effective approach, in which freshly isolated mammary epithelial cells (MECs) are cultured in non-adherent conditions, with the premise that only MaSCs will survive and form spheres in suspension while all the other cell types will die by anoikis. Moreover, the ability to form several generations of mammospheres in serial non-adherent passages is related to the self-renewal ability of the MaSCs6,9,11. Here, we describe a detailed protocol of a quantitative mammosphere assay, which was initially developed by Dontu and colleagues7 as a modification of the pioneering neurosphere assay12, enabling the growth of putative SCs in non-adherent, serum-free conditions with the addition of appropriate growth factors7,12.
Protocol
In vivo procedures were performed in accordance with EU directive 2010/63 and after approval from our institutional ethics committee (Organism for Animal Wellbeing–OPBA) and the Italian Ministry of Health (IACUC Numbers 762/2015 and 537/2017).
1. Murine Mammary Gland Collection and Digestion
- For a typical experiment, sacrifice 8-10 weeks-old virgin female mice by CO2 inhalation. Depending on the aims of the experiment, use 5-30 mice. Place the mice on dissection boards under a hood. Use needles to stretch the forelimbs and wash down the fur of the animal with ethanol.
- Lift the skin with forceps and perform a vertical incision starting at the level of the pelvic area and moving all the way to the cervix, leaving the peritoneum intact. To avoid rupturing the skin or the peritoneum, use round-edged scissors.
- Carefully detach the skin from the body with gentle movements of the scissors across the lateral axis of the body.
- Once the skin is fully detached from the thoracic and abdominal area, perform four incisions across the four limbs of the animal and pin down the skin with needles. Use the scissors and forceps to fully detach the body of the animal from the extended skin.
- Use the forceps to gently lift the mammary fat pads that are now fully exposed and carefully detach them from the skin with the aid of the scissors. Collect the lower thoracic and abdominal mammary glands of each mouse and immerse them in Dulbecco's phosphate buffered saline (DPBS), in a 50 mL conical tube. Collect up to 20 glands per tube. Keep the tissues on ice.
NOTE: If needed, the glands can remain on ice overnight. This is a safe stopping point. The following steps should be performed under sterile conditions. - Prepare and filter the digestion medium: Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mM glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 200 U/mL collagenase and 100 µg/mL hyaluronidase (see Table of Materials).
- Transfer up to 20 glands to a 100 mm Petri dish and mince the tissue using a scalpel or curved scissors. Avoid transferring large volumes of DPBS to the dish.
- Add 10 mL of digestion medium to each Petri dish and transfer the minced tissue to a 50 mL conical tube, using a 25 mL serological pipette.
- Seal the tubes with parafilm and place them on a rotator. Set the rotator at a low speed (0.03 x g) and incubate for 2.5 h at 37 °C in a humid atmosphere containing 5% CO2.
- Visually inspect the suspension before proceeding to the next steps. If large pieces of tissue remain undigested, prolong the incubation at 37 °C for another 30 min.
NOTE: As an alternative, the digestion can be performed overnight at 37 °C, 5% CO2, using a gentle collagenase/hyaluronidase enzyme mix13.
2. Isolation of Primary Murine MECs
- Stop the rotator and remove the tubes from the incubator. Adjust a P1000 tip at the opening of a 5 mL serological pipette. If the suspension passes through the P1000 tip, proceed to the next steps. Otherwise, prolong the incubation at 37 °C for another 30 min.
- Centrifuge at 100 x g, for 5 min at 4 °C. Carefully decant the supernatant and resuspend the pellet of each tube in 3 mL of DPBS.
NOTE: The low centrifugation speed allows the removal of lymphocytes and adipose tissue cells. Gentle manipulation is required to avoid dislodging the pellet at this step. - Filter the cell suspension in each tube separately, using 100, 70 and 40 µm cell strainers. At each filtering step, wash the strainers with 2 mL of DPBS before collecting the pass-through.
NOTE: From this point on, the cell suspensions can be pooled and processed together. - Centrifuge at 300 x g, for 5 min at 4 °C. Carefully decant the supernatant and resuspend the cells in the remaining volume of DPBS.
- Proceed to the red blood cell (RBC) lysis by adding an equal volume of ammonium-chloride-potassium (ACK) lysis buffer (see Table of Materials). Mix by pipetting and incubate on ice for up to 5 min.
- Add 10 mL of DPBS and centrifuge at 300 x g, for 5 min at 4 °C. Carefully decant the supernatant and visually inspect the pellet. If the pellet is white, proceed to the next step. Otherwise repeat the RBC lysis step (step 2.5).
- Resuspend the cell pellet in 1-5 mL of mammosphere media: mammary epithelial cell growth basal medium (MEBM), supplemented with 2 mM glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 5 µg/mL insulin, 0.5 µg/mL hydrocortisone, 2% B27, 20 ng/mL epidermal growth factor (EGF), 20 ng/mL basic fibroblast growth factor (bFGF) and 0.4 IU/mL heparin (see Table of Materials).
3. Serial Mammosphere Re-plating
- For the establishment of mammosphere cultures, plate the cells onto non-tissue culture treated (ultra-low adhesion) 6-well plates at a density of 200,000 viable cells/mL in mammosphere medium. Incubate the cells for 7-10 days at 37 °C, 5% CO2.
- Prepare and filter 1% poly-HEMA solution in 95% ethanol (see Table of Materials). Coat ultra-low adhesion 6-well plates for the following passages, by adding 400 µL of 1% poly-HEMA solution per well and allowing the ethanol to dry completely. For better results, repeat the coating twice.
NOTE: The use of non-coated plates during the first passage allows the selective removal of fibroblasts from the culture. - At the end of the 7-10 days culture, collect the primary mammospheres from all the wells and centrifuge at 300 x g, for 5 min at 4 °C.
- Carefully decant the supernatant and proceed to the mechanical dissociation of the mammospheres using a P200 pipette with filtered tips. Pipette for approximately 100 times and visually inspect the suspension for the presence of large spheroids.
NOTE: A short incubation (2-5 min) of the sphere pellet in a low volume (0.2-0.5 mL) of trypsin or Accutase at 37 °C, 5% CO2, can be used to facilitate mechanical dissociation. Prepare fresh mammosphere medium (step 2.7) for the plating of each passage. - Resuspend in 1-5 mL of fresh mammosphere medium and count the viable cells. If applicable, split the cells in treatment groups or culture conditions.
- Plate 20,000 viable cells/mL on poly-HEMA-coated ultra-low adhesion 6-well plates and incubate at 37 °C, 5% CO2.
NOTE: Mammospheres reach their maximum size within 5-7 days after cell plating. When possible, distribute the cells of each sample or condition to multiple wells to obtain technical replicates. Plating density should be kept low to avoid cell aggregation. A maximum of 20,000 cells/mL is recommended for 6-well plates and 5,000 cells/mL for 24-well plates. - After 5-7 days, count the number of spheres per well. This can be done either manually, using a microscope equipped with a 10x magnification lens, or using digital image analysis (see step 4).
- Collect the mammospheres of each well separately and centrifuge at 300 x g, for 5 min at 4 °C.
- Carefully decant the supernatant and proceed to the mechanical dissociation of the mammospheres using a P200 pipette with filtered tips. Pipette for approximately 100 times and visually inspect the suspension for the presence of large spheroids.
- Resuspend in 1 mL of fresh mammosphere medium and count the viable cells. Pool the cells of each sample or condition and repeat steps 3.6-3.10. Record the number of cells plated and the number of spheres and cells counted per well for each passage. Proceed to step 5 for the cumulative growth calculation.
4. Sphere Enumeration Using Digital Image Analysis (DIA)
- At the end of each passage, scan the entire surface of all the wells and acquire images of the spheres using a digital camera mounted on a stereoscope. Save the images as .tif files.
- Import the stereoscope images as an Image Sequence using ImageJ14. Set the scale and the type of image to 8-bit.
- Duplicate the stack of images and select Subtract Background from the tab Process on the menu bar. Check the options Light background and Sliding paraboloid and click on the button OK. Process all the images of the stack.
- Select Adjust and then Threshold from the tab Image of the menu bar. By clicking on Apply, a dialogue window titled Convert Stack to Binary will appear. Select Default as method and Light as background. Check the box Calculate threshold for each image and click on the button OK.
- Select sequentially the commands Watershed, Open and Erode from the Binary list under the tab Process of the menu bar. Process all images of the stack by clicking on the button Yes of the dialogue box that appears.
NOTE: These processes allow for visual segmentation of objects that touch, object smoothing and removal of pixels from the edge of the objects. - Select the function Analyze Particles from the menu Analyze. Set the minimum size threshold at 10,000 µm2 and circularity between 0.50 and 1.00. Select the option Ellipses from the Show drop-down menu. Check Summarize, Exclude on edges and In situ Show and click on the button OK. Process all images of the stack.
- Visually inspect the correspondence of the ellipses with spheres and, if needed, correct the particle count accordingly. Sum the counts from all the frames of each well to obtain the total count of mammospheres per well.
5. Cumulative Growth Curve Calculation
NOTE: The number of mammospheres counted in each well at the end of each passage (PN) reflects the number of mammosphere-initiating cells seeded at the beginning of PN.
- For each passage (PN), register the number of plated cells and the number of cells and spheres counted per well. Calculate the sphere size at the end of each passage:
sphere size PN (cells / sphere) = cells counted PN / spheres counted PN
NOTE: The sphere size at PN is a measure of the proliferative potential of each mammosphere-initiating cell seeded at PN. - Infer the number of plated spheres by dividing the number of cells plated for PN by the sphere size calculated at the end of the previous passage (PN-1):
spheres plated PN = cells plated PN / sphere size PN-1
NOTE: By convention, the sphere size is assumed stable during the first passage of the culture (i.e., sphere size P0 = sphere size P1). If multiple wells are used as technical replicates, use the average sphere size at PN-1 as denominator. - Calculate the cumulative cell and sphere number for each well per passage:
cumulative number PN = (count PN / plated PN) X cumulative number PN-1.
NOTE: By convention, cumulative number P0 = plated P1. If multiple wells are used as technical replicates, calculate the average cumulative number per sample or condition for each passage. - Plot the data points on a semi-logarithmic scale. Display the passage number (P0 to PN) on the x axis (linear scale) and the cumulative cell or sphere number on the y axis (logarithmic scale).
- Fit an exponential trend-line to the data points and calculate the coefficient of determination (R2) to measure the goodness of the fit.
NOTE: The trend-line fitted to the data points should approximate an exponential curve, as expected for a cell population that grows or dies with a constant rate. R2 takes values between 0 and 1, with values closest to 1 indicating a better fit. - Depict the equation of the trend-line as a natural exponential function to infer the growth rate (GR) of the culture:
y = y0 e(GR)x, where y0 is the value of y when x = 0.
Representative Results
Myc overexpression in normal MECs, leads to an increased frequency of mammosphere initiating cells. This is achieved through a double mechanism: Myc increases the rate of MaSC symmetric divisions and the frequency of progenitor reprogramming into new MaSCs11. To test the effect of low constitutive Myc expression, we used the Rosa26-MycER transgenic mouse model, in which Myc activity can be induced by 4-hydroxytamoxifen (4-OHT)15. We first plated the MECs on ultra-low adhesion 6-well plates to remove fibroblasts, in the absence of 4-OHT. After the first passage, we split the culture in two: two wells were kept untreated (control) and two wells were treated with 200 nM 4-OHT (MycER). We counted the sphere and cell numbers of 5 consecutive passages for three independent experiments (Table 1). The cumulative cell and sphere numbers per passage are shown on Table 2. Induction with 4-OHT leads to increased sphere and cell growth rates, as shown in Figure 1.
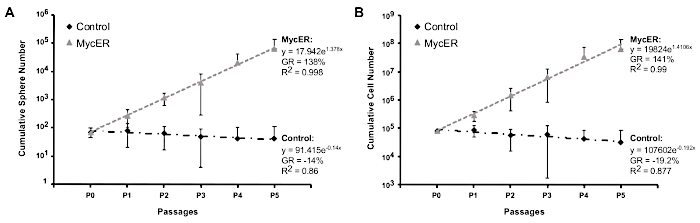
Figure 1: Representative results. Cumulative sphere (A) and cell (B) growth curves of control and MycER mammospheres. Mean and standard deviation of 3 independent experiments are shown. Please click here to view a larger version of this figure.
| 1st plating | 2nd plating | 3rd plating | 4th plating | 5th plating | ||||||||||||
| Cells Plated | Cell Count | Sphere Count | Cells Plated | Cell Count | Sphere Count | Cells Plated | Cell Count | Sphere Count | Cells Plated | Cell Count | Sphere Count | Cells Plated | Cell Count | Sphere Count | ||
| Exp1 | Control | 77,000 | 50,000 | 31 | 65,000 | 58,500 | 41 | 57,000 | 30,000 | 32 | 30,000 | 22,000 | 5 | 22,000 | 0 | 0 |
| 77,000 | 81,000 | 41 | 65,000 | 55,000 | 23 | |||||||||||
| MycER | 77,000 | 31,0000 | 193 | 80,000 | 375,000 | 323 | 80,000 | 380,000 | 217 | 80,000 | 220,000 | 223 | 80,000 | 170,000 | 155 | |
| 77,000 | 110,000 | 142 | 80,000 | 505,000 | 396 | 80,000 | 270,000 | 149 | 80,000 | 290,000 | 194 | 80,000 | 250,000 | 160 | ||
| Exp2 | Control | 75,000 | 75,000 | 71 | 60,000 | 17,000 | 34 | 28,000 | 45,000 | 29 | 45,000 | 13,000 | 2 | 13,000 | 0 | 0 |
| 75,000 | 47,000 | 45 | 60,000 | 11,000 | 47 | |||||||||||
| MycER | 75,000 | 200,000 | 188 | 80,000 | 225,000 | 277 | 80,000 | 230,000 | 155 | 80,000 | 210,000 | 211 | 80,000 | 100,000 | 95 | |
| 75,000 | 250,000 | 192 | 80,000 | 202,500 | 283 | 80,000 | 305,000 | 185 | 80,000 | 160,000 | 237 | 80,000 | 100,000 | 133 | ||
| Exp3 | Control | 82,500 | 130,000 | 121 | 80,000 | 45,000 | 105 | 80,000 | 110,000 | 86 | 80,000 | 58,500 | 75 | 58,500 | 58,500 | 78 |
| 82,500 | 125,000 | 177 | 80,000 | 71,250 | 42 | |||||||||||
| MycER | 82,500 | 325,000 | 457 | 80,000 | 610,000 | 327 | 80,000 | 367,000 | 309 | 80,000 | 500,000 | 260 | 80,000 | 115,000 | 146 | |
| 82,500 | 475,000 | 463 | 80,000 | 455,000 | 392 | 80,000 | 415,000 | 204 | 80,000 | 470,000 | 295 | 80,000 | 185,000 | 161 | ||
Table 1: Numbers of spheres counted and numbers of cells plated and counted at each passage.
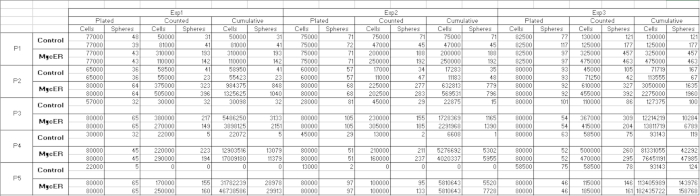
Table 2: Calculation of plated sphere numbers and cumulative sphere and cell numbers. Please click here to view a larger version of this table. (Right-click to download.)
Discussion
Here, we describe a protocol for the quantitative description of MaSC growth properties in vitro. As an example, we present the effect of low-level constitutive Myc expression on normal murine MaSCs. This approach, however, can be equally applied to various contexts. Human or murine primary cells, as well as established cell lines, can be cultured in anchorage independent conditions to establish mammosphere cultures that can be serially passaged. Gene overexpression and RNA interference can be easily introduced in the protocol with the addition of a viral transduction step at the end of the first passage (after step 3.5). Alternatively, cells can be infected in adhesion and then plated as mammospheres.
A critical aspect of the assay presented here is the seeding cell density, which should be low enough to avoid the generation of aggregates interfering with the interpretation of the results16,17. The morphology of the mammospheres can be informative to resolve this ambiguity. Only compact, round spheres should be enumerated at the end of each passage. Both the circularity of the spheroids and the size should be taken into consideration. Using the automated process of the DIA, this step is ensured with the appropriate thresholds in an objective and absolute manner. Often, progenitors will form acinar structures or smaller clusters of cells which should be excluded from the mammosphere counts. As a rule of thumb, we use a threshold of 100 µm diameter. Finally, care should be taken to avoid the transfer of intact or non-fully dissociated mammopsheres from one passage to the next. On the other hand, excess pipetting will lead to increased cell death. Thus, if such difficulties are encountered, we recommend using mild trypsinization or Accutase treatment and passing the dissociated spheres through a 40 µm strainer to ensure the generation of single-cell suspensions.
Sphere forming efficiency (SFE) has been used alternatively, as a surrogate for SC or CSC quantitation ex vivo in mammosphere cultures. SFE is indeed a measure of stem-like cells in a given cell population. However, it represents a less conscientious approach since it provides information only at distinct time points. The calculation of cumulative sphere numbers and the generation of cumulative growth curves, instead, enables the inference of the growth rate of the culture from the initial cell seeding step until the culture exhausts or, in the case of immortalized cultures, for the desired number of passages. The assessment of growth properties allows the evaluation of the deviation from the exponential growth through the coefficient R2 and, at a second step, the assessment of the GR value itself.
Importantly, cumulative mammosphere growth curves can be used to evaluate the effect of small molecule inhibitors or other chemotherapeutic drugs selectively at the CSC level6,11. Contrary to normal primary mammospheres, which functionally exhaust in 5-7 passages, tumor mammospheres tend to expand indefinitely. This feature is linked to the unlimited CSC self-renewal ability. The effects on proliferation and CSC self-renewal can be uncoupled through the generation of tumor cell and mammosphere growth curves, respectively. A CSC-specific effect is expected to result in a decrease in the cumulative mammosphere growth rate, with or without effect on the cumulative cell growth rate6,11.
Finally, another area of interest is the one of adult tissue SC reprogramming. Fully grown mammospheres consist of a phenotypically heterogeneous cell population, in which only a minor fraction retains stem-like features, including mammosphere-initiating ability and mammary gland regeneration upon transplantation in vivo6,9,11,18,19. Mammary progenitors can be thus isolated either using in vitro label-retaining assays6,9,11 or, ex vivo, using established surface markers2,3. Notably, mammary progenitors do not survive anoikis and are unable to form mammospheres. Enforced Myc expression has been shown to confer mammosphere initiation potential to mammary progenitors isolated as PKHneg11, resulting in the generation of a culture that can be passaged indefinitely. Similarly, interference of negative regulators of physiological reprogramming can be tested using the same assay. In this context, a common issue that may arise is the limited number of cell input. If the cell input is lower than 10,000 cells, we recommend seeding in 24-well plates (maximum 5,000 viable cells/mL). Nevertheless, anchorage independent culture conditions can be proven to be too harsh for scoring reprogramming, especially in cases where the reprogramming effect is not immediate. In such cases, the use of a supportive matrix and 3-dimensional organoid cultures could be more appropriate20.
Overall, the mammosphere assay is a cost-effective option that can be easily employed for scoring stem-like properties in normal and tumoral MEC populations. The quantitative approach taken in this protocol facilitates the comparisons between cultures carried out in different conditions or exposed to diverse stimuli. When followed rigorously, it provides a relatively simple ex vivo model system that permits uncoupling of the multiple players that define stem properties in vivo, offering the possibility of more detailed mechanistic studies.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Bruno Amati for the kind gift of the Rosa26-MycER transgenic mouse model. This work was funded by grants from WWCR, AIRC, ERC, and the Italian Ministry of Health to P.G.P. T.V. and X.A. were supported by FIRC and A.S. by a FUV grant.
Materials
| ACK lysis buffer | Lonza | 10-548E | Ammonium-chloride-potassium lysis bufer, 100 mL. |
| B27 | Invitrogen | 17504-044 | B27 supplement 50X (10 mL). Final concentration 2% v/v. |
| bFGF | Peprotech | 100-18B | Human recombinant fibroblast growth factor – basic, 50 μg. Stock solution 100 μg/μL in Tris 5 mM pH 7.6. Final dilution 0.02% v/v. |
| Collagenase | Sigma | C2674 | Collagenase from Clostridium histolyticum. Type I-A, lyophilized powder, 1 g. Stock 20,000 U/mL in DMEM. Final dilution 1% v/v. |
| DMEM | Lonza | 12-614F | Dulbecco's modified Eagle's medium |
| DPBS | Microgem | S17859L0615 | Dulbecco's phosphate buffered saline |
| EGF | Tebu-Bio | AF-100-15 | Recombinant human epithelial growth factor. Stock solution 100 μg/mL in sterile dH2O. Final dilution 0.02% v/v. |
| Glutamine | Lonza | 17-605E | L-Glutamine, 200 mM. Final dilution 1% v/v. |
| Heparin | PharmaTex | 34692032 | Stock concentration 5,000 IU/mL. Final dilution 0.008% v/v. |
| Hyaluronidase | Sigma | H4272 | Type IV-S, powder, 750-3,000 U/mg solid, 30 mg. Stock solution 10 mg/mL in sterile dH2O. Final dilution 1% v/v. |
| Hydrocortisone | Sigma | H0888 | Stock concentration 100 μg/mL. Final dilution 0.5% v/v. |
| Insulin | SAFCBiosciences | 91077C | Insulin, human recombinant, dry powder, 250 mg. Stock concentration 1 mg/mL. Final dilution 0.5% v/v. |
| Low attachment 6-well plates | Corning | 351146 | Sterile 6-well not treated cell culture plates with clear flat bottom and lid. |
| MEBM | Lonza | CC-3151 | Mammary epithelial cell growth basal medium |
| Penicllin-Streptomycin mixture | Lonza | 17-602F | Contains 10,000 U potassium penicillin and 10,000 µg streptomycin sulfate per mL in 0.85% saline. Final dilution 1% v/v. |
| Poly-HEMA | Sigma | P3932 | Dissolve in 95% EtOH overnight at 55 °C. Stock concentration 12% w/v. Final dilution 1% v/v in 95% EtOH. Filter (0.22 μm) before use. |
References
- Kordon, E. C., Smith, G. H. An entire functional mammary gland may comprise the progeny from a single cell. Development. 125 (10), 1921-1930 (1998).
- Shackleton, M., et al. Generation of a functional mammary gland from a single stem cell. Nature. 439 (7072), 84-88 (2006).
- Stingl, J., et al. Purification and unique properties of mammary epithelial stem cells. Nature. 439 (7079), 993-997 (2006).
- van Amerongen, R., Bowman, A. N., Nusse, R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 11 (3), 387-400 (2012).
- Van Keymeulen, A., et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 479 (7372), 189-193 (2011).
- Cicalese, A., et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 138 (6), 1083-1095 (2009).
- Dontu, G., et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & Development. 17 (10), 1253-1270 (2003).
- Grimshaw, M. J., et al. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Research. 10 (3), R52 (2008).
- Pece, S., et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 140 (1), 62-73 (2010).
- Ponti, D., et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Research. 65 (13), 5506-5511 (2005).
- Santoro, A., et al. p53 Loss in Breast Cancer Leads to Myc Activation, Increased Cell Plasticity, and Expression of a Mitotic Signature with Prognostic Value. Cell Reports. 26 (3), 624-638 (2019).
- Reynolds, B. A., Weiss, S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 255 (5052), 1707-1710 (1992).
- Gao, H., et al. Murine mammary stem/progenitor cell isolation: Different method matters?. Springerplus. 5, 140 (2016).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 9 (7), 671-675 (2012).
- Murphy, D. J., et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 14 (6), 447-457 (2008).
- Bailey, P. C., et al. Single-Cell Tracking of Breast Cancer Cells Enables Prediction of Sphere Formation from Early Cell Divisions. iScience. 8, 29-39 (2018).
- Lombardo, Y., de Giorgio, A., Coombes, C. R., Stebbing, J., Castellano, L. Mammosphere formation assay from human breast cancer tissues and cell lines. Journal of Visualized Experiments. (97), (2015).
- Peng, T., Qinghua, M., Zhenning, T., Kaifa, W., Jun, J. Long-term sphere culture cannot maintain a high ratio of cancer stem cells: a mathematical model and experiment. PLoS One. 6 (11), (2011).
- Smart, C. E., et al. In vitro analysis of breast cancer cell line tumourspheres and primary human breast epithelia mammospheres demonstrates inter- and intrasphere heterogeneity. PLoS One. 8 (6), (2013).
- Panciera, T., et al. De Novo Generation of Somatic Stem Cells by YAP/TAZ. Journal of Visualized Experiments. (135), (2018).

