Measuring Nucleotide Binding to Intact, Functional Membrane Proteins in Real Time
Summary
This protocol presents a method for measuring adenine nucleotide binding to receptors in real time in a cellular environment. Binding is measured as Förster resonance energy transfer (FRET) between trinitrophenyl nucleotide derivatives and protein labeled with a non-canonical, fluorescent amino acid.
Abstract
We have developed a method to measure binding of adenine nucleotides to intact, functional transmembrane receptors in a cellular or membrane environment. This method combines expression of proteins tagged with the fluorescent non-canonical amino acid ANAP, and FRET between ANAP and fluorescent (trinitrophenyl) nucleotide derivatives. We present examples of nucleotide binding to ANAP-tagged KATP ion channels measured in unroofed plasma membranes and excised, inside-out membrane patches under voltage clamp. The latter allows for simultaneous measurements of ligand binding and channel current, a direct readout of protein function. Data treatment and analysis are discussed extensively, along with potential pitfalls and artefacts. This method provides rich mechanistic insights into the ligand-dependent gating of KATP channels and can readily be adapted to the study of other nucleotide-regulated proteins or any receptor for which a suitable fluorescent ligand can be identified.
Introduction
Several important classes of protein are directly regulated by ligand binding. These range from soluble enzymes to membrane-embedded proteins including receptor tyrosine kinases, G protein-coupled receptors (GPCRs), and ion channels. GPCRs and channels account for ~34% and ~15% of all current drug targets, respectively1,2. Therefore, there is a considerable biochemical, as well as medical interest in developing methods that provide mechanistic insights into ligand-receptor interactions. Traditional methods for measuring ligand binding, including photoaffinity labeling and radioligand binding studies, require large amounts of partially purified protein and are typically performed under non-physiological conditions and time scales. An ideal method would require only small amounts of protein, could be performed on intact proteins expressed in a cellular or membrane environment, could be monitored in real time, and would be compatible with direct readouts of protein function.
Förster resonance energy transfer (FRET) is a method that detects the proximity between two fluorescently tagged molecules3. FRET occurs when an excited donor fluorophore transfers energy in a non-radiative fashion to an acceptor molecule (typically another fluorophore). Energy transfer results in quenching of the donor fluorescence emission and sensitization of the acceptor emission (if the acceptor is a fluorophore). Transfer efficiency is dependent on the 6th power of the distance between the donor and the acceptor. Furthermore, the donor and acceptor must be in proximity (usually less than 10 nm) for FRET to occur. As such, FRET can be exploited to measure the direct binding between a fluorescently labeled protein receptor and a fluorescent ligand.
Several different proteins are regulated or activated by binding intracellular or extracellular adenine nucleotides (ATP, ADP, AMP, cAMP). Many transporter proteins require ATP hydrolysis for their reaction cycle, including ATP-binding cassette transporters and P-type ATPases like the Na+/K+ pump4,5. ATP-sensitive K+ (KATP) channels, the cystic fibrosis transmembrane conductance regulator (CFTR), and cyclic-nucleotide regulated channels are all ion channels that are gated by the binding of intracellular adenine nucleotides, making them exquisitely sensitive to changes in cellular metabolism and signal transduction6,7,8. Purinergic P2X and P2Y receptors respond to changes in extracellular ATP, which can be released as a neurotransmitter or as a result of tissue damage9. We have developed a FRET-based assay for the measurement of adenine nucleotide binding to membrane proteins in real time. We have previously applied this method to study nucleotide binding to KATP channels10,11.
To measure nucleotide binding via FRET, a protein of interest must first be tagged with a fluorophore. The fluorescent tag must be inserted site-specifically in the protein of interest such that it is close enough to the ligand binding site for FRET to occur, with special care taken to ensure that the tag does not affect the protein’s overall structure and function. To accomplish this, we employ a technique developed by Chatterjee et al., using amber stop-codon suppression to insert a fluorescent non-canonical amino acid (l-3-(6-acetylnaphthalen-2-ylamino)-2-aminopropionic; ANAP) at the desired site12. We measure nucleotide binding as FRET between ANAP-labeled protein and fluorescent, trinitrophenyl (TNP) nucleotide derivatives (Figure 1A). The emission spectrum for ANAP overlaps with the absorbance spectrum of TNP-nucleotides, a condition necessary for FRET to occur (Figure 1B). Here we outline two different types of binding experiment. In the first, nucleotide binding to the intracellular side of ANAP-labeled KATP channels is measured in cells that have been unroofed by sonication leaving adherent fragments of plasma membrane on a glass cover slip10,11,13,14.
In the second method, nucleotide binding to ANAP-labeled KATP channels is measured in a membrane patch under voltage clamp, allowing for the simultaneous measurement of ionic currents and fluorescence. By combining these two experimental approaches, changes in binding can be directly correlated with changes in channel function11. Typical results, potential pitfalls, and data analysis are discussed.
Protocol
1. Preparation of cover slips
NOTE: These steps must take place in a sterile tissue-culture hood. Quantities are given for the preparation of 10 dishes.
- Place ten autoclaved, 30 mm borosilicate cover glass slips individually into ten 35 mm non-treated sterile dishes and rinse once with 2 mL of sterile, distilled water.
- Dilute 1 mL of 0.1% w/v poly-L-lysine solution into sterile, distilled water to a total volume of 10 mL (final concentration of 0.01% w/v). Mix well, then pipette 1 mL onto each cover slip and incubate at room temperature for 20 min.
- Aspirate the poly-L-lysine and wash each cover slip twice with at least 2 mL of sterile distilled water. Leave until completely dry i.e., at least 3 h.
2. Seeding HEK-293T cells
NOTE: These steps must take place in a tissue-culture hood. HEK-293T cells were chosen for their low current background and ease of growing in culture. This protocol may be adapted to other cell types.
- Rinse an 80-90% confluent T75 flask of HEK-293T cells once with 12 mL phosphate buffered saline (PBS) before incubating with 2 mL trypsin for 2-5 min, or until the cells are fully detached and almost completely dissociated.
- Resuspend the cells by adding 10 mL Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 µg/mL streptomycin. Pipette gently against the bottom of the flask to break up remaining clumps of cells.
- Add 2 mL of supplemented DMEM to the desired number of 35 mm dishes containing coated cover slips. Add 100 µL of resuspended cells to each dish. Incubate overnight at 37 °C.
3. Transfection
NOTE: These steps must take place in a tissue-culture hood. Quantities are given for the transfection of 10 dishes. For site-specific ANAP incorporation, the DNA codon at the position intended for labeling must be replaced with the amber (TAG) stop codon. This construct is co-transfected with two plasmids: pANAP and peRF1-E55D12,15. pANAP encodes several copies of an ANAP-specific tRNA/tRNA synthetase pair. In the presence of ANAP, transfection of this plasmid produces tRNA charged with ANAP that recognizes the amber stop codon. peRF1-E55D encodes a dominant negative ribosomal release factor that increases the yield of full-length, ANAP-tagged protein.
- Prepare a 1.5 mL tube with 10 µg pANAP, 10 µg peRF1-E55D, and DNA for the construct intended for labeling with ANAP. Bring to a final volume of 500 μL with unsupplemented DMEM.
- In a separate tube, prepare 3 µL of lipid-based transfection reagent (see Table of Materials) for each 1 µg of DNA and bring to a final volume of 500 μL with unsupplemented DMEM.
- Combine the DNA and transfection reagent mixtures in a single tube and incubate for 20 min at room temperature.
- Add 400 µL of the 1 mM ANAP stock (trifluoroacetate salt in 30 mM NaOH) to 20 mL supplemented DMEM for a final concentration of 20 µM ANAP. Replace the old media from the plated cells with 2 mL of the ANAP-containing media per dish.
- Pipette 10% of the DNA transfection mix onto each dish. Incubate at 33 °C for 2-4 days before experiments. Incubation at 33 °C slows the cell division and increases the protein yield per cell16.
4. Unroofed membrane experiments
- Use a pair of forceps to break a cover slip with transfected cells into smaller fragments.
- Follow one of the procedures below to unroof cells.
- If using pre-coated cover slips, rinse a fragment with PBS, then place it on the bottom of a 35 mm dish containing 2 mL PBS. Briefly sonicate using a probe sonicator (50 W, 20%-40% amplitude, 3 mm probe) positioned 3-5 mm above the sample to unroof cells and leave behind adherent plasma membrane fragments (Figure 2A,C).
NOTE: Sonicator power, duration, and probe height above the sample can all be varied to obtain a high yield of unroofed membranes without completely denuding the coverslip. - If not using pre-coated cover slips, rinse a cover slip fragment with PBS, then dip into a tube containing 0.1% w/v poly-L-lysine for ~30 s before briefly sonicating (as in step 4.2.1) to unroof cells and leave behind unroofed/partially unroofed plasma membrane fragments (Figure 2A,C,D). Brief exposures to poly-L-lysine have been demonstrated to improve adherence to the coverslip13.
- If using pre-coated cover slips, rinse a fragment with PBS, then place it on the bottom of a 35 mm dish containing 2 mL PBS. Briefly sonicate using a probe sonicator (50 W, 20%-40% amplitude, 3 mm probe) positioned 3-5 mm above the sample to unroof cells and leave behind adherent plasma membrane fragments (Figure 2A,C).
- Place the sonicated fragment into a cover glass-bottom 35 mm dish containing 2 mL bath solution and mount onto an inverted microscope equipped with a high NA, 60x water immersion objective. The camera port of the microscope is connected to a spectrograph in series with a high sensitivity CCD camera. Perfuse the bath chamber (0.5 – 1 mL/min) with buffer using a peristaltic pump. The composition of the buffer will vary depending on the protein under study.
NOTE: If the user does not have access to an objective with a long working distance, it may be impossible to focus on the unroofed membrane fragments due to the extra height of the cover slip. An alternative is to seed cells directly onto dishes with poly-L-lysine glass bottoms (see Table of Materials for an example). This will also reduce potential aberrations in the image associated with focusing through two pieces of glass. These aberrations do not affect the shape of the spectra acquired. - Identify unroofed membrane fragments expressing the ANAP-labeled channel by looking for channel fluorescence (Figure 2C,D).
NOTE: It is recommended to use an additional fluorescent label (where the emission spectrum is distinguishable from the ANAP emission spectrum) to help identify unroofed membranes containing the protein of interest. The experiments in Figure 2C,D were performed on ANAP-labeled channels with C-terminal fluorescent protein tags. - Partially engage the spectrometer mask (raise ~10%) between the camera port on the microscope and the spectrograph. The shadow of the mask will appear on the camera image. Align the unroofed membrane with the spectrometer mask, by adjusting the microscope stage. Acquire a bright field and fluorescence image of the unroofed membrane. These will be used to select a region of interest for analysis.
- Bring the tip of the microvolume perfusion system close to the unroofed membrane.
NOTE: To reduce the background fluorescence, the outflow of the perfusion system was replaced with a custom tip made from borosilicate glass. - To image fluorescence spectra, excite the membrane with a 385 nm LED through a 390/18 nm band-pass excitation filter and a 416 nm edge dichroic. Collect emitted light through a 400 nm long-pass emission filter (Figure 2B).
- Engage the spectrometer mask and ensure the emitted light is passed through. Engage the spectrometer gratings (300 grooves/mm). With the gratings in place, the light diffracted by the spectrometer will be projected onto the chip of the CCD camera to produce spectral images (Figure 3A). These images retain spatial information in the y dimension. The x dimension is replaced with wavelength.
- Optionally, if the protein of interest is tagged with a fluorescent protein, acquire a spectral image of the fluorescent protein using the appropriate filter set.
- Take one or more 0.1-10 s exposures at the start of the experiment while perfusing nucleotide-free buffer solution. These will be used to correct and normalize data throughout the rest of the experiment (see section 5 below).
NOTE: Choice of exposure time will depend on the expression level achieved, the brightness of the fluorophore, and the optics. Exposure time should be chosen to maximize signal and minimize the observed bleaching rate. The exposure time range given in 4.10 is suitable for equilibrium binding measurements but may be useful for measuring slower kinetic changes10. The ability to use short exposure times to track faster kinetics will be limited by protein expression levels and photobleaching, rather than hardware. - Apply a range of concentrations of TNP-ATP (usually prepared in bath solution) to establish a concentration-response curve. Perfuse each solution for at least 1 min to ensure that a steady state is reached and wash out each concentration with bath solution for at least 1 min.
NOTE: It is important to ensure that the perfusion system can rapidly reach equilibrium (Figure 2E) and achieve the correct local concentration of TNP-ATP (Figure 2F). - Take an exposure (with the same duration as used in step 4.10) at each concentration and at the end of each washout.
5. Spectral analysis
NOTE: These instructions are written for use with the analysis code “pcf.m”, which can be found at GitHub. https://github.com/mpuljung/spectra-analysis10. Additional and alternative code can be found at https://github.com/smusher/KATP_paper_201911. We have described the operations performed by the software here so that the user may create their own code or choose to analyze the data manually.
- Start the analysis program by typing the program’s name (“pcf”) in the command line.
- When an open file/folder dialog box opens with the prompt: “Select files for ROI”, select the file names associated with the brightfield and fluorescence images of the unroofed membrane. A prompt will appear in the command line to type the name of the output file.
- Type the file name and hit enter.
- When the software displays the brightfield and fluorescence images, select a region of interest (ROI) in the spectral image corresponding to the location of the unroofed membrane fragment or excised patch (see section 6) following the software prompts. Select a background region in the same spectral image (representing the same wavelength range as in the ROI) corresponding to a section of cover slip or dish with no membrane attached (Figure 3A). The software will prompt to click the Top of the ROI and press Enter, click the bottom of the ROI and press Enter and then repeat this process for the background region.
- When an open file/folder dialog box opens with the prompt: “Select File for FP Spectrum”, select the file name associated with the fluorescent protein (FP) spectrum (optional step 4.9). If no FP spectrum was acquired, select a different spectrum file. The FP spectrum serves as quality control to distinguish between tagged protein and background fluorescence.
- When an open file/folder dialog box opens with the prompt: “Select Files for Analyisis”, select all the files corresponding to the ANAP spectra (from steps 4.10 to 4.12), including the files needed for bleach correction.
- When an open file/folder dialog box opens with the prompt: “Select Files for Bleaching Collection”, select the subset of files from step 5.6 corresponding to initial spectra acquired in nucleotide-free solution at the beginning of the experiment or spectra acquired during washes in nucleotide-free solution to be used for correction (from steps 4.10 to 4.12).
- Line-average each image to produce spectra, i.e., average the intensity for all pixels in the y dimension of an ROI or background region at each wavelength. (Figure 3B). Subtract the resulting averaged background spectrum from the averaged spectrum acquired from the ROI to remove background fluorescence and fluorescence from unbound TNP-ATP (Figure 3C). These steps are performed automatically by the software.
- Determine the ANAP intensity for each exposure by averaging the intensity of a 5 nm window centered around the ANAP peak of the subtracted spectra (typically ~470 nm but may vary depending on the local microenvironment of the ANAP residue).
NOTE: Figure 3D shows 6 spectra obtained from consecutive 10 s exposures of an unroofed membrane fragment expressing ANAP-tagged channels. The inset shows the averaged intensity of the peak of each spectrum. The software will automatically find the peak wavelength in the first spectrum acquired and use this value throughout. The intensity will be calculated automatically by the software. - Normalize the ANAP intensities for each experiment by dividing the ANAP intensity of a given exposure (F) by the ANAP intensity of the first exposure in the time series, which was taken in step 4.10 (Fmax). Again, the software performs these calculations automatically.
- Perform the steps below to obtain data.
- To correct for ANAP photobleaching, first fit a single exponential decay, (F/Fmax) = A*exp(-t/τ)+(1-A), where t is the cumulative exposure time, τ is the time constant and A is the amplitude) to either the intermediate wash steps between TNP-ATP applications or to multiple initial exposures taken before washing on TNP-ATP (Figure 3D, inset).
NOTE: The software will display this fit and prompt to accept or reject it. If the fit is rejected, another opportunity will be provided to select files for bleaching correction. - Divide the normalized (in step 5.10) ANAP spectra by the predicted value of the exponential fit from step 5.11.1 at each time point (Figure 3E).
NOTE: For the example shown, the observed normalized peak fluorescence at 50 s is 0.65 and the predicted fluorescence from the exponential fit is 0.64. To correct for bleaching, divide the observed value (0.65, Figure 3E inset, empty circle) by the predicted value (0.64, Figure 3E inset, dashed line) to produce the corrected value (~1, Figure 3E inset, colored circle). If bleaching correction is adequate, the intensity of ANAP from all exposures acquired in the absence of nucleotides should be approximately equal (Figure 3E). These calculations are performed automatically by the software. - Obtain the output as an image plotting the data and a tabbed spreadsheet containing the raw spectra, subtracted spectra, spectra corrected for photobleaching, and the peak data for each file so that further analysis may be conducted.
- To correct for ANAP photobleaching, first fit a single exponential decay, (F/Fmax) = A*exp(-t/τ)+(1-A), where t is the cumulative exposure time, τ is the time constant and A is the amplitude) to either the intermediate wash steps between TNP-ATP applications or to multiple initial exposures taken before washing on TNP-ATP (Figure 3D, inset).
6. Patch-clamp fluorometry experiments
- Pull patch pipettes from thick-walled borosilicate glass capillaries to a resistance of 1.5 MΩ to 2.5 MΩ when filled with pipette solution. The composition of the pipette solution will vary depending on the protein under study.
- Transfer a cover slip with transfected cells onto a cover glass-bottom 35 mm dish containing 2 mL bath solution and mount onto an inverted microscope equipped with a high NA, 60x water immersion objective. Perfuse the bath chamber (0.5 – 1 mL/min) with bath solution using a peristaltic pump. As for the pipette solution, the bath solution will vary depending on the protein under study.
- Identify a cell expressing ANAP-labeled channels by looking for fluorescence at the cell membrane.
- Fill a patch pipette with pipette solution. Apply gentle positive pressure to the pipette and place in the bath chamber. Press the pipette against the membrane of the cell and apply gentle suction to achieve a GΩ seal (Figure 4A).
- Excise the patch by rapidly moving the pipette holder away from the cell (Figure 4A).
NOTE: Excising the patch in this fashion should form an inside-out patch, with the cytosolic domains of the protein exposed to the perfusion system. If the location of the nucleotide binding site under study is not cytosolic, it will be necessary to use outside-out patches or whole-cell recordings to perform PCF experiments. - Bring the tip of the patch pipette close to the tip of the perfusion system, and check that the patch is within the slit of the spectrometer mask (Figure 4A).
- Apply TNP-ATP and image spectra as in steps 4.10-4.12, while simultaneously recording the ionic current response to nucleotide application.
NOTE: The pipette glass may introduce spatial aberrations and reflections in the acquired images. However, these aberrations will not affect the shape of the acquired spectra and reflected excitation light is easily separated from fluorescence using either the spectrograph or a long-pass emission filter. - Analyze the spectra. Spectra imaged from excised patches can exhibit over-subtraction of unbound TNP-ATP fluorescence due to the exclusion of TNP-ATP from the glass of the patch pipette (Figure 4C-E). This over-subtraction does not affect the ANAP emission spectrum and so can be ignored.
NOTE: As the fluorescence signal in excised patches will be lower than in unroofed membranes, it is important to use an exposure time which gives high enough signal-to-noise without bleaching ANAP too rapidly.
Representative Results
Figure 2 depicts the basic experimental setup for measuring nucleotide binding to fluorescent proteins in unroofed membrane fragments obtained by sonication (Figure 2A,B). Two different approaches were used to obtain unroofed membranes, directly culturing cells on poly-L-lysine-coated cover slips or culturing cells on untreated glass and exposing them briefly to poly-L-lysine (0.1% in water) before unroofing. Figure 2C depicts a typical unroofed membrane fragment from an HEK-293T cell expressing KATP channels tagged with orange fluorescent protein (OFP). Unroofed membranes were virtually invisible in bright-field images and were identified by the fluorescence of tagged membrane proteins or by counter-staining with a membrane dye like octadecyl rhodamine B13. In addition to unroofed membranes, sonication of HEK-293T cells also produced partially unroofed cell fragments (Figure 2D)10,17. These fragments were visible in bright field. This might be the result of ruffled plasma membranes that are only poorly adherent to the cover glass. Alternatively, these fragments may contain vesicles and membranes from intracellular organelles. As such, it is preferable to acquire images only from “true” unroofed membranes, as labeled target protein associated with intracellular membranes may reflect intermediate stages of post-translational processing and assembly. Culturing cells on poly-L-lysine-coated glass is recommended as this resulted in a higher yield of “true” unroofed membranes upon sonication.
A microvolume perfusion system was applied to fluorescent nucleotides to minimize the quantities needed in a typical experiment (Figure 2B). The provided polyimide-coated glass tip was replaced with a hand-pulled borosilicate glass tip in our perfusion set up, which reduced the fluorescence background. To minimize nucleotide accumulation around the unroofed membranes being imaged, the entire bath chamber was slowly perfused with buffer. As such, we wished to measure the rate of solution change from our microvolume perfusion system and to verify that the we were able to achieve the intended ligand concentration in our region of interest, i.e., that the ligand from our perfusion system was not diluted directly into the bathing media before reaching the unroofed membrane. To control for these possibilities, the wash-in and wash-out of a 50 μM solution of tetramethylrhodamine-5-maleimide (TMRM) from our microvolume perfusion system directed at the surface of a cover glass-bottom dish perfused with water were measured (Figure 2E). Solution exchange kinetics were reproducible and well described by a single exponential decay with time constants less than 1 s for both wash-in and wash-out. Such solution exchange times limit our ability to measure kinetics of ligand binding and unbinding in our current setup. To verify that we were able to achieve the desired ligand concentration at the surface of the cover slip, we compared the fluorescence intensity of 50 μM TMRM delivered to the cover slip by our microvolume perfusion system to 50 μM TMRM in a still bath (Figure 2F). No difference in intensity was observed, verifying that appropriate ligand concentrations at the surface of the cover slip with our microvolume perfusion system can be achieved, even when the bath is perfused.
Figure 3A shows a spectral image obtained from ANAP-tagged KATP channels in an unroofed membrane from an HEK-239T cell exposed to 5 μM TNP-ATP. To obtain such images, emitted light from the unroofed membrane was directed through a spectrometer in series with a CCD camera. The emitted fluorescence was diffracted off gratings and projected onto the camera chip, producing spectra. The resulting images retain spatial information in the y dimension, but the x dimension was replaced with wavelength. The region of interest (ROI), corresponding to the unroofed membrane is outlined in orange. Two regions of high intensity are evident in the image, corresponding to the peak emission of ANAP and TNP-ATP. This was best appreciated in the wavelength-by-wavelength-averaged (over the entire ROI) spectrum shown in Figure 3B. The peak ~470 nm corresponds to ANAP incorporated into KATP; the peak ~535 nm corresponds to TNP-ATP. To correct for background fluorescence and direct excitation of TNP-ATP in solution, a background region (Figure 3A, gray) was selected from each image. The averaged background spectrum is shown in Figure 3B. The final spectrum was obtained by subtracting the averaged background spectrum from the averaged ROI spectrum (Figure 3C).
ANAP is prone to photobleaching artefacts. Figure 3D shows the reduction in peak ANAP fluorescence after multiple exposures. The peak fluorescence from several exposures in the absence of TNP-ATP (or from washes in between concentrations of TNP-ATP) were fitted to a single-exponential decay and this was used to correct for photobleaching artefacts (Figure 3E). Performing concentration-response experiments from both low-to-high and high-to-low nucleotide concentrations is recommended. If bleaching correction does not introduce any additional artefacts, the results should be comparable11.
Figure 5A shows representative spectral images from an unroofed membrane obtained from a cell expressing ANAP-tagged KATP channels in the absence and presence of TNP-ATP. The corrected spectra are shown in Figure 5B. Observing emission spectra, there was a clear separation between the donor and acceptor fluorescence emission. As some non-specific binding of TNP-ATP to naive plasma membranes from untransfected HEK-293T cells was observed, it is recommended to quantify FRET as a reduction in the donor (ANAP) fluorescence10,11. This peak was specific to the labeled receptor.
For ligands that induce a conformational change in their receptor, binding studies in isolation do not provide direct, mechanistically meaningful information about the ligand binding process18. The concentration-response relationship for ligand binding depends not only on the intrinsic binding affinity, but also the conformational change induced by ligand binding, and the inherent propensity of the receptor to change conformation in the absence of ligand. To better understand the processes underscoring ligand-receptor interactions, binding measurements can be paired with experiments that provide a readout of protein function. To this end, ion channels are an ideal model system, as their currents can be measured with sub-ms time resolution down to the single-molecule level using voltage clamp. Historically, paired current and fluorescence measurements have provided significant insights into the opening and closing (gating) of voltage- and ligand-gated ion channels19,20,21. Experiments have been conducted to simultaneously measure ionic currents and fluorescent cyclic nucleotide binding to various cyclic nucleotide-regulated channels22,23,24. These studies employed a ligand that increased its quantum yield upon binding. Fluorescence from unbound ligand in the volume of solution near the patch can be subtracted by imaging the patches using confocal microscopy22,23. In our studies, binding was measured using the reduction in ANAP fluorescence. As this signal is specific to the channel and FRET between ANAP and TNP-ATP is strongly distance dependent (half maximal at ~43 Å), contamination of our signal by non-specifically bound and unbound nucleotides was avoided.
Figure 4A shows a typical patch-clamp fluorometry (PCF) experiment. A high resistance (GΩ) seal was formed between a saline-filled borosilicate glass pipette (connected to a voltage clamp amplifier) and a cell expressing ANAP-tagged KATP. After the seal formation, the pipette was pulled away from the cell, allowing access to the intracellular nucleotide binding sites. The pipette was then positioned over the microscope objective, centered on the slit of the spectrometer mask and the outflow of the microvolume perfusion system (modified with a borosilicate glass tip) was brought close to the pipette (Figure 4D). Voltage was controlled and currents were measured from the channels in the patch. Representative currents and spectra from ANAP-tagged KATP channels are shown in Figure 4B, color coded to match the spectra to the currents. Emission spectra were corrected for background and bleaching as for unroofed membranes.
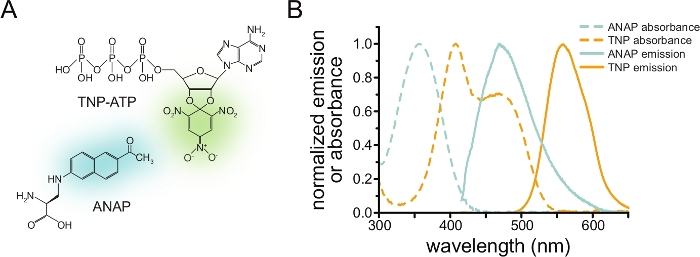
Figure 1: ANAP and TNP-ATP make a suitable FRET pair. (A) Structures of ANAP and TNP-ATP. The fluorescent moieties are highlighted. (B) Absorbance and fluorescence emission spectra of ANAP and TNP-ATP. Overlap between ANAP emission and TNP-ATP absorbance is required for FRET. Adapted from Puljung et al. (published under the Creative Commons Attribution License, https://creativecommons.org/licenses/by/4.0/)10. Please click here to view a larger version of this figure.
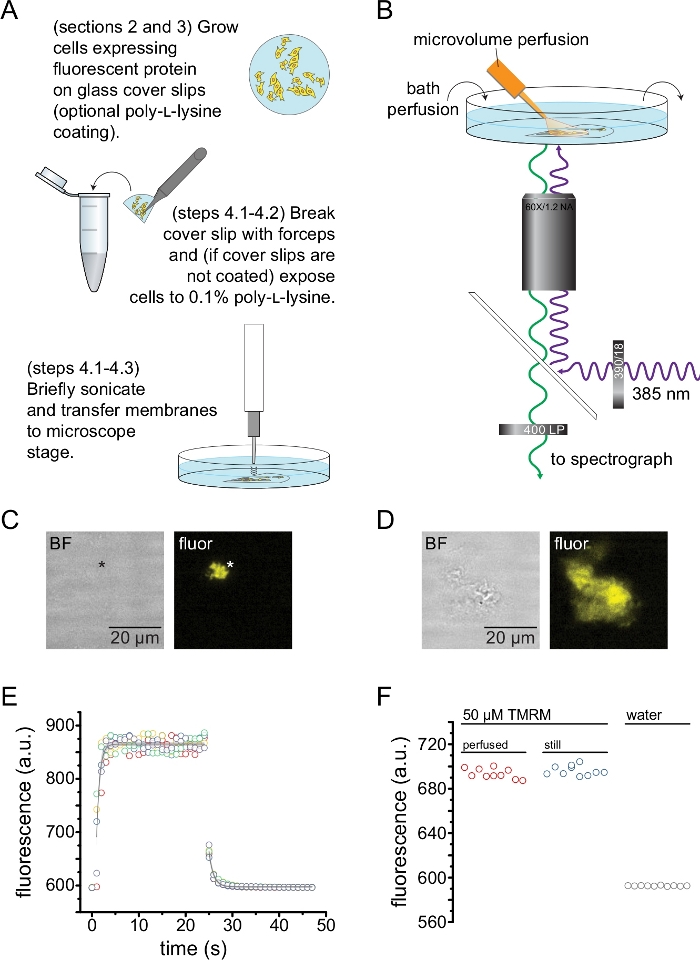
Figure 2: Measuring nucleotide binding in unroofed plasma membranes. (A) Schematic for the preparation of unroofed plasma membranes from adherent cells expressing a fluorescent membrane protein. Instructions are provided for cells grown on poly-L-lysine-coated or untreated cover slips. (B) Experimental setup for measuring nucleotide binding in unroofed membranes. (C) Bright field and fluorescent images of a completely unroofed plasma membrane derived from a cell expressing orange fluorescent protein (OFP) tagged KATP channels. The asterisk marks the position of the membrane, which is nearly invisible in the bright-field image. OFP was excited with a broad 565 nm LED through a 531/40 nm band-pass filter and 562 nm edge dichroic and emitted light was collected through a 593/40 nm band-pass filter. (D) Bright field and fluorescent images of a partially unroofed membrane fragment derived from a cell expressing orange fluorescent protein (OFP) tagged KATP channels. (E) Solution exchange time course acquired using the setup described in B. Five technical replicates are shown. The microvolume perfusion system was loaded with 50 μM tetramethylrhodamine-5-maleimide (TMRM). The bath was perfused with water at a rate of ~0.5 mL/min. Data from the wash-on (increasing fluorescence) and wash-out (decreasing fluorescence) time courses were fit with a single exponential decay of the form F = A*exp(-x/τ) + y0. The time constant (τ) for wash-in was ~0.6 s. The time constant for wash-out was ~1.0 s. TMRM was excited with a broad 565 nm LED through a 540/25 nm band-pass filter and 565 nm edge dichroic and emitted light was collected through a 605/55 nm band-pass filter. (F) Comparison of the fluorescence intensity of a 50 μM solution of TMRM applied using the microvolume perfusion system as in B and a still bath containing 50 μM TMRM. Please click here to view a larger version of this figure.
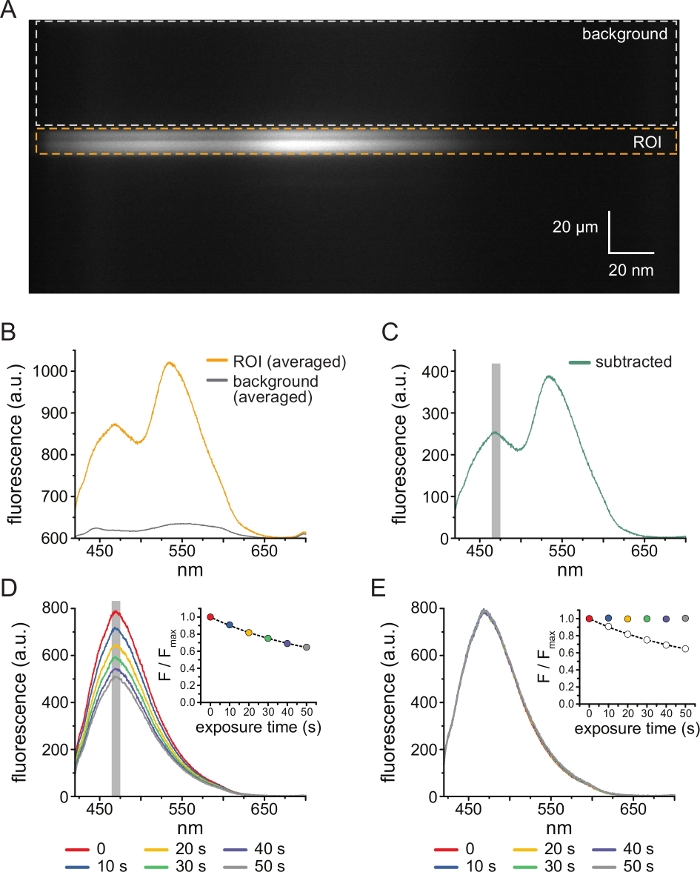
Figure 3: Background subtraction and bleaching correction. (A) Spectral image (spatial information in the y dimension, wavelength in the x dimension) of an unroofed plasma membrane from a cell expressing ANAP-labeled KATP channels. 5 μM TNP-ATP was applied using the setup described in Figure 2B. The orange box denotes the region of interest (ROI), corresponding to the unroofed membrane. The gray box denotes the background region used for correcting the spectrum. (B) Emission spectra derived from wavelength-by-wavelength averages of the ROI and background regions in A. (C) Spectrum derived by subtracting the averaged background spectrum from the averaged ROI spectrum in B. The 5 nm window around the ANAP peak used to determine the average intensity is shown as a gray shaded area. (D) Spectra acquired from six consecutive 10-s exposures of an unroofed plasma membrane from a cell expressing ANAP-labeled KATP channels. Note the decrement in fluorescence resulting from photobleaching. The inset shows the normalized peak fluorescence fit with a single exponential decay of the form F/Fmax = A*exp(-t/τ) + (1-A). The symbols in the inset are color-coded to match the spectra. (E) The same spectra as in D corrected for photobleaching. The inset shows the normalized peak fluorescence from D as open circles, with the corrected peak fluorescence shown using filled circles. Please click here to view a larger version of this figure.

Figure 4: Simultaneous measurements of nucleotide binding and channel currents using patch-clamp fluorometry (PCF). (A) Schematic showing the experimental setup for measuring nucleotide binding and ionic currents. (B) Example currents (left) and spectra (right) acquired from a membrane patch excised from a cell expressing ANAP-labeled KATP channels. Currents were recorded at a holding potential of -60 mV, digitized at 20 kHz, and filtered at 5 kHz. The gray shaded area corresponds to the wavelength range from which ANAP intensity was quantified. Adapted from Usher et al. (published under the Creative Commons Attribution License, https://creativecommons.org/licenses/by/4.0/)11. (C) Spectrum acquired from a membrane patch excised from a cell expressing ANAP-labeled KATP channels exposed to 1 mM TNP-ATP. Note the negative peak corresponding to the wavelength range over which TNP-ATP fluorescence is observed. The gray shaded area denotes the wavelength range used to quantify ANAP fluorescence as in B. Adapted from Usher et al. (published under the Creative Commons Attribution License, https://creativecommons.org/licenses/by/4.0/)11. (D) Bright field and fluorescent images of a patch pipette exposed to 1 mM TNP-ATP. The asterisk marks the tip of the pipette. (E) Spectral image of the same patch pipette in 1 mM TNP-ATP. The asterisk marks the position of the pipette. Please click here to view a larger version of this figure.

Figure 5: TNP-ATP binding to ANAP-labeled KATP channels. (A) Spectral images of an unroofed plasma membrane from a cell expressing ANAP-labeled KATP channels in the absence of TNP-ATP or in the presence of 50 μM or 1 mM TNP-ATP. Intensities are shown as a heat map. (B) Wavelength-by-wavelength-averaged spectra from the images in A showing quenching of ANAP fluorescence by TNP-ATP. The shaded areas represent two different band-pass filters that can be used to measure ANAP quenching if a spectrometer is not available. Please click here to view a larger version of this figure.
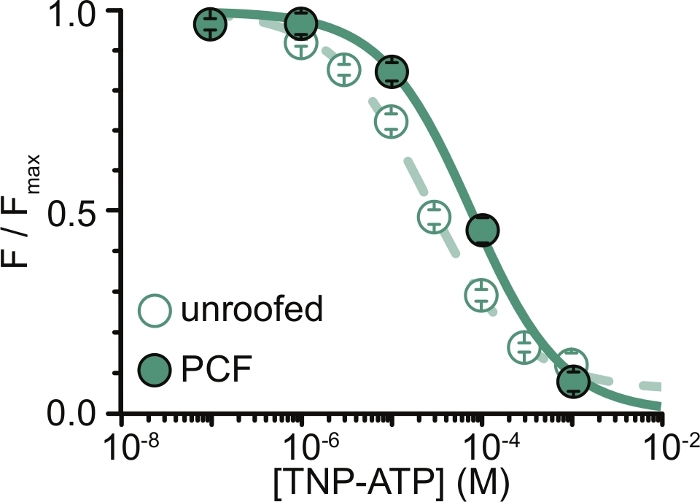
Figure 6: Quenching of ANAP-labeled KATP channels by TNP-ATP in unroofed membranes and PCF. Overlay of data from Usher et al. (published under the Creative Commons Attribution License, https://creativecommons.org/licenses/by/4.0/)11. Data were fit to the Hill equation: F / Fmax = Emax + (1 – Emax) / (1+10(EC50 – [TNP-ATP])*h). F is the measured fluorescence, Fmax is the maximal fluorescence in the absence of nucleotide, Emax is the maximal quenching at saturating nucleotide concentrations, and h is the Hill slope. EC50, (the nucleotide concentration at which quenching is half maximal) and [TNP-ATP] are log values. Unroofed membranes: EC50 = -4.59 (25.7 μM), h = 0.82, Emax = 0.93. PCF: EC50 = -4.11 (77.6 μM), h = 0.87, Emax = 1.00. Please click here to view a larger version of this figure.
Discussion
We have developed a method to measure adenine nucleotide binding in real time to intact membrane proteins. Our method builds on several other established techniques including labeling of proteins with ANAP using amber stop codon suppression12, cell unroofing14, and voltage-clamp fluorometry/PCF19,20,21,22,23,24,25. The synthesis of these approaches allows for measurement of nucleotide binding with high spatial and temporal resolution. Indeed, in our previous work, we were able to distinguish between different binding sites on the same protein complex using this approach10,11. Importantly, this technique can be directly applied to small amounts of protein in a cellular environment under conditions that preserve protein function. Using our binding method in conjunction with direct, electrophysiological readout of ion channel currents allows us to obtain rich insights into the molecular underpinnings of channel gating11.
As spectrometers are a non-standard piece of laboratory equipment, ANAP intensity can also be monitored in relative isolation using band-pass filters. Figure 5B depicts the spectral properties of two such filters. The 470/10 nm band-pass filter effectively screens out the fluorescence signal from TNP-ATP and overlaps well with the peak ANAP fluorescence. However, the peak transmittance of this filter is only around 50%, which may make it difficult to obtain good signals from dim membranes (or in excised membrane patches under voltage clamp). Another option is a 460/60 nm band-pass filter. There is slightly more overlap between the 460/60 nm filter and the foot of the TNP-ATP emission peak compared to the 470/10 nm filter. However, the 460/60 nm band-pass has a transmittance of 90-95% over a broad range of the ANAP peak, which would be expected to boost the fluorescence emission signal.
ANAP is an environmentally sensitive fluorophore12,26,27. The peak emission and quantum yield vary depending on the site of incorporation on the protein of interest and may change as the protein changes conformation. Such changes would be immediately evident from emission spectra but would not be as obvious when ANAP intensity is measured using filters. In any case, appropriate controls are required to demonstrate that the fluorescence signal does not vary because of changes in the local environment around ANAP subsequent to the nucleotide binding. Control experiments with unlabeled nucleotides can help verify that any changes in ANAP intensity are the result of FRET between ANAP and TNP-nucleotides. TNP-nucleotides can bind non-specifically to the membranes derived from untransfected cells (either to the plasma membrane or to native membrane proteins)10. We quantify binding as a decrement in the donor fluorescence, as this signal is specific to the labeled channel. However, we recommend performing additional control experiments for each agonist/receptor pair, for example mutating the nucleotide binding site if known, to verify that the change in donor fluorescence is really the result of direct binding to the labeled receptor11. Finally, working with constructs that contain a fluorescent protein tag in addition to the ANAP label is recommended. This helps to differentiate labeled receptor fluorescence from background/autofluorescence. Background fluorescence can be distinguished from ANAP by the peak and shape of emission spectra10, but such determinations can be very difficult when only filter sets are used. Additionally, cells and unroofed membranes expressing fluorescent receptors can be identified using the fluorescent protein tag without having to excite ANAP and risk excessive photobleaching.
In many of our PCF records, we observed a strong negative peak in our spectra at high TNP-ATP concentrations (Figure 4C). This negative peak is an artefact of our background subtraction protocol. Figure 4D shows bright-field and fluorescent images of a patch pipette exposed to 1 mM TNP-ATP. A shadow at the pipette tip is evident, resulting from exclusion of TNP-ATP from the volume of the pipette walls, which is most evident within the plane of focus. The spectral image in Figure 4E shows a dark band, corresponding to this shadow. When a region above or below this dark band is used for background subtraction, it produces a negative peak. Importantly, this peak occurred over a wavelength range corresponding to TNP-ATP emission and did not affect our measurements of ANAP quenching.
The major limitation of our experiments was in obtaining adequate plasma membrane expression of ANAP-tagged constructs to measure fluorescence. It was generally easier to acquire high quality spectra from unroofed membranes than in PCF, due to their larger size and our ability to quickly scan an entire dish of unroofed membranes, unlike in PCF where patches can only be obtained one at a time. In our experiments, the data from unroofed membranes and PCF experiments were similar but not equivalent (Figure 6)11. However, there is no a priori reason why this should be a universal observation as proteins in a patch pipette may be in a different functional state than those in unroofed membranes.
Here, attempts have been made to maximize expression of our ANAP-tagged constructs, in particular lowering the cell culture temperature to 33 °C10,11,16. In our experience, attempting to identify sites in the protein at which ANAP would be a conservative substitution did not consistently result in constructs that expressed well. We had more success systematically scanning entire protein regions for ANAP incorporation sites and screening candidates for surface expression10. The ANAP labeling system also works in Xenopus laevis oocytes, which allows for much larger membrane patches to be excised, thus increasing signal to noise26,27,28.
Whereas larger levels of expression are expected to result in brighter signals, the minimum number of channels required to measure fluorescence depends on several factors, including the brightness of the fluorophore, the degree of photobleaching, the intensity of the excitation light, and the plane of focus. In theory, estimates could be made by correlating the fluorescence intensity and the channel current as has been shown previously28,29. However, the reliability of such estimates requires some knowledge of the single-channel conductance and the channel’s open probability. In addition to the factors listed above, the fluorescence signal will also be affected by channels associated with vesicles or sections of the plasma membrane stuck to the pipette glass that are not under voltage clamp.
This method is readily adapted to the study of other nucleotide-sensitive ion channels. CFTR is structurally similar to the accessory sulfonylurea receptor subunit of KATP30,31. Like KATP CFTR gating is controlled by nucleotide binding, making it an obvious future target of our method7. Purinergic P2X receptors are ion channels gated by extracellular ATP9. TNP-ATP acts as an antagonist for P2X receptors32,33. Therefore, it will not be useful for studying P2X activation, although it may be used in competition assays with P2X agonists. Alternatively, other fluorescent ATP derivatives with sufficient spectral overlap with ANAP emission may be used to study activation. Alexa-647-ATP is a fluorescent P2X agonist34. The calculated R0 between Alexa-647 and ANAP is ~85 Å, which means that direct binding to P2X should result in substantial quenching of ANAP incorporated into the channel. However, such a long R0 will also result in quenching from Alexa-647-ATP bound to neighboring subunits and increases the likelihood that non-specific nucleotide binding will result in FRET. As the ligand binding site in P2X receptors is extracellular, binding measurements would be performed on intact cells, in whole-cell voltage clamp, or in outside-out membrane patches. Our method can also be extended to study binding and activation of electrogenic and non-electrogenic transporters and pumps that depend on ATP for their reaction cycle as well as G protein coupled P2Y receptors. Finally, even though we have developed this method to measure adenine nucleotide binding (TNP-ATP, TNP-ADP, TNP-AMP), the same approach can be used to study binding to virtually any receptor for which a suitable, fluorescent ligand has been identified.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We wish to thank Raul Terron Exposito for excellent technical assistance. This work was funded by the Biotechnology and Biological Sciences Research Council (BB/R002517/1; MCP and FMA) and the Wellcome Trust (203731/Z/16/A; SGU)
Materials
| T75 tissue-culture treated flask | StarLab | CC7682-4875 | |
| 0.1% w/v poly-L-lysine | Sigma-Aldrich | P8920 | |
| 30 mm borosilicate cover glass slips | VWR | 631-0174 | |
| 35 mm non-treated sterile dishes | CytoOne | CC7672-3340 | |
| 35 mm cover glass bottom dish | WPI | FD35-PDL-100 | |
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | 31966021 | |
| Foetal bovine serum (FBS) | Gibco | 10500-064 | |
| Penicillin/Streptomycin | Gibco | 15140-122 | |
| TrypLE select (tryosin) | Gibco | 12563-011 | Trypsin/EDTA reagent |
| Phosphate buffered saline (PBS) | Gibco | 14040-091 | |
| UltraPure distilled water | Invitrogen | 10977-035 | |
| HEK293T cells | ATTC | CRL-3216 | Used between passages 5-30 |
| ANAP-TFA | AsisChem | ASIS-0014 | Reconstituted in 30 mM NaOH to a final concentration of 1 mM |
| pANAP expression plasmid | Addgene | Plasmid #48696 | Encodes tRNA/tRNA synthetase pair for expression of ANAP-tagged protein |
| peRF1-E55D | Chin Lab (MRC Laboratory of Molecular Biology, Cambridge, UK) | Jason Chin: DOI: 10.1021/ja5069728 | Encodes dominant-negative eukaryotic ribosomal release factor |
| TransIT-LT1 | Mirus Bio | MIR 2300 | Lipopolyplex transfection reagent |
| Thick-walled borosilicate glass capillaries | Harvard Apparatus | GC150F-15 | |
| Tetramethylrhodamine-5-maleimide | Sigma-Aldrich | 94506 | |
| TNP-ATP | Jena Bioscience | NU-221L | Delivered at 10 mM in water |
| Nikon Eclipse TE2000-U inverted microscope microscope | Nikon | ||
| 60x water immersion objective (1.4 NA) | Nikon | MRD07602 | |
| 4-Wavelength High-Power LED Head | ThorLabs | LED4D245 | 385/490/565/625 nm LEDs |
| Four-Channel LED Driver | ThorLabs | DC4100 | |
| 390/18 nm band-pass excitation filter | ThorLabs | MF390-18 | For ANAP excitation |
| 400 nm long-pass emission filter | ThorLabs | FEL0400 | For imaging ANAP spectra |
| 416 nm edge dichroic | ThorLabs | MD416 | For imaging ANAP spectra |
| 460/60 nm band-pass emission filter | ThorLabs | MF460-60 | Suggested wide band-pass filter for imaging ANAP fluorescence (Figure 4B) |
| 470/10 nm band-pass emission filter | ThorLabs | FB470-10 | Suggested narrow band-pass filter for imaging ANAP fluorescence (Figure 4B) |
| 531/40 band-pass excitation filter | Brightline | FF01-531/40-25 | For orange fluorescent protein (OFP) excitation |
| 540/25 nm band-pass excitation filter | Chroma | D540/25X | For tetramethylrhodamine-5-maleimide (TMRM) excitation |
| 562 nm edge dichroic | Semrock | FF562-Di03 | For imaging OFP fluorescence |
| 565 nm edge dichroic | Chroma | 565DC | For imaging TMRM fluorescence |
| 593/40 nm band-pass excitation filter | Brightline | FF01-387/11-25 | For imaging OFP fluorescence |
| 605/55 nm band-pass emission filter | Chroma | D605/55M | For imaging TMRM fluorescence |
| IsoPlane-160 Imaging Spectrometer | Princeton Instruments | IsoPlane-160 | |
| PIXIS 400BR_eXcelon Camera | Princeton Instruments | PIXIS: 400BR_eXcelon | |
| Axopatch 200B amplifier | Molecular Devices | Axopatch 200B-2 | |
| Digidata 1440A digitizer | Molecular Devices | Digidata 1440A | |
| Probe sonicator | Sonics & Materials | VC-50 | For unroofing |
| REGLO digital peristaltic pump | Ismatec | ISM 832 | For bath perfusion |
| Microvolume perfusion system | ALA Scientific Instruments | ALA μFlow-8 | For TNP-ATP perfusion |
| pClamp 10.6.2 | Molecular Devices | Recording and analysing currents | |
| Lightfield 5.20.1507 | Princeton Instruments | Acquisition software for images and spectra | |
| Matlab | Mathworks | For data analysis | |
| Python 3.8.1 | Python Software Foundation | For data analysis |
References
- Garcia, M. L., Kaczorowski, G. J. Ion channels find a pathway for therapeutic success. Proceedings of the National Academy of Sciences of the United States of America. 113 (20), 5472-5474 (2016).
- Hauser, A. S., Attwood, M. M., Rask-Andersen, M., Schioth, H. B., Gloriam, D. E. Trends in GPCR drug discovery: new agents, targets and indications. Nature Reviews Drug Discovery. 16 (12), 829-842 (2017).
- Lakowicz, J. R. . Principles of fluorescence spectroscopy. 3rd edn. , (2006).
- Higgins, C. F., Linton, K. J. The ATP switch model for ABC transporters. Nature Structural & Molecular Biology. 11 (10), 918-926 (2004).
- Toyoshima, C., Cornelius, F. New crystal structures of PII-type ATPases: excitement continues. Current Opinion in Structural Biology. 23 (4), 507-514 (2013).
- Craven, K. B., Zagotta, W. N. CNG and HCN channels: two peas, one pod. Annual Review of Physiology. 68, 375-401 (2006).
- Csanady, L., Vergani, P., Gadsby, D. C. Strict coupling between CFTR’s catalytic cycle and gating of its Cl- ion pore revealed by distributions of open channel burst durations. Proceedings of the National Academy of Sciences of the United States of America. 107 (3), 1241-1246 (2010).
- Vedovato, N., Ashcroft, F. M., Puljung, M. C. The Nucleotide-Binding Sites of SUR1: A Mechanistic Model. Biophysical Journal. 109 (12), 2452-2460 (2015).
- Burnstock, G. Introduction to the Special Issue on Purinergic Receptors. Advances in Experimental Medicine and Biology. 1051, 1-6 (2017).
- Puljung, M., Vedovato, N., Usher, S., Ashcroft, F. Activation mechanism of ATP-sensitive K(+) channels explored with real-time nucleotide binding. Elife. 8, 41103 (2019).
- Usher, S. G., Ashcroft, F. M., Puljung, M. C. Nucleotide inhibition of the pancreatic ATP-sensitive K+ channel explored with patch-clamp fluorometry. Elife. 9, 52775 (2020).
- Chatterjee, A., Guo, J., Lee, H. S., Schultz, P. G. A genetically encoded fluorescent probe in mammalian cells. Journal of the American Chemical Society. 135 (34), 12540-12543 (2013).
- Gordon, S. E., Senning, E. N., Aman, T. K., Zagotta, W. N. Transition metal ion FRET to measure short-range distances at the intracellular surface of the plasma membrane. Journal of General Physiology. 147 (2), 189-200 (2016).
- Heuser, J. The production of ‘cell cortices’ for light and electron microscopy. Traffic. 1 (7), 545-552 (2000).
- Schmied, W. H., Elsasser, S. J., Uttamapinant, C., Chin, J. W. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. Journal of the American Chemical Society. 136 (44), 15577-15583 (2014).
- Lin, C. Y., et al. Enhancing Protein Expression in HEK-293 Cells by Lowering Culture Temperature. PloS One. 10 (4), 0123562 (2015).
- Usukura, J., et al. Use of the unroofing technique for atomic force microscopic imaging of the intra-cellular cytoskeleton under aqueous conditions. Journal of Electron Microscopy. 61 (5), 321-326 (2012).
- Colquhoun, D. Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. British Journal of Pharmacology. 125 (5), 924-947 (1998).
- Mannuzzu, L. M., Moronne, M. M., Isacoff, E. Y. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 271 (5246), 213-216 (1996).
- Zheng, J., Zagotta, W. N. Gating rearrangements in cyclic nucleotide-gated channels revealed by patch-clamp fluorometry. Neuron. 28 (2), 369-374 (2000).
- Zheng, J., Zagotta, W. N. Patch-clamp fluorometry recording of conformational rearrangements of ion channels. Science’s STKE. 2003 (176), 7 (2003).
- Biskup, C., et al. Relating ligand binding to activation gating in CNGA2 channels. Nature. 446 (7134), 440-443 (2007).
- Kusch, J., et al. Interdependence of receptor activation and ligand binding in HCN2 pacemaker channels. Neuron. 67 (1), 75-85 (2010).
- Wu, S., et al. State-dependent cAMP binding to functioning HCN channels studied by patch-clamp fluorometry. Biophysical Journal. 100 (5), 1226-1232 (2011).
- Cha, A., Bezanilla, F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 19 (5), 1127-1140 (1997).
- Kalstrup, T., Blunck, R. Dynamics of internal pore opening in K(V) channels probed by a fluorescent unnatural amino acid. Proceedings of the National Academy of Sciences of the United States of America. 110 (20), 8272-8277 (2013).
- Kalstrup, T., Blunck, R. S4-S5 linker movement during activation and inactivation in voltage-gated K(+) channels. Proceedings of the National Academy of Sciences of the United States of America. 115 (29), 6751-6759 (2018).
- Dai, G., Aman, T. K., DiMaio, F., Zagotta, W. N. The HCN channel voltage sensor undergoes a large downward motion during hyperpolarization. Nature Structural & Molecular Biology. 26 (8), 686-694 (2019).
- Liu, C., et al. Patch-clamp fluorometry-based channel counting to determine HCN channel conductance. Journal of General Physiology. 148 (1), 65-76 (2016).
- Hwang, T. C., et al. Structural mechanisms of CFTR function and dysfunction. Journal of General Physiology. 150 (4), 539-570 (2018).
- Puljung, M. C. Cryo-electron microscopy structures and progress toward a dynamic understanding of KATP channels. Journal of General Physiology. 150 (5), 653-669 (2018).
- Kasuya, G., et al. Structural insights into the competitive inhibition of the ATP-gated P2X receptor channel. Nature Communications. 8 (1), 876 (2017).
- Virginio, C., Robertson, G., Surprenant, A., North, R. A. Trinitrophenyl-substituted nucleotides are potent antagonists selective for P2X1, P2X3, and heteromeric P2X2/3 receptors. Molecular Pharmacology. 53 (6), 969-973 (1998).
- Bhargava, Y., Nicke, A., Rettinger, J. Validation of Alexa-647-ATP as a powerful tool to study P2X receptor ligand binding and desensitization. Biochemical and Biophysical Research Communications. 438 (2), 295-300 (2013).

