Hand Dissection of Caenorhabditis elegans Intestines
Summary
The present protocol describes a procedure for isolating intestines from adult Caenorhabditis elegans nematode worms by hand for input in genomics, proteomics, microbiome, or other assays.
Abstract
Comprised of only 20 cells, the Caenorhabditis elegans intestine is the nexus of many life-supporting functions, including digestion, metabolism, aging, immunity, and environmental response. Critical interactions between the C. elegans host and its environment converge within the intestine, where gut microbiota concentrate. Therefore, the ability to isolate intestine tissue away from the rest of the worm is necessary to assess intestine-specific processes. This protocol describes a method for hand dissecting adult C. elegans intestines. The procedure can be performed in fluorescently labeled strains for ease or training purposes. Once the technique is perfected, intestines can be collected from unlabeled worms of any genotype. This microdissection approach allows for the simultaneous capture of host intestinal tissue and gut microbiota, a benefit to many microbiome studies. As such, downstream applications for the intestinal preparations generated by this protocol can include but are not limited to RNA isolation from intestinal cells and DNA isolation from captured microbiota. Overall, hand dissection of C. elegans intestines affords a simple and robust method to investigate critical aspects of intestine biology.
Introduction
The Caenorhabditis elegans nematode worm, with a mere 959 cells and a 4 day egg-to-egg life cycle, is an ideal model system for many genetics, genomics, and developmental studies1,2. The ease of forward and reverse genetic screening, the prevalence of engineered fluorescent markers, the capacity to perform nucleotide-specific genome editing, and the numerous community-wide resources have all contributed to major discoveries and insights in the C. elegans system. However, a significant drawback is the difficulty of obtaining pure populations of cells, tissues, or organs, which are small, fragile, and can be interconnected. As pure populations of cells are important for genomics assays such as RNA-seq, ChIP-seq, and ATAC-seq, several approaches have emerged to obtain pure preparations of C. elegans cells, tissues, and organs. Here, a method for hand dissecting intestines, in large sections, out of adult C. elegans worms is described. The resulting preparations are suitable for downstream genomics assays (Figure 1).
The fine-scale tissue dissection method described here (Figure 2) is just one approach. Other alternative techniques-such as molecular tagging, disaggregating worms, and purifying cell types of interest with fluorescence-activated cell sorting (FACS) and post hoc analysis-have also been successfully used to survey the tissue-specific features of C. elegans molecular biology. An advantage of hand dissection over these other approaches, however, is that it can be used to simultaneously explore the features of the C. elegans intestine and its bacterial contents3,4,5. This enables 16S rRNA gene sequencing and facilitates microbiome studies within the C. elegans system. An important limitation, however, is that intestinal cells are not individually isolated.
Molecular tagging imparts a cell type-specific tag to molecules only within the specified tissue or cells of interest. These tags can then be isolated from total worm preparations. In this way, tissue-specific promoters driving a tagged polyA-binding protein or spliced leader have enabled tissue-specific transcriptome profiling6,7,8,9,10 and 3'UTR mapping11,12. Similarly, tissue-specific transcription factor profiles have been conducted using ChIP-seq and DamID, in which promoter-specific transcription factor variants were appended with tags or enzyme fusions13,14.
FACS allows for isolating cell types of interest from dissociated worms based on their intrinsic cellular characteristics and fluorescent properties. This approach has generated tissue-specific transcriptomes from diverse organs8,15,16 and individual neuronal cell types8,9,15,16,17,18 and has been used to create an expression map of the entire C. elegans nervous system19,20. FACS, and its cousin fluorescence-activated nuclei sorting (FANS), have also been used to generate cell-specific chromatin profiles21,22.
Finally, post-hoc analysis can be performed in single-cell resolution assays. In this method, all individual cells are surveyed, the cell type of each is ascribed in the analysis stage, and the cell types of interest are selectively filtered for further study. Post-hoc analysis has been successfully used to obtain transcriptomes of developing cells with both high spatial and temporal resolution in C. elegans embryos23,24,25,26,27 and L128 stage worms. Chromatin accessibility has also been characterized using ATAC-seq instead of RNA-seq using a similar strategy29.
Each approach has its advantages and limitations. For the C. elegans intestine, worm disaggregation and FACS isolation of intestinal cells is achievable in the embryo and larval stages30 but is challenging in adults. This is thought to be due to the intestine's large, endo-reduplicated, and strongly adherent cells making them difficult to dissociate undamaged. The hand dissection method described here circumvents these challenges, allowing for the isolation of large sections of the adult worm's intestine. The practice of hand dissecting gonads from this same stage is widespread and straightforward. Intestine dissection is similar to gonad dissection but less commonly performed32. The protocol presented here is adapted from a longer, unpublished protocol developed by Dr. James McGhee and Barb Goszczynski. This streamlined protocol borrows techniques for isolating blastomeres from early-stage embryos23,33,34,35. Though hand dissection is not feasible for isolating most cell or tissue types in C. elegans, it is ideal for isolating intestines from adult worms. Therefore, hand dissection complements other means for obtaining intestine-specific cell preparations.
Protocol
CL2122 worms were used for the present study. The worms were obtained through the Caenorhabditis Genetics Center (CGC, see Table of Materials), funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
1. Growing of worms for dissection
- Grow one large plate of mixed stage CL2122 worms for synchronization following standard culturing procedures (i.e., NGM plates seeded with E. coli OP50)36,37.
NOTE: It generally takes ~96 h to reach maximal egg laying capacity.- Embryo prep the worms within the 72-96 h period. After embryo prepping, allow the embryos to hatch in M9 (see Table 1) for 48 h. This will yield a synchronous population of L1 stage worms.
NOTE: CL2122 worms harbor an integrated transgene-GFP (green fluorescent protein) driven off the intestine-specific mtl-2 (MeTaLlothionein 2) promoter38. This promoter is specific to the intestinal cell cytoplasm and allows the intestine to be visualized on a fluorescent dissection microscope. Once trained, users may not find fluorescence guidance necessary.
- Embryo prep the worms within the 72-96 h period. After embryo prepping, allow the embryos to hatch in M9 (see Table 1) for 48 h. This will yield a synchronous population of L1 stage worms.
- Plate synchronized L1 worms on a minimum of two small plates. Grow worms on NGM plates with sufficient food until they reach the adult stage (identified by the presence of embryos capable of egg laying). This takes between 38-46 h at 20 °C.
- Take care to ensure that the same developmental stage is harvested across replicates and across comparative strains.
NOTE: The length of time to grow the worms is dependent on the strain, temperature, food source, and developmental stage targeted39. Other stages near the adult stage can also be used, such as the L4 and older adult stages. For planning microbiome experiments, different bacterial food sources beyond the traditional food source (E. coli OP50) can be used to grow worms, such as the CeMbio strains4 or a pathogen of interest40.
- Take care to ensure that the same developmental stage is harvested across replicates and across comparative strains.
2. Preparation of stock solutions and microcapillary pipettes
NOTE: Table 1 provides the details of all the buffers and solutions used for the present study.
- Prepare 50 mL of Egg salts (a.k.a., Egg buffer) in a nuclease-free conical tube (see Table of Materials). Once made, store at room temperature.
- Prepare 10 mL of 100 mM levamisole stock solution in a nuclease-free conical tube. Once prepared, make 500 µL aliquots and store them at −20 °C. Aliquots can be defrosted and stored at 4 °C for 1 week at a time.
- Prepare 1 mL of 20 mg/mL stock acetylated bovine serum albumin (BSA) solution in a nuclease-free microcentrifuge tube. Once prepared, make 50 µL single-use aliquots and store them at −20 °C.
NOTE: It is important to use the acetylated version of BSA because this is the only form of BSA that is nuclease-free. Non-acetylated BSA can be a significant source of nucleases and, thus, lead to the degradation of samples. - Prepare at least five 50 µm and 100 µm microcapillary pipettes each following the steps below.
- First, pull standard glass capillaries (4 in long and 1.2 mm outer diameter) into an injection needle shape using a needle puller (see Table of Materials) and conditions for "Adherent Cell, C. elegans, & Drosophila" from the Sutter Instruments Pipette Cookbook41 (Conditions: Heat = Ramp + 5; Pull = 100; Vel. = 75; Delay = 90; Pressure = 500; 2.5 mm x 2.5 mm Box).
- Next, forge the microcapillary pipettes to either 50 µm (three tick marks as indicated by the microforge ocular ruler under the M5/0.1 objective) or 100 µm (five tick marks under the same conditions) size using a microforge (see Table of Materials). These sizes represent the estimated opening diameter of the microcapillary pipette (Figure 3).
- Then, affix a mouth aspirator tube to the microcapillary pipettes.
NOTE: Aspirator pipettes are traditionally controlled by mouthing pipetting, but many modern safety protocols disallow this method. As such, users can control aspiration with the mouth aspirator tubing by pinching the tubing between the finger and thumb. Additionally, a syringe filter can be installed within the mouth aspirator tubing system for added safety.
3. Experimental preparation
- Prepare 5 mL of dissection buffer in a nuclease-free conical tube. Store at room temperature.
- Prepare 1 mL of working acetylated BSA solution in a nuclease-free microcentrifuge tube. Keep on ice.
- Make 350 µL of working levamisole solution in a nuclease-free microcentrifuge tube. Prepare in triplicate (one tube for about 20 worms). Keep on ice.
- Prepare chelation buffer in a nuclease-free microcentrifuge tube (Table 1). Make in replicate (one tube for about 10 dissected worms). Keep on ice. Using chelation buffer during hand dissections improves RNA quality and quantity (Figure 4).
- Prepare one microcentrifuge tube per experimental group that contains 500 µL of nucleic acid isolation reagent or kit-specified isolation reagent (see Table of Materials) in a nuclease-free tube. Keep on ice. This tube will be used to collect the final, isolated intestines for storage or later use.
- Prepare one M9 bath and one dissection buffer bath. To do so, obtain two 35 mm diameter sterile Petri dishes. Then, add 2 mL of M9 to one dish and 2 mL of dissection buffer to the other. Finally, add 100 µL of working BSA solution to each bath. Swirl to mix. Adding BSA to the baths will prevent the worms from sticking to the plastic.
- Prepare the dissection array. Obtain a 2-well concavity slide (see Table of Materials) and add 150 µL of working levamisole solution to the first well. Then, add 150 µL of dissection buffer to the second well. Finally, add 20 µL of working BSA solution to each well. Adding BSA to each well will prevent the worms from sticking to the slide.
4. Hand dissection of the C. elegans intestine
- Using a worm pick, move 20 adult worms from the NGM plate into the M9 bath (step 3.6). This will wash external bacteria from the worms.
- Then, move all 20 worms from the M9 bath to the dissection buffer bath (step 3.6). This will further wash away external bacteria and equilibrate the worms in the dissection buffer.
- Next, move batches of worms (i.e., in sets of 10) from the dissection buffer bath into the well containing levamisole solution (step 3.7).
NOTE: Levamisole will temporarily paralyze the worms.- Once the worm movements slow down, quickly move them from the levamisole well to the well containing the dissection buffer (step 3.7). Take care not to over-paralyze the worms.
NOTE: The number of worms moved in batches into the levamisole solution can vary based on the user's comfort. This is also true for the number of worms picked initially into the M9 bath and then moved into the dissection buffer bath. It is good practice to have additional worms available for dissection, especially during training, as the user learns and gets comfortable with the protocol.
- Once the worm movements slow down, quickly move them from the levamisole well to the well containing the dissection buffer (step 3.7). Take care not to over-paralyze the worms.
- Then, allow the worms to start moving a bit in the dissection buffer prior to starting dissections.
NOTE: Intestines do not extrude well from overly paralyzed worms (i.e., worms that are not moving at all). - When ready, dissect the worms under a fluorescent dissecting scope using a hypodermic needle (i.e., 27 G x 1/2 in) (see Table of Materials) by making one cut either just behind the pharynx (Figure 5Aa) or just in front of the rectum (Figure 5Ab). This will produce two large sections of the intestine, an anterior-mid half and a mid-posterior halfI. Continue this pattern for the rest of the worms, keeping the number of intestine sections obtained from the anterior-mid and mid-posterior sections equivalent until the desired total number of intestines is acquired.
NOTE: Affixing the hypodermic needle to an empty 1 mL syringe barrel can aid in its manipulation during hand dissections. Depending on the comfort and experience of the user, it is not uncommon to make a dissection cut that yields no intestinal fragment. However, with practice, this becomes far less common. - Wait about 1 min for the intestines to maximally extrude from the body. They typically take on a loop shape and may be stuck to a portion of the gonads (Figure 5A). While waiting, add 50 µL of chelation buffer to the well to help reduce RNA degradation (Figure 4).
- While waiting and to further help facilitate intestine extrusion, use the 100 µm microcapillary pipette attached to a mouth aspirator and draw the intestine/worm in and out of the pipette (Figure 5B). This will also help to liberate the intestine from the gonad. Take care not to lose the intestine by accidentally sucking it up completely into the microcapillary pipette.
NOTE: The user can alternate between the 100 µm and 50 µm microcapillary pipettes to liberate the intestine from the rest of the body and gonad. The smaller diameter opening of the 50 µm microcapillary pipette may afford a benefit for removing sticky pieces from the intestine. Aspirator pipettes are traditionally controlled by mouthing pipetting, but many modern safety protocols disallow this method. As such, users can control aspiration with the mouth aspirator tubing by pinching the tubing between the finger and thumb. Additionally, a syringe filter can be installed within the mouth aspirator tubing system for added safety. - Once an intestine is sufficiently extruded, use the 27 G hypodermic needle to cut it away from the rest of the body and any remaining gonad. The size of the intestine section isolated can vary greatly depending on the experience of the harvester, the quality of the cut, and the level of paralysis in the worm.
NOTE: The integrity of the intestine and intestinal fragments can be easily monitored throughout the protocol via their GFP fluorescence. - Now, use the microcapillary pipette to aspirate the intestine section and move it out of the well and into the microcentrifuge tube containing nucleic acid isolation reagent or kit-specified isolation reagent. Keep the isolated intestines in reagent on ice and repeat for the remaining intestines.
NOTE: Multiple intestines can be added to the microcentrifuge tube containing nucleic acid isolation reagent or another kit-specified isolation reagent throughout the day. Keep on ice. If all the intestines cannot be isolated in 1 day, those that are already isolated and stored in a nucleic acid isolation reagent (which preserves the nucleic acids) can be kept at −80°C until the isolations resume and/or can be completed. Here, the samples are stable long-term (i.e., several months to 1 year) and can remain until ready to isolate nucleic acids. Intestines can be harvested into water or any kit-specific lysis buffer. However, consideration must be given when determining how long intestines can stay in a given buffer on ice (or other desired temperature) prior to moving on to downstream applications.
5. RNA isolation from dissected intestines
- Homogenize the acquired tissues stored in the nucleic acid isolation reagent by performing three freeze/thaw/vortex cycles. To do so, use a 37 °C bead bath and liquid nitrogen (or other comparable means).
- Then, add 0.2 volumes of phenol:chloroform:IAA reagent (see Table of Materials) to the sample and vortex briefly. For example, for a starting sample volume of 500 µL, 0.2 volumes of chloroform would be 100 µL.
- Shake the tube by hand for 20 s, and then incubate at room temperature for 3 min.
- Separate the sample phases by centrifugation (10,000 x g, 18 min, 4 °C).
- Remove the aqueous phase and transfer to a new nuclease-free microcentrifuge tube.
NOTE: Take care not to aspirate or disturb the interface. - Then, add an equal volume of 100% ethanol to the aqueous phase and shake the sample by hand for 20 s.
- Transfer 700 µL of the sample to the spin column (see Table of Materials). Then, adhere RNA to the column with centrifugation (≥8,000 x g, 30 s, RT [room temperature]). Discard the flow-through. Repeat for any additional remaining sample.
- Add 350 µL of RW1 buffer (see Table of Materials) to the column to wash the sample. Then, centrifuge (≥8,000 x g, 30 s, RT) and discard the flow-through.
- Perform on-column DNA digestion. Add 80 µL of DNase I in RDD buffer (see Table of Materials) to the sample column. Then, incubate for 15 min at RT.
- Then, add 350 µL of RW1 buffer to the column to wash the sample. Centrifuge (≥8,000 x g, 30 s, RT) and transfer the column to a new collection tube. Discard the flow-through and old collection tube. This wash removes the DNase.
- Add 500 µL of RPE (see Table of Materials) to the sample column. Then, centrifuge (≥8,000 x g, 30 s, RT) and discard the flow-through. Repeat for a second wash.
- Then, centrifuge for an additional 1 min to further dry the column membrane (≥8,000 x g, 1 min, RT). Next, transfer the sample column to a fresh nuclease-free microcentrifuge collection tube with a lid. Discard the flow-through and old collection tube.
- Add 14 µL of nuclease-free water directly to the membrane of the sample column. Then, incubate the sample for 2 min at RT. Next, centrifuge (≥8,000 x g, 1 min, RT) to elute the RNA.
- Store the sample on ice. Next, assess the sample's RNA quality and quantity using commercially available assay kits (see the Table of Materials). Once done, store the sample in a −80 °C freezer.
NOTE: RNA is generally stable at −80 °C for up to 1 year without degradation.
Representative Results
The present protocol was used to isolate large sections of the intestine from adult C. elegans by hand (Figure 2). The final intestine sample for each experimental group shown is comprised of an equal collection of anterior-mid and mid-posterior intestine sections. However, depending on the experimental question, it could also comprise a collection of only anterior, mid, or posterior intestine sections. Collectively, three representative results are presented for this protocol. The first depicts the successful dissection and isolation of intestines (Figure 6). The second reports the results of RNA isolation from isolated intestines (Figure 7). The third shows the results of microbial surveillance from isolated intestines (Figure 8).
For the first result, Figure 6A displays what an extruded intestine looks like after making a primary incision at site "a" in Figure 5A within adult CL2122 worms. As CL2122 worms harbor the intestine-specific mtl-2 promoter fused to GFP (mtl-2p::GFP), isolated intestines will glow green under the fluorescent dissecting scope. The successful dissection of an intestinal segment is then shown in Figure 6B. This intestine section is free from visible contaminants such as debris from the gonad or carcass. In contrast, Figure 6C displays intestines that are not successfully dissected, as gonad and carcass are still visibly attached to the intestinal segment.
For the second result, the intestines were harvested into a nucleic acid isolation reagent and processed the next day using a low-input total RNA extraction protocol (see Table of Materials). A final intestine sample of 60 total intestine sections from the anterior-mid and mid-posterior sections yields roughly 15 ng of high-quality total RNA (Figure 7A). This amount of total intestines can easily be obtained in a single day but can also be broken up over several days if needed. RNA yields from hand dissection are more efficient than worm disaggregation and FACS isolation of intestinal cells in that hundreds of thousands of intestinal cells are required to obtain commensurate quantities of total RNA (Figure 7B). Importantly, the RNA yields generated by hand dissection are more than sufficient as input for commercial RNA-seq library kits (i.e., NEBNext Ultra II and NEBNext Single Cell/Low Input), which can take as little as 2 pg of RNA.
For the third result, the intestines were harvested into sterile ddH20 and processed the same day using a commercial microbial DNA isolation kit (see Table of Materials). A final intestine sample of 40 total intestine sections from the anterior-mid and mid-posterior sections yields around 0.009 pg of total microbial DNA using a pan-bacterial detection assay (see Table of Materials) (Figure 8). These quantities are too low for traditional quantification methods and must be extrapolated from qPCR standard curves. Ideally, users should target a final total amount of intestines >40 as this increases the detection limit and boundary of signal-to-noise regarding reagent contamination levels.
When considering experimental design, collecting appropriate control samples is always necessary. For transcriptomics experiments, a suitable experimental control can include preparations of the whole worm collected in the same manner as intestines. During the dissection and isolation of intestines from C. elegans, however, it is common to see bits of carcass and gonad clinging to the intestine sections. While ideally these contaminating tissues will be removed from the intestines prior to storage for downstream use, additional experimental controls can include the collection of the leftover worm carcasses post intestine dissection and/or the collection of dissected gonads. For microbiome experiments, controls can also include preparations of the whole worm collected in the same manner as intestines, in addition to conventional positive (bacterial culture) and negative (water) controls.
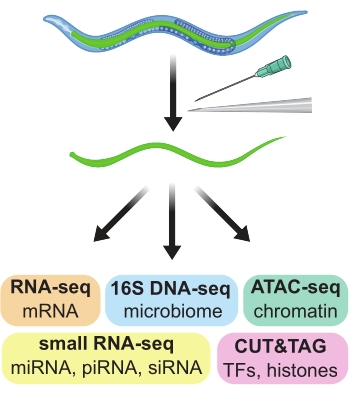
Figure 1: Hand dissection of intestines from adult C. elegans used to generate tissue-specific preparations for use in a wide variety of downstream omics assays. Please click here to view a larger version of this figure.
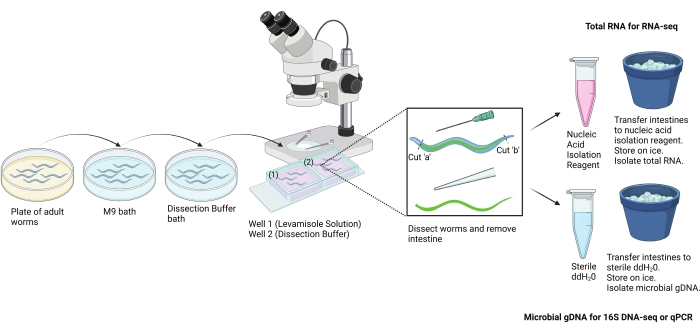
Figure 2: Scheme for the hand dissection protocol. This protocol was used to isolate large sections of the intestine from adult C. elegans by hand. Intestines can be isolated for different downstream assays. Shown here is the use of intestines for RNA isolation and microbial DNA isolation. Image created with BioRender.com. Please click here to view a larger version of this figure.
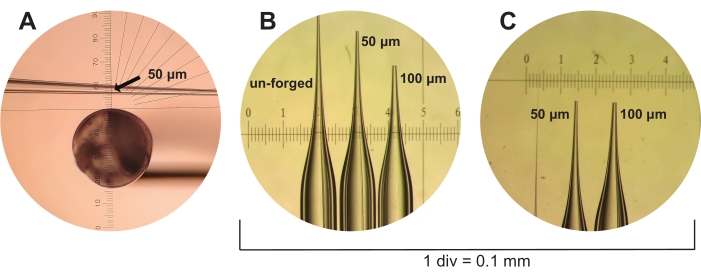
Figure 3: Microcapillary pipette fabrication. (A) A freshly pulled but unforged microcapillary pipette is shown through the ocular piece of a microforge. An ocular ruler is used to measure 50 µm sized microcapillary pipettes to an estimated inner diameter of three tick marks (shown, indicated by arrow). (B) Unforged, 50 µm and 100 µm microcapillary pipettes are shown under the dissecting scope alongside a calibration ruler with 1 div = 0.1 mm. (C) The 50 µm and 100 µm microcapillary pipettes from (B) are shown again but from a different vantage point. Please click here to view a larger version of this figure.
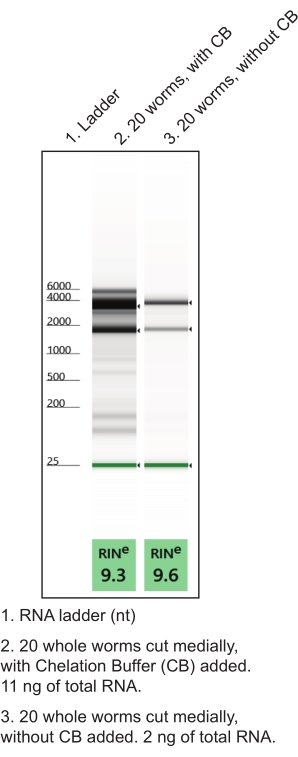
Figure 4: The use of chelation buffer (CB) during hand dissection to improve RNA yield. A low input total RNA extraction protocol generated RNA preparations from named tissues. Representative gel electrophoresis runs characterizing isolated total RNA quality (RIN, RNA integrity number) and quantity are shown. Please click here to view a larger version of this figure.
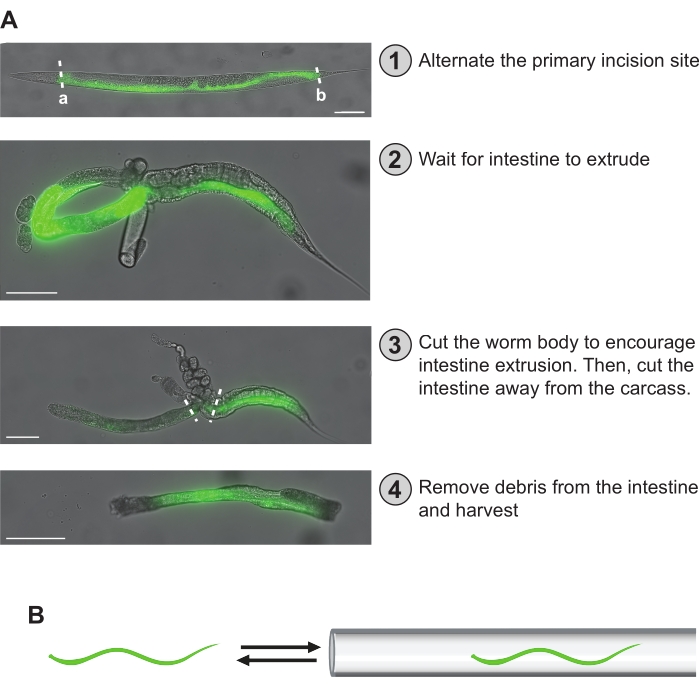
Figure 5: Hand dissection steps. (A) The dissection steps described in the protocol are outlined here. In (1), the intestine of the worm is visualized by GFP fluorescence. The primary incision site can be evenly distributed among the worms to ensure even coverage across the intestine's entire length. Alternatively, either "a" or "b" primary incision sites can be selected to obtain an anterior-mid or mid-posterior intestinal fragment-specific preparation. In (2), the intestine has extruded, taking on a loop shape. In (3), the intestine is first cut away from the worm body while attempting to liberate it from the carcass and gonad. The intestine is then removed from any remaining carcass or gonad that cannot be removed. In (4), a cleaned section of the intestine is isolated and ready for storage. (B) A critical step is to dislodge any clinging debris (i.e., gonad, carcass) from the intestine by passing it in and out of the microcapillary pipette fashioned at the end of the mouth aspirator. Scale bars = 100 µm. Please click here to view a larger version of this figure.
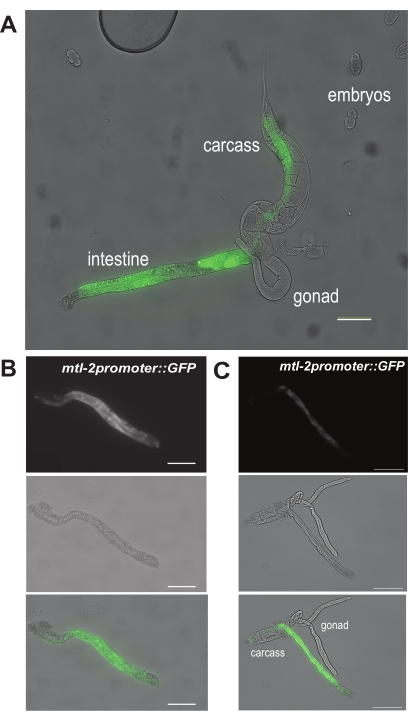
Figure 6: Representative results of hand dissection steps. A representative image showing (A) the extrusion of the intestine from the worm. These are worms of the CL2122 genotype, expressing GFP under the intestinal cell-specific mtl-2 promoter. Two representative images show (B) a cleaned and fully isolated section of the intestine and (C) an isolated intestine with contaminating gonad and carcass tissues. Scale bars = 100 µm. Please click here to view a larger version of this figure.
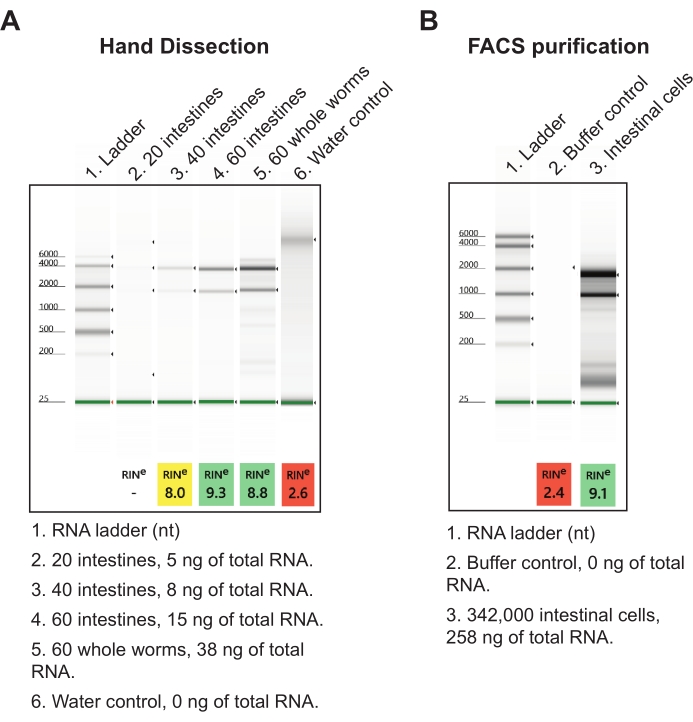
Figure 7: Representative results of total RNA extraction from isolated intestines. (A) A representative image of RNA preparations resolved by agarose gel electrophoresis is shown, characterizing the quality (RINs, RNA integrity Number) and quantity of isolated total RNAs. (B) A representative gel of total RNA isolated from intestinal cells harvested from L1 stage worms via fluorescence-activated cell sorting (FACS) is shown for comparison. At 20 cells per intestine, it can be inferred that the hand dissection method yields more total RNA per intestine than FACS-purified intestinal cells. Please click here to view a larger version of this figure.
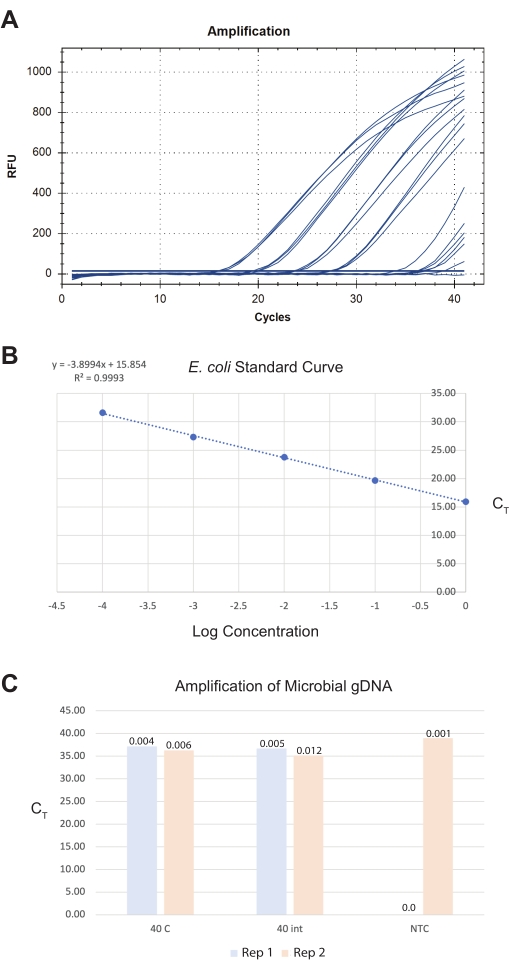
Figure 8: Representative results for total microbial DNA extraction from isolated intestines. A commercial microbial DNA isolation kit generated DNA preparations from intestines, whole worms, and controls. A pan-bacterial gene assay was used to quantify the number of bacteria in the samples. A representative image of (A) the qPCR amplification curves generated, (B) the E. coli OP50 standard curve used to quantify the samples, and (C) the samples are shown. In (B), the log starting concentrations of E. coli standards are graphed against their CT values. In (C), two replicates (reps) of 40 whole worms were cut medially, and 40 isolated intestinal sections were processed for microbial gDNA isolation. The No-Template-Control (NTC) from the qPCR run is also shown. The Y-axis represents the sample CT values. The amount of DNA quantified from PCRs is shown as total picograms (pg) values above the sample bars. Please click here to view a larger version of this figure.
Table 1: Compositions of the buffers and solutions used in this study. Please click here to download this Table.
Discussion
This article describes the step-by-step protocol for hand dissecting intestines from adult C. elegans, generating pure preparations for downstream assays. Critical steps in this protocol include (1) ensuring not to over-paralyze the worms, (2) making accurate dissection cuts, (3) forging appropriately sized micro-pipettes for dissection, and (4) ensuring the speedy recovery of healthy intestines during the final harvest. For these reasons, care must be taken when exposing worms to the levamisole solution, and the hypodermic needles need to be refreshed frequently to ensure maximum sharpness. Handling the intestine using the microcapillary pipette and mouth aspirator is another step that will take practice. Properly forged micro-pipettes of the appropriate size also make a substantial difference in isolating large sections of the intestine during dissections, in addition to reducing the risk of losing intestines within the micro-pipette. New protocol users commonly lose intestines on the inner edge of the microcapillary pipette before they can be ejected into the isolation reagent. This problem can be amended with practice and properly forged microcapillary pipettes.
The protocol described herein was designed for use in adult worms. Preliminary trials support that this protocol is also effective for use in L4 worms and older adult worms. However, the efficacy of this protocol has not yet been evaluated in early larval stage worms. A limitation of this approach is the small amount of material it yields. Though the quantities are sufficient for RNA-seq and PCR, they may not be adequate for other assays. As such, users need to determine if the minimum required input for an assay can be feasibly collected with this protocol.
Our lab routinely utilizes FACS to purify intestinal cells post isolation30, post-hoc analysis methods for intestinal cell identification, and this hand dissection method30,42. Hand dissection has the advantage of being amenable for use in adult worms when worm disaggregation and cell isolation are less successful. Furthermore, the efficiency and quality of total RNA extracted from hand dissection preparations are high, likely because the tissues are rapidly plucked from the worms and then quickly deposited in a nucleic acid isolation reagent, reducing RNA degradation. Another benefit of the hand dissection method is that it is low cost, easy to learn, and does not require specialized equipment. Finally, this approach allows for the harvest and isolation of gut bacteria from worm intestines, enabling downstream microbiome studies.
The hand dissection protocol described here for isolating intestines from adult C. elegans represents a powerful tool for studying various aspects of C. elegans biology. For example, with a pure preparation of intestines, researchers can investigate the intersection between immunity, aging, metabolism, and the microbiome.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We are indebted to the pioneering work of James McGhee and Barb Goszczynski, who initially developed the intestine dissection method from which this protocol is adapted. Our work is supported by a MIRA (R35) Award overseen by the National Institute of General Medical Sciences (National Institutes of Health, R35GM124877 to EON) and an NSF-CAREER Award overseen by the NSF MCB Div Of Molecular and Cellular Bioscience (Award #2143849 to EON).
Materials
| Acetylated Bovine Serum Albumin (BSA) | VWR | 97061-420 | Nuclease free BSA |
| CL2122 worm strain | CGC (Caenorhabditis Genetics Center) | CL2122 | dvIs15 [(pPD30.38) unc-54(vector) + (pCL26) mtl-2::GFP]. Control strain for CL2120. Phenotype apparently WT. |
| Calcium Chloride Dihydrate | Fisher | C79 | needed for making Egg Salts |
| 50 mL Centrifuge Tubes, Bulk | Olympus Plastics | 28-108 | Nuclease free conical tube needed for solution making. |
| 15 mL Centrifuge Tubes, Bulk | Olympus Plastics | 28-103 | Nuclease free conical tube needed for solution making. |
| Concavity slide (2-well) | Electron Microscopy Sciences | 71878-08 | 12-pk of 2-well concavity slides |
| Ethylene glycol-bis(2-amino-ethylether)-N,N,N',N'-tetraacetic acid (EGTA) | Millipore Sigma | E3889 | needed for making chelation buffer |
| Fluorescent Dissection Microscope | Leica | M205 FCA | This is an optional piece of equipment that can be used with fluorescent C. elegans strains to help guid users during hand dissections |
| N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid), 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | Millipore Sigma | H4034 | needed for making dissection buffer |
| High Sensitivity RNA ScreenTape | Agilent | 5067-5579 | for assesment of total RNA quality and quantity |
| High Sensitivity RNA ScreenTape Ladder | Agilent | 5067-5581 | for assesment of total RNA quality and quantity |
| High Sensitivity RNA ScreenTape Sample Buffer | Agilent | 5067-5580 | for assesment of total RNA quality and quantity |
| HostZERO Microbial DNA Kit | Zymo Research | D4310 | Isolation of microbial DNA from worm intestines/worms |
| Hypodermic Needle (27G x 1/2") | BD Scientific | 305109 | needed for hand dissection of intestines |
| Levamisole (a.k.a. (-)-Tetramisole hydrochloride) | Millipore Sigma | L9756 | used to temporarily paralyze worms prior to hand dissection of intestines. |
| Luer-Lok General Use Disposable Syringe (1 mL) | BD Scientific | 309628 | Optional. Can be used to affix the hypodermic needle to, allowing easier manipulation of the needle during dissection. Remove the plunger. |
| Magnesium Chloride Hexahydrate | Fisher | M33 | needed for making Egg Salts |
| Magnesium Sulfate Hpetahydrate | Sigma-Aldrich | 230391-500G | needed for making M9 buffer |
| MF-900 Microforge | Narishige | MF-900 | Used to forge the microcapillary pipettes. Available through Tritech Research. |
| 1.7 mL Microtubes, Clear | Olympus Plastics | 22-282 | Nuclease free microfuge tube needed for solution making and sample storage. |
| Mouth Aspirator Tube | Millipore Sigma | A5177 | Mouth aspirator tube is needed in combination with the microcapillary pipette to allow aspiration of dissected intestines. |
| 16S Pan-Bacterial Control TaqMan Assay | Thermo Fisher | A50137 | Assay ID: Ba04930791_s1. Assay used for gut microbial detection via qPCR. |
| P-1000 Micropipette Puller | Sutter Instruments | Model P-1000 | Used to pull the microcapillary pippettes prior to forging. |
| Petri Dish (35 x 10 mm) | Genesee Scientific – Olympus Plastics | 32-103 | Used to make M9 bath and Disseciton Buffer bath for washing worms prior to dissection. |
| Phenol:Chloroform:IAA | Ambion | AM9730 | Used in the isolation of total RNA |
| Potassium Chloride | Millipore Sigma | 529552 | needed for making Egg Salts |
| Potassium Phosphate Monobasic | Sigma-Aldrich | P0662-500G | needed for making M9 buffer |
| Qubit 3 Fluorometer | Invitrogen | Q33216 | Accompanies the Qubit RNA HS Assay Kit. Can be used to quantify RNA prior to running sample on the Agilent ScreenTape. |
| Qubit RNA HS Assay Kit | Invitrogen | Q32852 | Can be used to quantify RNA prior to running sample on the Agilent ScreenTape. |
| RNasin Ribonuclease Inhibitor | Promega | N2111 | Broad spectrum inhibition of common eukaryotic Rnases |
| RNase-Free DNase Set | Qiagen | 79254 | used for on-column DNA digestion during RNA isolation protocol. |
| RNeasy Micro Kit | Qiagen | 74004 | Used for isolation of total RNA from worm intestines/worms |
| Standard Glass Capillaries | World Precision Instruments | 1B100F-4 | 4 in OD 1.2 mm standard borosilicate glass capillaries used to make microcapillary pipettes for dissection |
| Sodium Chloride | Fisher | S271 | needed for making Egg Salts |
| Sodium phosphate dibasic heptahydrate | Fisher Scientific | S373-500 | needed for making M9 buffer |
| Syringe filter (0.2 micrometer SCFA) | Thermo Fisher | 72302520 | Optional for use with the mouth aspirator tube when mouth pipetting. |
| 4150 TapeStation System | Agilent | G2992AA | Accompanies the RNA ScreenTape reagents for assessing RNA quality and quantity |
| TaqPath BactoPure Microbial Detection Master Mix | Applied Biosystems | A52699 | master mix used for qPCR |
| TRIzol Reagent | Thermo Fisher Scientific | 15596026 | Nucleic acid isolation and preservation. QIAzol (Qiagen; 79306) can be substituted if preferred. |
| Worm Pick | NA | NA | Made in house from a pasteur pipette and a platinum wire. See wormbook for details. |
References
- Riddel, D. L., Blumenthal, T., Meyer, B. J., Priess, J. . C. elegans II. , (1997).
- Corsi, A. K., Wightman, B., Chalfie, M. A transparent window into biology: A primer on Caenorhabditis elegans. Genetics. 200 (2), 387-407 (2015).
- Zhang, F., et al. Natural genetic variation drives microbiome selection in the Caenorhabditis elegans gut. Current Biology. 31 (12), 2603-2618 (2021).
- Dirksen, P., et al. CeMbio – The Caenorhabditis elegans microbiome resource. G3: Genes, Genomes, Genetics. 10 (9), 3025-3039 (2020).
- Zhang, F., et al. Caenorhabditis elegans as a model for microbiome research. Frontiers in Microbiology. 8, 485 (2017).
- Roy, P. J., Stuart, J. M., Lund, J., Kim, S. K. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 418 (6901), 975-979 (2002).
- Pauli, F., Liu, Y., Kim, Y. A., Chen, P. -. J., Kim, S. K. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 133 (2), 287-295 (2005).
- Spencer, W. C., et al. A spatial and temporal map of C. elegans gene expression. Genome Research. 21 (2), 325-341 (2011).
- Spencer, W. C., et al. Isolation of specific neurons from C. elegans larvae for gene expression profiling. PLoS One. 9 (11), 112102 (2014).
- Ma, X., et al. Analysis of C. elegans muscle transcriptome using trans-splicing-based RNA tagging (SRT). Nucleic Acids Research. 44 (21), 156 (2016).
- Blazie, S. M., et al. Comparative RNA-Seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biology. 13 (1), 4 (2015).
- Blazie, S. M., et al. Alternative polyadenylation directs tissue-specific miRNA targeting in Caenorhabditis elegans somatic tissues. Genetics. 206 (2), 757-774 (2017).
- Gómez-Saldivar, G., et al. et al.Tissue-specific transcription footprinting using RNA PoI DamID (RAPID) in Caenorhabditis elegans. Genetics. 216 (4), 931-945 (2020).
- Katsanos, D., Barkoulas, M. Targeted DamID in C. elegans reveals a direct role for LIN-22 and NHR-25 in antagonizing the epidermal stem cell fate. Science Advances. 8 (5), 3141 (2022).
- Kaletsky, R., et al. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 529 (7584), 92-96 (2015).
- Kaletsky, R., et al. Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genetics. 14 (8), 1007559 (2018).
- Mathies, L. D., et al. mRNA profiling reveals significant transcriptional differences between a multipotent progenitor and its differentiated sister. BMC Genomics. 20 (1), 427 (2019).
- Liang, X., Calovich-Benne, C., Norris, A. Sensory neuron transcriptomes reveal complex neuron-specific function and regulation of mec-2/ Stomatin splicing. Nucleic Acids Research. 50 (5), 2401-2416 (2021).
- Glenwinkel, L., et al. In silico analysis of the transcriptional regulatory logic of neuronal identity specification throughout the C. elegans nervous system. eLife. 10, 64906 (2021).
- Taylor, S. R., et al. Molecular topography of an entire nervous system. Cell. 184 (16), 4329-4347 (2021).
- Charest, J., et al. Combinatorial action of temporally segregated transcription factors. Developmental Cell. 55 (4), 483-499 (2020).
- Steiner, F. A., Talbert, P. B., Kasinathan, S., Deal, R. B., Henikoff, S. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Research. 22 (4), 766-777 (2012).
- Tintori, S. C., Nishimura, E. O., Golden, P., Lieb, J. D., Goldstein, B. A transcriptional lineage of the early C. embryo. Developmental Cell. 38 (4), 430-444 (2016).
- Hashimshony, T., Wagner, F., Sher, N., Yanai, I. CEL-Seq: Single-cell RNA-Seq by multiplexed linear amplification. Cell Reports. 2 (3), 666-673 (2012).
- Hashimshony, T., Feder, M., Levin, M., Hall, B. K., Yanai, I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 519 (7542), 219-222 (2015).
- Packer, J. S., et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 365 (6459), 1971 (2019).
- Warner, A. D., Gevirtzman, L., Hillier, L. W., Ewing, B., Waterston, R. H. The C. elegans embryonic transcriptome with tissue, time, and alternative splicing resolution. Genome Research. 29 (6), 1036-1045 (2019).
- Cao, J., et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 357 (6352), 661-667 (2017).
- Durham, T. J., et al. Comprehensive characterization of tissue-specific chromatin accessibility in L2 Caenorhabditis elegans nematodes. Genome Research. 31 (10), 1952-1969 (2021).
- King, D. C., et al. The transcription factor ELT-2 positively and negatively impacts direct target genes to modulate the Caenorhabditis elegans intestinal transcriptome. bioRxiv. , (2021).
- Kocsisova, Z., Mohammad, A., Kornfeld, K., Schedl, T. Cell cycle analysis in the C. elegans germline with the thymidine analog EdU. Journal of Visualized Experiments. (140), e58339 (2018).
- Han, S., et al. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature. 544 (7649), 185-190 (2017).
- Edgar, L. G., Wolf, N., Wood, W. B. Early transcription in Caenorhabditis elegans embryos. Development. 120 (2), 443-451 (1994).
- Edgar, L. G., Goldstein, B. Culture and manipulation of embryonic cells. Methods in Cell Biology. 107, 151-175 (2012).
- Nishimura, E. O., Zhang, J. C., Werts, A. D., Goldstein, B., Lieb, J. D. Asymmetric transcript discovery by RNA-seq in C. elegans blastomeres identifies neg-1, a gene important for anterior morphogenesis. PLoS Genetics. 11 (4), 1005117 (2015).
- Stiernagle, T. Maintenance of C. elegans. WormBook. , 1-11 (2006).
- Porta-de-la-Riva, M., Fontrodona, L., Villanueva, A., Cerón, J. Basic Caenorhabditis elegans methods: Synchronization and observation. Journal of Visualized Experiments. (64), e4019 (2012).
- Fay, D. S., Fluet, A., Johnson, C. J., Link, C. D. In vivo aggregation of β-amyloid peptide variants. Journal of Neurochemistry. 71 (4), 1616-1625 (1998).
- Altun, Z. F., Hall, D. H. WormAtas Hermaphrodite Handbook – Introduction to C. elegans Anatomy. WormAtlas. , (2006).
- Tan, M. -. W., Mahajan-Miklos, S., Ausubel, F. M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 96 (2), 715-720 (1999).
- Oesterle, A. Pipette Cookbook 2018: P-97 & P-1000 Micropipette Pullers. Sutter Instrument Company. , (2018).
- Dineen, A., Nishimura, E. O., Goszczynski, B., Rothman, J. H., McGhee, J. D. Quantitating transcription factor redundancy: The relative roles of the ELT-2 and ELT-7 GATA factors in the C. elegans endoderm. Developmental Biology. 435 (2), 150-161 (2018).

