Quantification of Interbacterial Competition using Single-Cell Fluorescence Imaging
Summary
This manuscript describes a method for using single-cell fluorescence microscopy to visualize and quantify bacterial competition in coculture.
Abstract
Interbacterial competition can directly impact the structure and function of microbiomes. This work describes a fluorescence microscopy approach that can be used to visualize and quantify competitive interactions between different bacterial strains at the single-cell level. The protocol described here provides methods for advanced approaches in slide preparation on both upright and inverted epifluorescence microscopes, live-cell and time-lapse imaging techniques, and quantitative image analysis using the open-source software FIJI. The approach in this manuscript outlines the quantification of competitive interactions between symbiotic Vibrio fischeri populations by measuring the change in area over time for two coincubated strains that are expressing different fluorescent proteins from stable plasmids. Alternative methods are described for optimizing this protocol in bacterial model systems that require different growth conditions. Although the assay described here uses conditions optimized for V. fischeri, this approach is highly reproducible and can easily be adapted to study competition among culturable isolates from diverse microbiomes.
Introduction
This article outlines a method for quantifying bacterial competition at the single-cell level using fluorescence microscopy. The structure and function of microbial communities is often shaped by competitive interactions among microbes, and in many cases characterizing these interactions requires observing different bacterial strains in coincubation1,2,3,4,5,6,7,8. Traditionally, bacterial competition is quantified at the population level by counting colony forming units (CFUs) of inhibitor and target strains before and after a coincubation period2,9. Mechanisms for microbial competition are broadly distributed among bacteria and may rely on either diffusion or cell-cell contact to inhibit target cells10,11,12,13,14,15,16,17,18,19.
Although bacterial strains are often observed in coincubation at the population level, this manuscript outlines an assay for single-cell quantification of bacterial competition. Further, this work includes suggestions for adapting the protocol for the use with other bacterial species. While the specific techniques in this article are used to study contact-dependent intraspecific competition between strains of the symbiotic bacterium Vibrio fischeri20,21,22, they can be adapted for competition between many organisms. This article provides instructions for slide setup on both upright and inverted microscopes, and all analysis is described using the open-source software FIJI23 so that the method can be used by researchers with access to different imaging setups and analysis programs. Given the importance of studying microbial competition at both the population and single-cell level, this method will be a valuable resource for researchers to quantify competitive interactions, particularly those that do not have access to proprietary analysis software.
Protocol
1. Optimization of bacterial strains
- Choose two bacterial strains for single-cell bacterial competition assays. Here, two strains of V. fischeri are used: a target strain (ES11424) and an inhibitor strain (MJ1125) that is known to kill the target strain using the type VI secretion system on chromosome II (T6SS2)1, which is a contact-dependent killing mechanism.
- Determine the appropriate controls for the experiment. In this example, the appropriate control is to incubate both the wild-type and the T6SS mutant inhibitor strains with the target strain to quantify the effect of T6SS-mediated killing.
NOTE: Additional controls can include a target strain that expresses the necessary immunity gene(s) to prevent T6SS-dependent killing or an inhibitor mutant strain expressing wild-type copies of the mutated genes in trans to restore T6SS activity1. - When possible, transform strains with stable plasmids encoding genes for different fluorescent proteins (e.g., GFP or RFP) to visually distinguish strain types on the microscope. Here, the inhibitor strain is tagged with a GFP-encoding plasmid (pVSV102), and the target strain is tagged with a dsRed-encoding plasmid (pVSV208)26.
NOTE: If it is not possible to use stable plasmids, fluorescent tags can be introduced onto the bacterial chromosome for visualization27,28. - During the initial optimization period, image clonal cultures of the tagged strains under each of the fluorescent filters that will be used during the experiment to ensure that cells are only visible in the intended channel. For example, ensure that a GFP-tagged strain is only visible in the FITC channel.
2. Agarose pad preparation
- Prepare agarose pad solution by dissolving 2% low-melt agarose (w/v) into mPBS. Heat the solution briefly in the microwave and vortex until the agarose is completely dissolved. Keep this solution warm by placing it in a 55°C water bath until ready to use. See the Discussion section for more information about preparing agarose pads.
NOTE: Here, mPBS was prepared by adding 20 g/L NaCl to standard 1x PBS. - Wrap a piece of lab tape around a glass slide five times. Repeat this process a second time on the same slide so that the distance between the two pieces of tape is slightly smaller than the width of a coverslip (Figure 1A). For example, if using 25 mm2 coverslips, the pieces of tape should be spaced approximately 20 mm apart.
NOTE: While the number of times the tape is wrapped around the slide can be modified to adjust the thickness of the agarose pad, it is important that the layers of tape are the same height on both sides of the slide so that the agarose pad remains flat. - Pipette warm agarose solution between the two pieces of tape and immediately top with a coverslip so that it rests on the pieces of tape. This will ensure that the surface of the agarose pad remains flat. The volume of agarose solution pipetted in this step should be enough that the coverslip makes contact with the liquid and pushes out any bubbles in the agarose solution. For this particular setup, 200 µL of warm agarose is sufficient.
- Let the agarose pad solidify at room temperature for at least 1 h prior to the coincubation assay. Step 2.2 will produce an agarose pad of approximately 20 mm2.
- Cut this agarose pad with a razor blade into four, 5 mm2 pads to be used for imaging.
NOTE: Agarose pads can be made up to one week prior to the experiment and stored at 4°C in an empty, sterile Petri plate sealed with parafilm to prevent drying.
3. Prepare strains for co-incubation
- Streak out each strain to be used in the coincubation assay from -80 °C stocks onto LBS agar plates supplemented with the appropriate antibiotics and incubate overnight at 24°C. For this example, three strains are used: the wild-type inhibitor strain, the type VI secretion system mutant, and the target strain.
- The next day, start overnight cultures in biological duplicate by picking two colonies from each strain and resuspending them in LBS medium supplemented with the appropriate antibiotics and incubate overnight at 24°C with shaking at 200 rpm.
- On the following morning, subculture each biological replicate 1:1000 into fresh LBS medium without antibiotics and incubate at 24°C with shaking for 4-5 h or until cells reach an OD600 of ~1.5.
NOTE: The timing of steps 3.1, 3.2, and 3.3 may need to be optimized for different bacterial species as their growth rate may vary substantially. For this assay, cells were aimed to be in mid-log phase at the start of the coincubation assay.
4. Coincubate bacterial strains
- Starting with mid-log cultures from step 3.3, measure and record the optical density at 600 nm (OD600) for all samples.
- Normalize each sample to an OD600 = 1.0, which corresponds to approximately 109 CFU/mL for V. fischeri, by diluting the culture with LBS medium.
- Mix the two competing strains together at a 1:1 ratio based on volume by adding 30 µL of each normalized strain to a labeled 1.5 mL tube. Vortex the mixed-strain culture for 1-2 s.
NOTE: In some cases, it may be appropriate to mix cocultures in different ratios. For example, when one strain grows much faster than the other, it may be necessary to start the slower growing strain at a numerical advantage in order to observe the competition. Optimization may also be required if OD600 does not correspond to similar CFU/mL for both strains. - Repeat step 4.3 for each biological replicate and treatment. In the example shown here, this will result in a total of four mixed-strain tubes: two biological replicates with the wild-type inhibitor strain mixed with the target strain and two biological replicates with the type VI secretion system mutant strain mixed with the target strain.
- To ensure competing cells are sufficiently dense for contact-dependent killing in the coincubation on the agar pad, concentrate each mixed culture 3-fold by centrifuging the mixed culture in a standard 1.5 mL centrifuge tube for 1 min at 21,130 x g, discarding the supernatant, and resuspending each pellet in 20 µL LBS medium. Repeat for each sample.
NOTE: Some bacterial cells are sensitive to damage by centrifugation at high rcf; in such cases the mixed culture can be centrifuged for 3 min at 4600 x g29. Additionally, when quantifying contact-dependent competition, it is important to ensure sufficient cell density on the slide to observe killing. In this article, "crowded" treatments, where killing is observed, had approximately 10 cells/20 µm2; see the Discussion section for more information.
5. Slide setup
- When using an upright microscope, place a ~5 mm2 agarose pad onto a standard 1 mm glass slide. Spot 2 µL of a mixed culture onto the agarose pad and place a #1.5 coverslip (25 mm2) over the spot. See Figure 1B for an example.
- When using an inverted microscope, spot 2 µL of a mixed culture onto the #1.5 coverslip bottom of a 35 mm Petri dish and place a ~5 mm2 agarose pad over the coincubation spot. Place a 12 mm circular glass coverslip over the agarose pad. See Figure 1C for an example.
- Repeat step 5.1 or 5.2, depending on the microscope setup used, for the remaining three mixed cultures, resulting in four slides or dishes to be imaged.
- Allow slides to sit on the benchtop for approximately 5 min before proceeding to step 6. This allows cells to settle on the agar pad and eliminates movement during the imaging process.
6. Fluorescence microscopy
- Begin by focusing on cells using white light (phase contrast or DIC) to minimize the effects of photo-bleaching. Based on the average size of a single bacterial cell, use a 60x or 100x oil objective.
- Adjust the exposure time and acquisition settings for each channel so that cells are visible in the appropriate channel with minimal background detection.
NOTE: It is appropriate to use different exposure times for different channels, but the same exposure time should be used across all biological replicates and treatments for a given channel. - For each sample, select at least five fields of view (FOV) and acquire images in each appropriate channel using the acquisition settings from step 6.2 (See examples in Figure 2). Save the XY points from each FOV so that the same FOV can be imaged during the final time point. Imaging the same FOV at each time point is necessary to determine the proportion of area occupied by target or inhibitor cells during the analysis steps.
NOTE: In this example, the fluorescence of GFP is detected using a filter with an excitation wavelength of 467 – 498 nm and an emission filter of 513 – 556 nm and is false-colored green. Fluorescence of dsRed is detected using a filter with an excitation wavelength of 542 – 582 nm and an emission filter of 603 – 678 nm and is false-colored magenta. - After 2 h, repeat step 6.3 for each sample using the previously saved XY points (Fig 2).
NOTE: The timing of subsequent images may need to be optimized for organisms with different growth rates or competitive mechanisms.
7. Image analysis in FIJI
- Download and install the FIJI image processing software using the instructions found here: https://imagej.net/Fiji/Downloads
- Open FIJI and import image files for analysis.
NOTE: In most cases .TIFF files will be used for image analysis, although some image acquisition software will export using proprietary file types. FIJI can recognize most proprietary file types and images can be imported and analyzed as follows. - For each image acquired in steps 6.3 and 6.4, convert the image to grayscale, separate the channels, and begin by thresholding (Ctrl + Shift + T) and creating a binary mask of the preprocessed image (Figure 3A,B).
NOTE: Here, the default thresholding settings in FIJI are used. In some cases, it may be necessary to change those settings, in which case the same settings should be used for all images in that experiment. - Set scale on the image (Analyze | Set Scale) using the appropriate values for the microscopy setup23.
- Set measurements (Analyze | Set Measurements) and select Area.
NOTE: Other measurements can be added if they are appropriate for the experiment. Only the object Area measurement is required for the example analysis shown here. - Analyze particles (Analyze | Analyze Particles) using the default settings (Figure 3C). If there are debris in the sample, it may be necessary to adjust the size or circularity to filter out non-cell particles. Select Show | Outlines so that the output of this analysis will include a numbered outline of all particles analyzed (Figure 3D).
NOTE: Comparing the outline in Figure 3D to the initial image is especially important in the optimization step to ensure that (1) all cells are being analyzed, and (2) that any debris is excluded from the analysis. - Export the measurements from step 7.4 (Figure 3E) into a spreadsheet software for further analysis and graphing.
- Repeat steps 7.1 – 7.5 for all channels and images acquired during the experiment.
8. Calculating the percent of initial target area over time
- For each field of view analyzed in section 7, ensure that the exported file contain an individual area measurement for each particle that was analyzed. Beginning with the target strain's fluorescence channel, calculate the sum particle area for each individual field of view. For two biological replicates with five FOV each, this should result in ten sum areas per treatment at each time point.
- Calculate the percent of initial target area over time for each FOV using the following equation: (
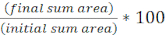 )
) - Repeat this calculation for all treatments and graph the percent of initial target area (result of the equation from step 8.2) for each treatment (Figure 4A).
- Determine whether there is a net increase in the target population (indicating growth), a net decrease in the target population (indicating death), or no change (indicating no growth or death) for each treatment.
NOTE: Percent of initial target area with values greater than 100 indicate net target growth, and values lower than 100 indicate net target death. Percent of initial target values that remain at 100 indicate no net change in target population. See discussion for suggested follow-up experiments.
9. Calculating the percent of initial inhibitor area over time
- Repeat steps 8.1 to 8.3, this time using the measurements collected from the inhibitor strain's fluorescence channel in section 7 (Figure 4B).
- Determine whether there was a net increase in inhibitor population (growth); a net decrease in inhibitor population (death), or no change for each treatment. Values greater than 100 indicate net inhibitor growth, and values lower than 100 indicate net inhibitor death.
Representative Results
To visualize and quantify competitive interactions between bacteria at the single-cell level, a protocol was developed and optimized for V. fischeri by modifying our well-established CFU-based assay1,2. This method utilizes GFP- and dsRed-encoding stable plasmids to visually distinguish different strains of V. fischeri. The competitive outcome of these interactions can be quantified by analyzing the images acquired from this assay using the open-source software FIJI. As an example, the following experiment was performed using V. fischeri isolates. An inhibitor strain harbored a plasmid that encodes GFP, and a target strain harbored a plasmid that encodes dsRed. Given that the T6SS2 encoded by the inhibitor is a contact-dependent killing mechanism, treatments were included where cells were either crowded (high cell-cell contact) or disperse (low cell-cell contact) on a slide to highlight the impact of experimental setup on the final results of this assay. In the sample data, competing strains were mixed at a 1:1 ratio and incubated on an agarose pad for 2 h, and both initial and final (2 h) images were taken. As a control, a T6SS2 mutant strain was also coincubated with the target strain in both crowded and disperse conditions. Cultures of each strain were prepared and coincubated as described above and slides were prepared as shown in Figure 1.
Figure 2 shows representative fluorescence microscopy images of each experimental treatment with the same field of view imaged at an initial and final time point. For each treatment, either a wild-type inhibitor or T6SS mutant strain harboring a GFP-encoding plasmid was mixed at a 1:1 ratio with the target strain harboring a dsRed-encoding plasmid. During a 2 h coincubation period with this experimental setup, growing V. fischeri cells may go through 1-2 divisions (Figure 2; gray arrows). In Figure 2A, cell-cell contact was forced between the target and inhibitor by concentrating the mixed culture before spotting onto the slide. Multiple target cells are observed to become rounded and/or disappear over the course of 2 h, consistent with target cells being eliminated by the inhibitor (Figure 2; white arrows). See the Discussion section for more information on interpreting rounding or lysing target cells. In Figure 2B, the same coincubation was spotted onto a slide, this time without concentrating the mixed culture so that the cells remained disperse and there was minimal contact between strains on the slide. Here, no target cells are observed to disappear or round, suggesting that the target strain was not inhibited in this treatment. Figure 2C and Figure 2D show the same crowded and disperse treatments described above, this time using a T6SS mutant as the inhibitor strain. Target cells were not observed to disappear or round when coincubated with a T6SS mutant in either crowded or disperse conditions, again suggesting that the target was not inhibited in either treatment.
Figure 3 shows the FIJI analysis workflow used to quantify competition in this protocol. A representative image from the target channel was selected (Figure 3A) and a binary mask was created using the default threshold settings in FIJI (Figure 3B). The image scale was set appropriately for this microscopy setup. Particles were analyzed using the size parameter = 0 – infinity, circularity parameter = 0.00 – 1.00, and Show Outlines was selected (Figure 3C). The results of this particle analysis are shown as both a numbered outline of each particle (Figure 3D), and as a table with columns for the particle number, file name (label), and particle area in µm2 (area) (Figure 3E).
In Figure 4, data obtained from Figure 3E is graphed and analyzed. In Figure 4A, the percent of initial target area at the final timepoint is presented for each treatment according to step 8.2. If the percent of initial target area is greater than 100, this represents net increase in target (i.e., growth) and is observed in conditions where the target population is not significantly inhibited. However, if the percent of initial target area is lower than 100, this result indicates a net decrease in the target (i.e., death) and is observed in conditions where the target population is significantly inhibited. When the target was coincubated with a wild-type inhibitor in crowded conditions, the data show a net decrease in the target area. By contrast, when the target was coincubated with either a wild-type inhibitor in disperse conditions or a T6SS mutant in crowded or disperse conditions, the data show a net increase in the target area. The percent of initial target area when the target was coincubated with a wild-type inhibitor in crowded conditions was below 100 and significantly lower than all other treatments according to a one-way ANOVA followed by a Tukey’s multiple comparisons test across all treatments (p < 0.0001). These data indicate that target cell death is dependent on a functional T6SS in the inhibitor and underscores the importance of an experimental setup that allows sufficient cell-cell contact, in order to detect cell death from a contact-dependent killing mechanism.
Figure 4B presents the percent of initial inhibitor area at the final timepoint for each treatment. In this example, net growth of the inhibitor strain was observed across all treatments. However, the percent of initial inhibitor area was significantly higher when a wild-type inhibitor was coincubated with the target in crowded conditions compared to all other treatments according to a one-way ANOVA followed by a Tukey's multiple comparisons test across all treatments (p < 0.0001). Initially, we considered that the net increase in inhibitor area may be driven by the increase in available space to grow into as target cells are eliminated. However, this same increase in inhibitor growth was not observed in disperse treatments, where inhibitor cells had room to grow from the beginning of the coincubation. Alternatively, this result could suggest that nutrients released from lysing target cells allow for a greater increase in the inhibitor population. Taken together, these results suggest that the inhibitor strain eliminates the target in a T6SS-dependent manner only when high cell-cell contact is forced by crowding cells on the slide.
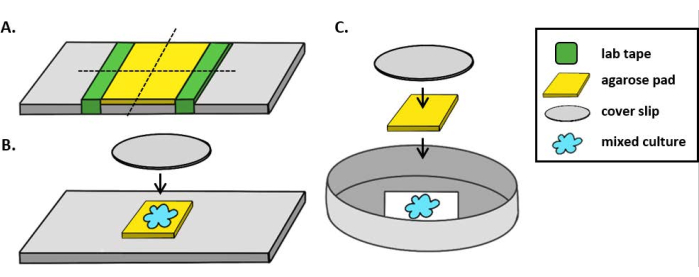
Figure 1: Agarose pad preparation and slide setup for coincubation assays. (A) Setup for making 2% agarose pads. Five layers of lab tape (green) are wrapped around a cover slip at two points approximately 20 mm apart. Next, warm 2% agarose in mPBS (yellow) is pipetted between the pieces of tape and immediately covered with a 25 mm2 cover slip and allowed to solidify for at least 1 h at room temperature. Use a razor blade to cut the agarose pad into ~5 mm2 pieces and use tweezers to transfer the pad to a new slide for imaging. (B) When imaging on an upright microscope, place the 5 mm2 agarose pad directly onto the slide, followed the mixed culture (blue) and a 12 mm circular #1.5 cover slip. (C) When imaging on an inverted microscope, spot the mixed culture directly onto the #1.5 glass cover slip bottom of a 35 mm Petri dish, and place an agarose pad on top of the culture followed by a second 12 mm circular cover slip to flatten the agarose pad. Please click here to view a larger version of this figure.
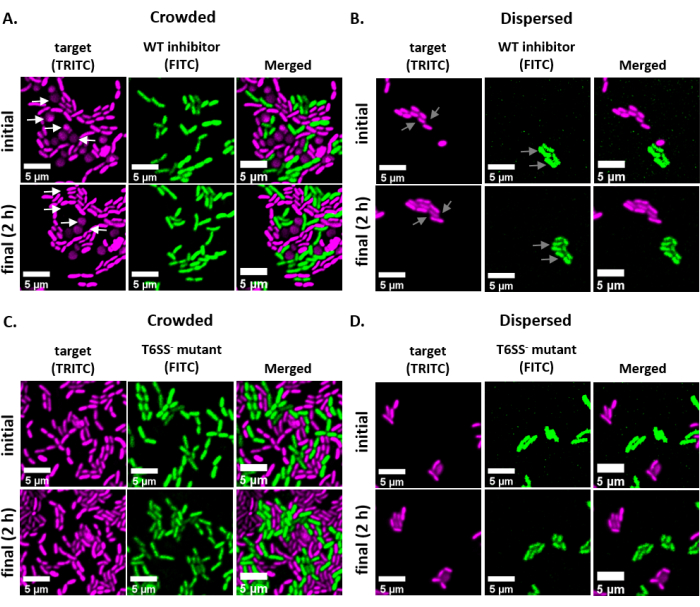
Figure 2: Time lapse images of coincubation spots in either crowded or disperse conditions. (A) Representative images at initial and final time points where a mixed culture of target and wild-type inhibitor was concentrated 3x prior to spotting on the slide to force cell-cell contact between strains. White arrows in TRITC channel indicate examples of target cells that round or lyse throughout the course of the experiment. (B) Representative images where a mixed culture of target and wild-type inhibitor was spotted without concentrating so that cells are disperse and there is minimal cell-cell contact between strains. Gray arrows in FITC and TRITC channels indicate examples of cell division throughout the course of the experiment. (C) Representative images where mixed culture of target and T6SS– mutant was concentrated 3x prior to spotting on the slide to force cell-cell contact between strains. (D) Representative images where mixed culture of target and T6SS– mutant was spotted without concentrating so that cells are disperse and there is minimal cell-cell contact between strains. Scale bars = 5 µm and are consistent across all images; TRITC channel is false-colored magenta, FITC channel is false-colored green. Deconvolution was performed on all images; background was subtracted, and brightness/contrast adjusted uniformly across all images. Please click here to view a larger version of this figure.
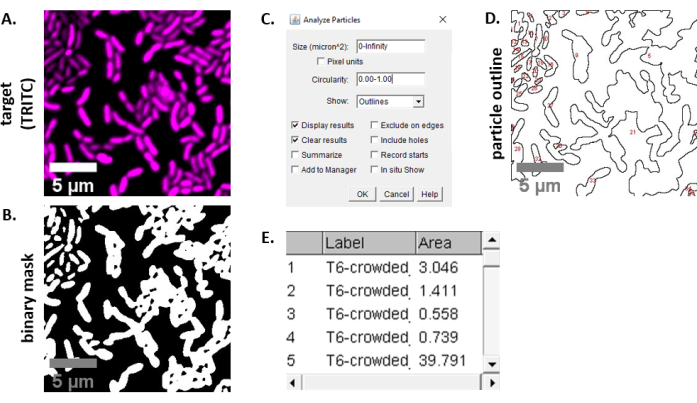
Figure 3: FIJI analysis workflow. (A) Representative image for analysis. This workflow is repeated for both channels across all fields of view and samples. Scale bars = 5 µm and are consistent across all images; TRITC channel is false-colored magenta, FITC channel is false-colored green. (B) Binary mask created by thresholding the image using the default settings in FIJI. (C) Example of settings for particle analysis used in this manuscript. Size range = 0 – infinity µm2; circularity = 0.00 – 1.00; show = outlines. (D) Particle outline created as an output of particle analysis in (C). The particle outline in (D) should be compared to the original image (A) to ensure that all cells were captured in the particle analysis. (E) Results table created as an output from particle analysis in (C). Object number (column 1) corresponds to individual particles (one or more cells) outlined and labeled in red in panel (D). Label = file name of analyzed image; Area = total particle area in µm2. Please click here to view a larger version of this figure.
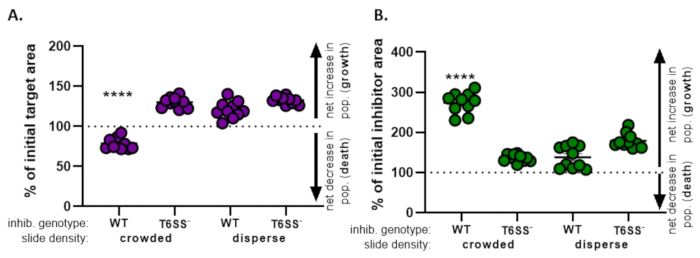
Figure 4: Sample data for assessing whether target strain is inhibited. The percent of initial area at the final time points for the target strain (A) and inhibitor strain (B), at different initial cell densities. Slide density indicates either a starting cell density that is crowded (high cell contact between strains), or more disperse (low cell contact between strains) as described in Figure 2. Inhibitor genotype indicates that either a wild-type or the T6SS mutant (T6SS–) strain was coincubated with the target strain. Asterisks indicate a significant difference in % change comparing all treatments (one-way ANOVA followed by a Tukey's multiple comparisons test comparing all treatments; (p < 0.0001). Dashed line indicates no net change in strain area between the initial and final timepoint; a % change > 100 indicates net increase (i.e., growth) and % change < 100 indicates net decrease (i.e., cell death). Please click here to view a larger version of this figure.
Discussion
The protocol described above provides a powerful tool for quantifying and characterizing interbacterial competition at the single-cell level. This assay, which was developed by modifying our CFU-based competition assay on agar plates1,2, allowed for the visualization of single-cell competition among V. fischeri isolates and suggestions are provided for optimizing the method for a wide range of systems and microscopy setups. Although the method described here was optimized for the light-organ symbiont V. fischeri, it can be easily modified to accommodate many diverse, culturable microbes. It is important to note that competitive mechanisms can be regulated by any number of environmental variables, including temperature, salinity, and viscosity30,31,32,33,34. Previous work has confirmed that V. fischeri competes using a contact-dependent Type VI Secretion System that is active on surfaces30, making the conditions described in this assay suitable for studying competition between the example strains. It is also important to consider the initial density of cells on the slide when quantifying bacterial competition. Given that contact between target and inhibitor cells is often required for killing to occur, the mixed culture should be concentrated such that cell-cell contact is maximized and cells remain in a single plane on the slide. Cell cultures should be grown to a similar optical density (mid-log phase) and then concentrated to force contact rather than simply growing cultures to a higher optical density due to the physiological changes of cells in different growth phases. In other systems, culture conditions and the experimental setup may need to be modified to ensure that the competitive mechanism is active and can be detected in the coincubation condition.
The agarose pads used in this assay provide several benefits: they provide stabilization so that cells do not move around freely, and they prevent the culture from drying out over the course of the experiment. Additionally, if chemical inducers, such as isopropyl-β-D-thiogalactoside (IPTG), are required for the experiment, they can easily be added to the agarose solution. However, it is important to note that the agarose preparation will likely need to be adjusted for different systems. In the example described above, the agarose pad was prepared by dissolving 2% agarose (w/v) into 20 psu mPBS, which is the standard salinity used in V. fischeri growth medium. Furthermore, in some cases a carbon source may need to be added to the agarose pad in order for cells to grow and compete over longer experiments. In such a case, the mPBS in agarose pads can be replaced with any growth medium, although the nutrients in growth medium may come with the tradeoff of additional background fluorescence.
Without proprietary image analysis software, it can be very difficult to get individual cell counts when cell-cell contact is high, which as we show here is required to observe contact-dependent killing. This assay was designed to provide an alternative method for quantification that does not rely on individual cell counts. Instead, the total cell area for each fluorescence channel is used to quantify the extent of killing between coincubated strains. Because this method relies on area rather than individual cell counts, default thresholding settings are typically sufficient for outlining the total cell area. The accuracy of thresholding can be verified by dividing the total object area for a representative field of view by the average cell size for the model organism and comparing this estimated cell number to a manual cell count for the same image.
In coincubations between one inhibitor and one target (non-killer) strain, net growth of the inhibitor is predicted. As seen in Figure 4, inhibitor growth may be significantly higher in treatments where killing is observed, compared to treatments where killing is not observed, perhaps because nutrients released by lysing target cells allow the inhibitor strain to grow more quickly. In the example shown here, net target death is observed because T6SS-mediated competition results in target cell lysis where the target is physically eliminated. However, it is important to note that not all competitive mechanisms result in the physical elimination of target cells. If a target is incapacitated by a toxin that causes growth inhibition, the protocol outlined here may result in the visible target population remaining stable over time as target cells no longer grow but also do not lyse. In such a case, it would be appropriate to compare the results of this assay with follow up tests for target cell viability, such as plating for colony forming units (CFUs) or by performing live-dead assays by staining with propidium iodide or SYTOX green35,36.
Compared to coincubation assays that rely on CFU counts, this assay makes it possible to observe and quantify the spatial structure of competition between strains and track changes in target cell morphology over time. For example, inhibitor cells that kill using a T6SS are known to encode LysM-domain proteins that degrade the target cell wall, resulting in initial cell rounding and then lysis13, which we observed in the example shown in Figure 2A. Further, this protocol can be used to track competition at high resolution over very short time scales. In the example shown here, a significant decrease in the target area is observed after only two hours when cells are crowded and cell-cell contact is forced between strains (Figure 4). The image analysis described here could also be performed using confocal microscopy, which would make it possible to study bacterial competition in vivo or in complex biofilms, without disrupting the spatial distribution of coincubated strains.
In summary, the assay described here aims to provide an accessible and easily modified approach for visualizing and quantifying bacterial competition at the single-cell level using fluorescence microscopy. This method can be applied to diverse bacterial isolates and can be used to visualize bacterial competition even in complex environments such as within a host or biofilm matrix.
Disclosures
The authors have nothing to disclose.
Acknowledgements
A.N.S was supported by NIGMS grant R35 GM137886 and S.N.S was supported by the National Defense Science and Engineering Graduate Fellowship Program.
Materials
| 1.5 mL Microcentrifuge tube | Fisher | 05-408-129 | |
| 10 uL single channel pipette | |||
| 1000 uL single channel pipette | |||
| 20 uL single channel pipette | |||
| 200 uL single channel pipette | |||
| Agarose | Fisher | BP165-25 | Low melting agarose |
| Calculator | |||
| Cellvis 35 mm Dish | Fisher | NC0409658 | #1.5 cover glass bottom |
| Chloramphenicol | Sigma | C0378 | stock (20 mg/mL in Ethanol); final concentration in media (2 μg /mL LBS) |
| DAPI Nucleic Acid Stain | Fisher | EN62248 | optional (if not using stable plasmids) |
| FIJI image analysis sofware | ImageJ | https://imagej.net/Fiji/Downloads | open-source software |
| Fisherbrand Cover Glasses: Circles | Fisher | 12-545-81P | #1.5 cover glass; 12 mm diameter |
| Kanamycin Sulfate | Fisher | BP906-5 | stock (100 mg/mL in water, filter sterilize); final concentration in media (1 μg/mL LBS) |
| Lens Cleaning Tissue Paper | Fisher | S24530 | |
| Parafilm | Fisher | 13-374-12 | |
| Petri Plates | Fisher | FB0875713 | sterile with lid |
| Razor Blades | Fisher | S65921 | |
| Semi-micro Cuvettes | VWR | 97000-586 | |
| Spectrophotometer | |||
| SYBR Green Nucleic Acid Stain | Fisher | S7563 | optional (if not using stable plasmids) |
| Thermo Scientific Gold Seal Plain Microscope Slides | Fisher | 12-518-100B | |
| Thermo Scientific Richard-Allan Scientific Cover Glass | Fisher | 22-050-235 | #1.5 cover glass, 25 mm2 |
| Type F Immersion Oil | Fisher | NC0297589 | |
| Upright or inverted fluorescence microscope with camera and imaging software | Images in this article were acquired on a Nikon TI-2 inverted fluorescent microscope outfitted with an ORCA-Fusion Digital CMOS camera using NIS-Elements software. | ||
| Vortex | |||
| Water bath | Used to keep agarose warm prior to pipetting | ||
| LBS media | |||
| 1M Tris Buffer (pH ~7.5) | 50 mL 1 M stock buffer (62 mL HCl, 938 mL DI water, 121 g Trizma Base) | ||
| Agar Technical | Fisher | DF0812-17-9 | 15 g (Add only for plates) |
| DI water | 950 mL | ||
| Sodium Chloride | Fisher | S640-3 | 20 g |
| Tryptone | Fisher | BP97265 | 10 g |
| Yeast Extract | Fisher | BP9727-2 | 5 g |
| mPBS (marine PBS) | Phosphate buffered saline with marine salts added; used for making agarose pad | ||
| 10X PBS | Fisher | ICN1960454 | |
| Instant Ocean Sea Salt | Instant Ocean | SS1-160P | Adjust concentration to appropriate salinity; 20 psu used here |
| Sterile Vacuum Filter Units | Fisher | SCGVU01RE | Used to filter-sterilize mPBS |
| Vacuum pump | Used to filter-sterilize mPBS |
References
- Speare, L., et al. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proceedings of the National Academy of Sciences. 115 (36), 8528-8537 (2018).
- Speare, L., Septer, A. N. Coincubation assay for quantifying competitive interactions between Vibrio fischeri isolates. Journal of Visualized Experiments. (149), e59759 (2019).
- Frost, I., et al. Cooperation, competition and antibiotic resistance in bacterial colonies. The ISME journal. 12 (6), 1582-1593 (2018).
- Stubbendieck, R. M., Vargas-Bautista, C., Straight, P. D. Bacterial communities: interactions to scale. Frontiers in Microbiology. 7, 1234 (2016).
- Souza, D. P., et al. Bacterial killing via a type IV secretion system. Nature Communications. 6 (1), 1-9 (2015).
- Anderson, M. C., Vonaesch, P., Saffarian, A., Marteyn, B. S., Sansonetti, P. J. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host and Microbe. 21 (6), 769-776 (2017).
- Basler, M., Ho, B., Mekalanos, J. Tit-for-tat: Type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 152 (4), 884-894 (2013).
- Guillemette, R., Ushijima, B., Jalan, M., Häse, C. C., Azam, F. Insight into the resilience and susceptibility of marine bacteria to T6SS attack by Vibrio cholerae and Vibrio coralliilyticus. PloS One. 15 (1), 0227864 (2020).
- Hachani, A., Lossi, N. S., Filloux, A. A visual assay to monitor T6SS-mediated bacterial competition. Journal of Visualized Experiments. (73), e50103 (2013).
- Hibbing, M. E., Fuqua, C., Parsek, M. R., Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nature Reviews Microbiology. 8 (1), 15-25 (2010).
- Ruhe, Z. C., Low, D. A., Hayes, C. S. Bacterial contact-dependent growth inhibition. Trends in Microbiology. 21 (5), 230-237 (2013).
- Wood, D. W., Pierson, L. S. The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene. 168 (1), 49-53 (1996).
- Smith, W. P., et al. The evolution of the type VI secretion system as a disintegration weapon. PLoS Biology. 18 (5), 3000720 (2020).
- Chen, L., Zou, Y., She, P., Wu, Y. Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiological Research. 172, 19-25 (2015).
- Sana, T. G., Lugo, K. A., Monack, D. M. T6SS: The bacterial “fight club” in the host gut. PLoS Pathogens. 13 (6), 1006325 (2017).
- Basler, M. Type VI secretion system: secretion by a contractile nanomachine. Philosophical Transactions of the Royal Society B: Biological Sciences. 370 (1679), 20150021 (2015).
- Joshi, A., et al. Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends in Microbiology. 25 (4), 267-279 (2017).
- Nadell, C. D., Drescher, K., Foster, K. R. Spatial structure, cooperation and competition in biofilms. Nature Reviews Microbiology. 14 (9), 589-600 (2016).
- Stubbendieck, R. M., Straight, P. D. Multifaceted interfaces of bacterial competition. Journal of Bacteriology. 198 (16), 2145-2155 (2016).
- Septer, A. N. The Vibrio-squid symbiosis as a model for studying interbacterial competition. Msystems. 4 (3), (2019).
- Tischler, A. H., Hodge-Hanson, K. M., Visick, K. L. Vibrio fischeri-squid symbiosis. eLS. , 1-9 (2019).
- Mandel, M. J., Dunn, A. K. Impact and influence of the natural Vibrio-squid symbiosis in understanding bacterial-animal interactions. Frontiers in Microbiology. 7, 1982 (2016).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Boettcher, K., Ruby, E. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna Scolopes. Journal of Bacteriology. 172 (7), 3701-3706 (1990).
- Doino, J. A., McFall-Ngai, M. J. A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. The Biological Bulletin. 189 (3), 347-355 (1995).
- Dunn, A. K., Millikan, D. S., Adin, D. M., Bose, J. L., Stabb, E. V. New rfp-and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Applied and Environmental Microbiology. 72 (1), 802-810 (2006).
- Lambertsen, L., Sternberg, C., Molin, S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environmental Microbiology. 6 (7), 726-732 (2004).
- Koch, B., Jensen, L. E., Nybroe, O. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. Journal of Microbiological Methods. 45 (3), 187-195 (2001).
- Peterson, B. W., Sharma, P. K., Van Der Mei, H. C., Busscher, H. J. Bacterial cell surface damage due to centrifugal compaction. Applied and Environmental Microbiology. 78 (1), 120-125 (2012).
- Speare, L., Smith, S., Salvato, F., Kleiner, M., Septer, A. N. Environmental viscosity modulates interbacterial killing during habitat transition. MBio. 11 (1), (2020).
- Salomon, D., Gonzalez, H., Updegraff, B. L., Orth, K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PloS One. 8 (4), 61086 (2013).
- Sana, T. G., et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proceedings of the National Academy of Sciences. 113 (34), 5044-5051 (2016).
- Bachmann, V., et al. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Neglected Tropical Diseases. 9 (8), 0004031 (2015).
- Ishikawa, T., et al. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infection and Immunity. 80 (2), 575-584 (2012).
- Johnson, M. B., Criss, A. K. Fluorescence microscopy methods for determining the viability of bacteria in association with mammalian cells. Journal of Visualized Experiments. (79), e50729 (2013).
- Stiefel, P., Schmidt-Emrich, S., Maniura-Weber, K., Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiology. 15 (1), 36 (2015).

