Isolation of Nuclei from Human Intermuscular Adipose Tissue and Downstream Single-Nuclei RNA Sequencing
Summary
The biology of intermuscular adipose tissue (IMAT) is largely unexplored due to the limited accessibility of human tissue. Here, we present a detailed protocol for nuclei isolation and library preparation of frozen human IMAT for single nuclei RNA sequencing to identify the cellular composition of this unique adipose depot.
Abstract
Intermuscular adipose tissue (IMAT) is a relatively understudied adipose depot located between muscle fibers. IMAT content increases with age and BMI and is associated with metabolic and muscle degenerative diseases; however, an understanding of the biological properties of IMAT and its interplay with the surrounding muscle fibers is severely lacking. In recent years, single-cell and nuclei RNA sequencing have provided us with cell type-specific atlases of several human tissues. However, the cellular composition of human IMAT remains largely unexplored due to the inherent challenges of its accessibility from biopsy collection in humans. In addition to the limited amount of tissue collected, the processing of human IMAT is complicated due to its proximity to skeletal muscle tissue and fascia. The lipid-laden nature of the adipocytes makes it incompatible with single-cell isolation. Hence, single nuclei RNA sequencing is optimal for obtaining high-dimensional transcriptomics at single-cell resolution and provides the potential to uncover the biology of this depot, including the exact cellular composition of IMAT. Here, we present a detailed protocol for nuclei isolation and library preparation of frozen human IMAT for single nuclei RNA sequencing. This protocol allows for the profiling of thousands of nuclei using a droplet-based approach, thus providing the capacity to detect rare and low-abundant cell types.
Introduction
Intermuscular adipose tissue (IMAT) is an ectopic adipose depot residing between and around muscle fibers1. As described in detail in a recent review by Goodpaster et al., IMAT can be detected using high-resolution computed tomography (CT) and magnetic resonance imaging (MRI) (Figure 1A,B) and is found around and within muscle fibers throughout the entire body1. The quantity of IMAT varies greatly between individuals and is influenced by BMI, age, sex, race, and sedentariness2,3,4. Moreover, IMAT deposition is commonly seen in pathological conditions associated with muscle degeneration5, and numerous studies have documented increased IMAT mass in individuals with obesity, type 2 diabetes, metabolic syndrome, and insulin resistance6,7,8,9. Nonetheless, the cellular and biological properties of IMAT are only beginning to be unraveled. The limited accessibility and the variation in IMAT locations and content throughout the body have challenged the collection of samples from this unique adipose depot2. Moreover, samples are easily 'contaminated' with skeletal muscle (SM) upon collection, making the separation between the biological contribution from the different tissues difficult to decipher (Figure 1C). To this end, single nuclei RNA sequencing (snRNA-seq), which has gained considerable attention during the last decade, serves as an ideal methodology to allow for the separation of IMAT- and SM-derived gene expression patterns with single-cell resolution. Moreover, nuclei isolation is particularly useful for adipose tissue due to the large lipid-laden adipocytes, which are impossible to dissociate into single-cell suspension without compromising the integrity of the cells. Lastly, this technology holds the potential to discover novel markers of IMAT-specific adipocytes and uncover the composition and presence of different progenitor cell populations, as well as study the variation of the cell composition in pathological and normal conditions.
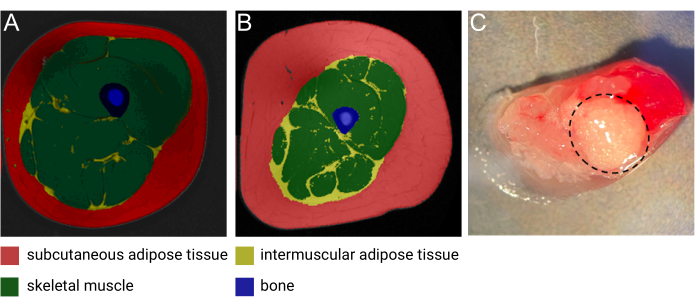
Figure 1: Images of IMAT. Representative magnetic resonance (MRI) image of IMAT from (A) a middle-aged lean female and (B) a middle-aged male with obesity. Red: subcutaneous adipose tissue, yellow: intermuscular adipose tissue, green: skeletal muscle, blue: bone. Image courtesy of Heather Cornnell, AdventHealth Translational Research Institute. (C) Fresh tissue sample with IMAT (encircled by dashed black line). Image courtesy of Meghan Hopf, AdventHealth Translational Research Institute and Bryan Bergman, University of Colorado. This figure has been modified with permission from Goodpaster et al.1. Please click here to view a larger version of this figure.
A number of studies have been published from the livestock industry investigating the marbling of meat (IMAT in particular) in pigs, chickens, and cattle using single-cell (sc) and snRNA-seq10. These studies have identified several subpopulations of adipocytes and markers of potential progenitor cells of IMAT11,12,13; however, whether these cellular compositions translate to human IMAT is unknown. To our knowledge, only one study has looked into the cellular heterogeneity of human muscle with fatty infiltration, obtained from male patients with hip osteoarthritis, using snRNA-seq14. The investigators reported a small adipocyte population and several fibro-adipogenic progenitor (FAP) subpopulations within the large population of myonuclei14. Our study is the first to develop a method to directly interrogate IMAT manually dissected from human muscle for cellular composition using snRNA-seq.
Importantly, protocols for snRNA-seq need to be customized for the specific tissue studied, as the amount of tissue available and the physical properties of the specific tissue will dictate the optimal processing steps. The tissue yield for IMAT is typically small, often not exceeding 50 mg, even when performing ultrasound-guided biopsies. Hence, proper processing of this scarce tissue is essential. We believe that this protocol will serve as a valuable resource for researchers studying human IMAT.
Protocol
The sample used for this protocol was part of the Study of Muscle, Mobility, and Aging (SOMMA)15, which was approved by the Western IRB-Copernicus Group (WCG) Institutional Review Board and was carried out in accordance with the Declaration of Helsinki. Participants provided written informed consent for their participation in the study.
NOTE : This protocol is adapted from a previous protocol using 100 mg of human abdominal subcutaneous adipose tissue on a nanowell-based platform16. The current protocol is optimized for 50 mg of human IMAT and library preparation using a droplet-based platform. Further optimization of this protocol for nuclei isolations from non-human IMAT or other adipose depots may be required.
1. Preparation of buffers and reagents (Table 1 and Table 2)
NOTE: Prepare buffers fresh on the day of the experiment and do not re-use.
- Pre-cool a centrifuge to 4 °C.
- Prepare homogenization buffer and nuclei isolation medium.
- Obtain two buckets of ice and pre-cool 2 x 15 mL conical tubes.
- Mix all reagents for the homogenization buffer (HB) in a 15 mL conical tube in the order listed in Table 1. Keep on ice. Mix by vortexing.
- Mix all reagents for nuclei isolation medium (NIM) in a 15 mL conical tube in the ordered list in Table 2. Keep on ice. Mix by vortexing.
- Prepare 10% Triton-X by adding 100 µL of Triton X-100 to 900 µL of Nuclease-free water. Vortex to ensure proper mixing. Keep at room temperature (RT).
| Reagent | Volume (μL) | Final concentration (mM) | |
| 1x | 2x | ||
| 1 M MgCl2 | 10 | 20 | 5 |
| 1 M Tris Buffer, pH 8.0 | 20 | 40 | 10 |
| 2 M KCl | 25 | 50 | 25 |
| 1.5 M Sucrose (-4oC) | 334 | 668 | 250 |
| 1mM DTT | 2 | 4 | 0.001 (~1 µM) |
| 100x protease inhibitor | 20 | 40 | 1x |
| Superasin 20 U/µL | 40 | 80 | 0.4 U/µL |
| Nuclease-free water | 1549 | 3098 | – |
| Total Volume | 2000 | 4000 | – |
Table 1: Homogenization buffer (HB). Keep on ice. Mix by vortexing.
| Reagent | Volume (μL) | Final concentration (mM) | |
| 1x | 2x | ||
| EDTA | 0.4 | 0.8 | 0.1 |
| Ribolock RNAse inhibitor (40U/µL) | 40 | 80 | 0.8 U/µL |
| 1% BSA-PBS (-/-) | 1959.6 | 3919.2 | – |
| Total Volume | 2000 | 4000 | – |
Table 2: Nuclei isolation medium (NIM). Keep on ice. Mix by vortexing.
2. Pulverization of frozen tissue ( Figure 2A)
- Set up the workstation for homogenization.
- Fill up the cannister with liquid nitrogen (LN2).
CAUTION: when working with LN2, always wear goggles and cryo gloves. - Obtain 2x mortars, 1x pestle, 1x micro-scoop spatula, 1x glass dounce, and 1x stainless-steel pestle for the automatic douncer.
- Set up the automatic douncer.
- Fill a beaker with ice and pre-cool the glass dounce.
- Fill up the cannister with liquid nitrogen (LN2).
- Fill the 2 mortars (containing the pestle and spatula) with LN2 to cool down the instruments. Let the LN2 evaporate and repeat.
- While instruments are cooling, add 1 mL of HB to the glass dounce.
- Fill both mortars with LN2 one last time and pour the 50 mg IMAT sample into one of the mortars.
- Pulverize the IMAT using the pestle by gently pressing it down on the piece of tissue to break it into small pieces. Make sure all pieces are pulverized.
- The LN2 will slowly evaporate while pulverizing the tissue. When tissue is properly pulverized, and there is still ¼ – ½ of a mortar of LN2 left, tilt the mortar towards the lip of the mortar to collect the pulverized tissue by the lip. Let the LN2 evaporate completely.
- Immediately after the last LN2 has evaporated, scoop the pulverized tissue into the glass dounce containing 1 mL of HB.
3. Homogenization of pulverized tissue
- Homogenize the pulverized tissue using the automatic douncer. Bring the glass dounce up and down the stainless-steel pestle for 10 strokes in the forward direction, followed by 10 strokes in the reverse direction.
- Ensure that the solution is cloudy after homogenization and contains no visible pieces of tissue. A light pink color is often expected due to contamination with muscle tissue.
- Transfer the homogenate to a pre-cooled 1.7 mL low-bind tube on ice.
- Use 400 µL of HB to rinse the dounce to make sure all the material is transferred and add it to the tube.
NOTE: Two samples can be processed at a time. To do this, double the amount of HB and NIM. Pulverize and homogenize one tissue sample and immediately thereafter pulverize and homogenize the second tissue sample to be able to perform the isolation and clean-up steps in parallel.
4. Nuclei isolation and clean-up ( Figure 2B)
- Add 14 µL of Triton-X (10%) to the homogenate for a 0.1% concentration.
- Keep the tube on ice and in the dark for 10-15 min while vortexing every 3 min.
- Pre-wet one 100 µm and one 40 µm cell strainer (per sample) with 100 µL of RT DPBS for each in a 50 mL conical tube.
- Filter the homogenate through the 100 µm cell strainer.
- Rinse the 1.7 mL tube with 400 µL of HB and filter through the 100 µm cell strainer.
- Next, filter the solution through the 40 µm cell strainer.
- Transfer an equal amount of solution into two pre-cooled 1.7 mL low-bind tubes corresponding to ~900 µL in each tube.
- Centrifuge the tubes for 10 min at 2700 x g at 4 °C. There should be a small pellet visible after centrifugation.
- Remove and discard the top lipid layer and the remaining supernatant, leaving ~50 µL solution from the first tube.
- Repeat for the second tube.
- Thoroughly resuspend the pellet in the first tube by gently pipetting up and down 20x and transfer to a new 1.7 mL low-bind tube. Avoid creating bubbles.
- Repeat this procedure for the second tube and transfer the resuspended solution to the same tube.
- Add 500 µL of NIM and mix with pipetting.
- Centrifuge the tube with a balance at 1000 x g for 10 min at 4 °C.
- Remove supernatant, leaving ~50 µL, and gently pipette up and down until the pellet is resuspended. Optionally, transfer the resuspended pellet to a new clean tube if some lipid leftover remains on the side of the tube.
- Add 200 µL of NIM and mix by pipetting.
5. Nuclei staining and counting (Figure 2C and Figure 3)
NOTE : To facilitate the counting, set up a 'nuclei counting' protocol on an automated cell counter, as adjusting the bright field and DAPI channels can affect the count greatly. Adjust the channels so that only nuclei and not debris are captured. Make sure the bright field channel only marks 'objects' that also have a DAPI stain.
- Add 1 drop of the live cell staining solution and leave it in the dark, on ice, for 15 min.
- Filter the solution through a 30 µm cell strainer.
- Mix the nuclei solution by pipetting and add 10 µL of the solution to a cell counting chamber slide.
- Count nuclei using an automated cell counter.
NOTE: The optimal concentration is 1000 nuclei/µL corresponding to 1.0 x 106/mL.- Ensure no clumps of nuclei are present, as this could clog the chip for generating single nuclei droplets (Figure 3).
- If the nuclei concentration is not high enough, spin down the solution at 1000 x g for 10 min at 4 °C to gain a pellet, remove the supernatant, and resuspend in a smaller volume.
- If the degree of debris in the solution is high, resuspend the nuclei solution in a larger volume of NIM (i.e., 1 mL) and filter through a 30 µm cell strainer again. Then spin down at 1000 x g for 10 min at 4 °C and resuspend in appropriate volume in relation to the concentration of nuclei.
- After obtaining the nuclei concentration, proceed directly to the first step in the library preparation.
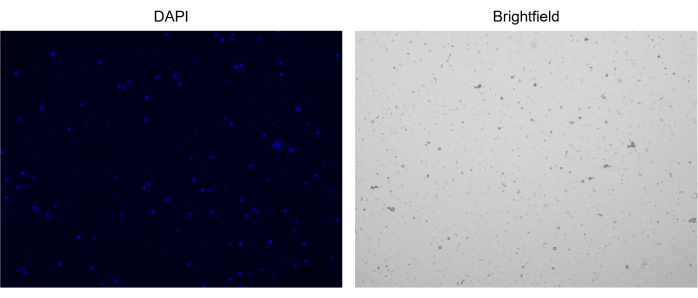
Figure 3: Staining of isolated nuclei. Image from cell counter of nuclei stained with NucBlue/DAPI (left image) and the corresponding bright field image (right image). The presence of minor amounts of debris is evident in the bright field image. The automated cell counter used here does not have an option to include scale bars. Please click here to view a larger version of this figure.
6. Library preparation and sequencing parameters
- Refer to a thorough protocol for library preparation using the droplet-based single nuclei approach available on the provider's webpage17.
- Aim for a targeted nuclei recovery of 10,000. However, for samples with a high level of debris or fragile nuclei, a lower number of nuclei recovered would be anticipated.
- Store the samples at 4°C for up to 72 h after step 2.3. in the library preparation protocol to combine the processing of more samples in parallel. Do this by processing two samples until step 2.3 on two consecutive days, and on the third day, process the 4 samples together from step 3 and forward in the library preparation protocol.
- Sequencing parameters: Sequence on a sequencing platform aiming for 50,000 paired-end reads per nuclei.
NOTE: The data presented in this protocol was sequenced on the NovaSeq 6000 platform, aiming for 50,000 paired-end reads per nuclei.
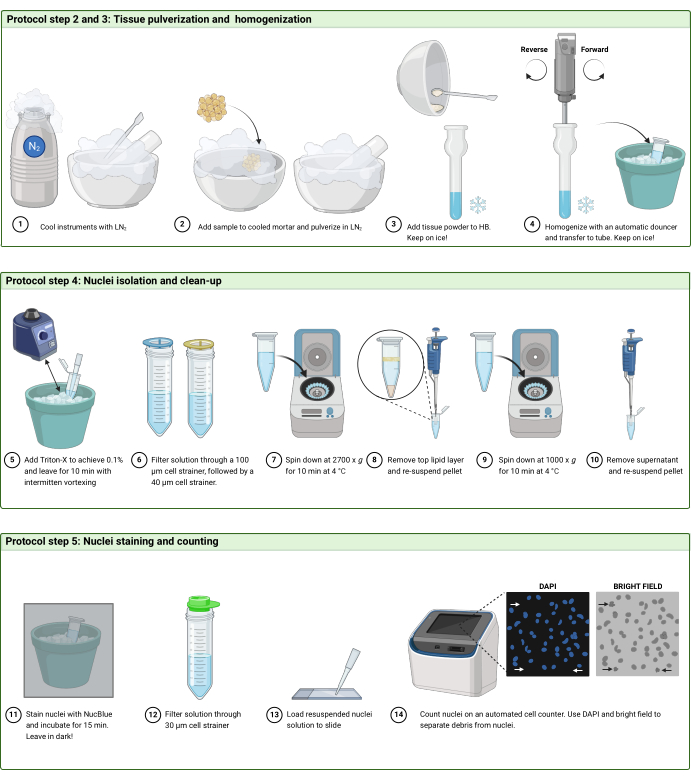
Figure 2: Protocol workflow. Schematic illustration of the workflow in (A) steps 2 and 3, (B) step 4, and (C) step 5 of the protocol. The figure was created with BioRender.com. Please click here to view a larger version of this figure.
7. Data processing and analysis
NOTE : In this protocol, some of the recommended software and R packages used to process the resulting sequencing data are briefly introduced, focusing on the steps after the initial pre-processing (Table 3). This study provides general quality control (QC) metrics and an example uniform manifold approximation and projection (UMAP) in Figure 4. However, an in-depth description of the bioinformatic analysis is out of the scope of this protocol. Therefore, readers can refer to the recent review on best practices for single-cell analysis by Heumos et al.18.
- Pre-processing of sequencing data
- Map the single nuclei reads to the human reference genome GRCh38.
- Include intron reads in the count.
- Perform QC and filtering of the data using the Seurat R package19.
- Compute a cell complexity score by dividing the log(10) number of genes detected by the log(10) number of reads detected.
- Plot out the most important QC metrics using a histogram or violin plot, including the number of genes detected per nuclei, mitochondrial read percentage, and cell complexity score.
- Filter out nuclei with less than 200 or more than 10,000 genes per nuclei, greater than 10% mitochondrial reads, and a complexity score below 0.8.
- Normalize data and perform dimensionality reduction.
- Use the SCTransform function from Seurat to normalize the data using 2000 variable features.
- Cluster data using the following functions from the Seurat R package: RunPCA, FindNeighbors, FindClusters, and RunUMAP.
- Plot a UMAP to visualize the clustering of the data.
- Filter out predicted doublets using the DoubletFinder R package20 and re-cluster data.
- Annotate clusters using known gene markers of the cell types expected to be present in the tissue (supervised approach) or based on the top 5 differentially expressed genes between the clusters (unsupervised approach).
- Use decontX21 to determine the degree of ambient RNA contamination and to adjust the gene expression matrix for ambient RNA.
- Include the raw gene matrix as background.
- Save the Seurat object for future exploration of the data.
NOTE: The code for QC and clustering analysis is available in Supplementary File 1.
| Software/R packages used in data workflow | Alternative software/packages | Processing step |
| CellRanger | STARsolo, kallisto | Trimming, alignment, mapping |
| Seurat | SingleCellExperiment, Cellranger | QC, analysis and data exploration |
| DoubletFinder | scds, scdblFinder, Scrublet | Doublet detection |
| DecontX | SoupX, CellBender | Ambient RNA adjustment |
Table 3: Software/tools for data workflow.
Representative Results
This workflow was designed to guide the processing of frozen human IMAT samples to obtain gene expression profiles at single nuclei resolution, enabling cell type identification. Here, one representative IMAT sample from a participant in the SOMMA study is presented.
The first step of any analysis of snRNA-seq data is to evaluate the quality of the data to identify poor-quality nuclei, which should potentially be removed from the dataset. Importantly, the filtering steps and thresholds should be determined for the specific type of sample and dataset you have in hand, as the commonly evaluated metrics can differ among tissues and cell types22,23. Figure 4A provides visuals of some of the key metrics used to assess the quality of the generated snRNA-seq data. The number of genes detected per nuclei is dependent on sequencing depth and cell type but would be expected to be above 200 for good-quality nuclei18,23. It was found that the data generated using this protocol is within the expected range with a median of 1134 genes per nuclei, from a total of 4662 nuclei.
The percentage of mitochondrial reads is evaluated since a high degree of mitochondrial contamination can arise from damaged nuclei or ambient RNA attaching to the nuclei, indicating poor-quality nuclei. In the dataset presented here, a median mitochondrial read percentage of 2.65 was found, which is well below the 5%-20% threshold commonly used in the literature24,25,26. The percentage of ribosomal reads differs among cell types and tissues. However, as large proportions of ribosomal genes can influence the clustering of the data, it is recommended to check the ribosomal read percentage and potentially remove ribosomal genes or nuclei with high levels of ribosomal genes from the dataset before clustering. The data generated with this protocol showed a low level of ribosomal reads with a median of 2.46% and a maximum of 16.5%, and therefore, we did not filter based on this metric. Lastly, a cell complexity score was calculated based on the log(10) number of genes detected divided by the log(10) number of reads detected. Good-quality nuclei are expected to be above 0.8, and a median of 0.92 was obtained in the sample used in this study. Based on these QC metrics, one can decide which nuclei to filter out of the dataset. For analysis, we chose to filter out nuclei with less than 200 or more than 10,000 genes per nuclei, greater than 10% mitochondrial reads, and a complexity score below 0.8.
Following the initial quality assessment and filtering step, a UMAP can be generated to visualize clustering of the nuclei. Clustering was performed based on the 2000 most variable genes using SCT transformation. The initial clustering steps can be used to check if any of the QC features cluster together, e.g., nuclei with high mitochondrial reads. Moreover, clustering information is required for some doublet detection methods, including DoubletFinder20, which was used in this protocol. DoubletFinder was used with an expected multiplet rate set to 4.8% as suggested by the providers of the droplet-based platform. After doublet removal, the level of ambient RNA contamination was estimated, which is particularly common in single nuclei preparations as RNA is released from the cytoplasm upon cell lysis and gets dispensed into the Gel Beads-in-emulsion (GEMs) and amplified in the following library preparation steps. Hence, several tools have been developed to correct the inherent problem of ambient RNA contamination (see Table 3). We used the R package decontX21, in which the raw background matrix (including only empty droplets) is used to adjust the gene expression matrix, enhancing the real gene expression signature.
The clustering and ability to detect low abundant cell types depend on the number of nuclei. This study detected all expected major cell types in IMAT (Figure 4B) from a total of 3817 nuclei after QC filtering, doublet removal, and ambient RNA adjustment. These included stem cells, fibro-adipogenic progenitors (FAPs), and mature adipocytes, as well as pericytes, smooth muscle cells, immune cells, muscle progenitor cells, and myonuclei from skeletal muscle cell contamination.
Overall, we have demonstrated that this protocol produces high-resolution single nuclei data enabling detection of cell type annotation important for unravelling the biology and cellular origins of IMAT.
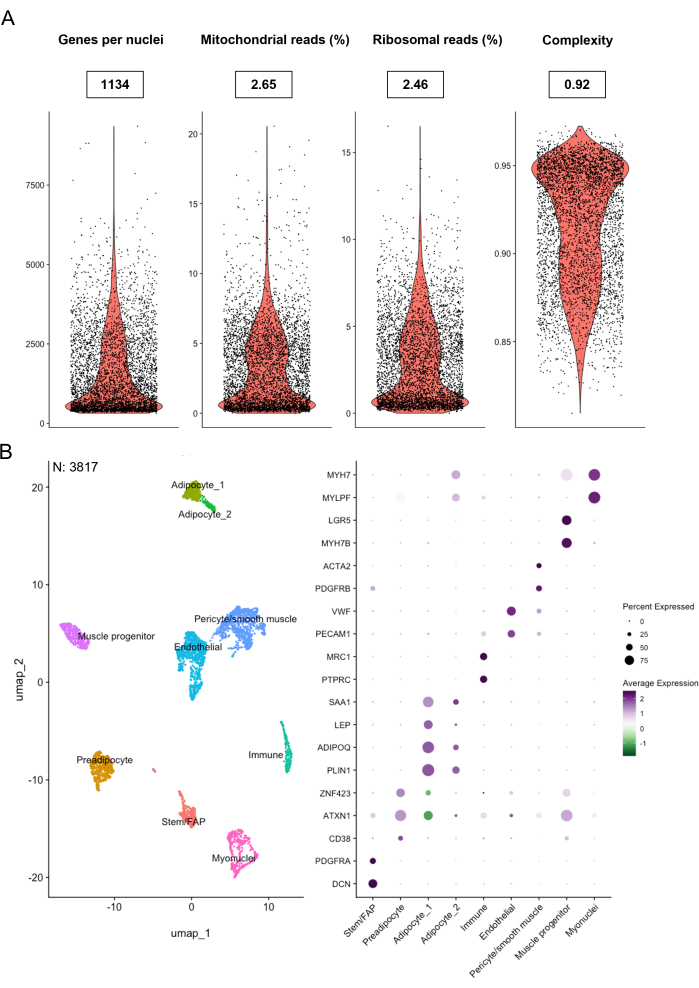
Figure 4: Quality assessment, clustering, and cell type annotation of sequencing data. (A) Violin plots of essential metrics for evaluation of the sample and sequencing performance, including the number of genes detected per nuclei, percentage of mitochondrial reads, percentage of ribosomal reads, and cell complexity measured as the log(10) number of genes detected divided by the log(10) number of reads detected. Median values for each metric are given in closed boxes. Total number of nuclei: 4662. (B) UMAP displaying clustering of individual nuclei and corresponding DotPlot showing relative gene expression of cell type marker genes for each cluster after filtering. Number of nuclei: 3817. Please click here to view a larger version of this figure.
Supplementary File 1: The code for QC and clustering analysis. Please click here to download this File.
Discussion
There are several inherent challenges to working with IMAT. In addition to its limited accessibility, the yield of sample material is often very scarce, and "contamination" of skeletal muscle is almost impossible to avoid. To obtain the best quality sample, one should penetrate the muscle fascia when inserting the biopsy needle (to make sure not to collect subcutaneous adipose tissue) and remove as much muscle tissue as possible by dissecting the sample under a microscope immediately after collection, followed by subsequent processing for histology, snap-freezing, etc. This is the first method to utilize IMAT samples prospectively collected and dissected from skeletal muscle. Comparatively, previous snRNA-seq data in IMAT has been through nuclei isolations of a mixture of skeletal muscle biopsies with known fatty infiltration and not the IMAT alone.
IMAT is denser in nature compared to subcutaneous adipose tissues, making proper pulverization of the tissue during nuclei isolation essential to release nuclei from the tissue structure. Improper pulverization can lead to a lower nuclei yield, which is unfavorable with a limited amount of tissue. Moreover, calculating the correct number of nuclei in the prepared nuclei solution is a critical step to ensure enough (but not too many) nuclei to load onto the chip for GEM generation. Too many nuclei, or the presence of large nuclei clusters or debris, can result in a clog, affecting the final cDNA concentration and, ultimately, the sequencing output. Circumventing nuclei clumping and debris in the single nuclei preparation is difficult and affected by the quality of the sample. Nuclei clumping and/or the presence of debris can be reduced by including an extra filtering step with the 30 µm cell strainer.
It is important to process samples quickly to reduce the risk of RNA degradation. Keeping solutions on ice or cooled during all possible steps of nuclei isolation, as well as the addition of a sufficient amount of RNAse inhibitors, can minimize degradation. Due to the importance of processing time for each sample, we recommended running a maximum of two samples at a time. Moreover, the nuclei solution must be handled with care to not make the nuclei too fragile. Fragile nuclei are susceptible to bursting upon GEM generation, which can result in large amounts of ambient RNA in the sample. In this protocol, the filtering step using a syringe (described in the STAR protocol16) was excluded to preserve the nuclei integrity.
The choice of library preparation method depends on the research question. We have worked with both a droplet-based and nano-well-based single nuclei approach. The nano-well-based approach profiles 1200-1600 nuclei and approximately 3000-6000 detected genes per nuclei16. Moreover, this approach supports full-length transcriptomics, which allows for the investigation of structural variation, pseudo-genes, and splice variants27. In comparison, more than 10,000 nuclei can be profiled using the droplet-based approach, but at the expense of a lower number of genes detected per nuclei (2-3-fold lower). For this protocol, we have chosen to use one of the droplet-based platforms as the larger number of nuclei profiled gives us a greater capacity to detect rare cell types with a low abundance28.
The major limitation of snRNA-seq is the loss of cytosolic content, restricting the expression analysis to nuclear transcripts. The nucleus contains 10-100-fold less mRNA than that of the entire cell29, which could be of concern for uncovering all cell types in a tissue. However, a recent study by Gupta et al. found that nuclear transcriptomics – compared to whole-cell transcriptomics29– detected similar cell populations and expression profiles of biologically relevant genes in cultured preadipocytes and mature adipocytes. Similar results have been found in other cell types30,31.
Single nuclei isolation has its advantages in providing single cell (sc) resolution gene expression patterns of fragile cells incompatible with scRNA-seq. Moreover, frozen biobank samples can be used, and nuclei isolation does not require enzymatic digestion of the tissue, which can induce a stress response in the cells, affecting the transcriptome32. By using snRNA-seq, we were able to transcriptionally profile adipocytes and other cell types present in IMAT, which would not be possible with scRNA-seq due to adipocytes being lipid-laden and incompatible with the droplet-based approach.
An important application of snRNA-Seq of IMAT is to decipher the cell composition and transcriptional differences of IMAT in varying metabolic states. Furthermore, snRNA-seq datasets can be used to deconvolute bulk RNA-seq on IMAT to assess cell compositional differences on a larger scale33. Additionally, pseudo-time analysis of snRNA-seq data can serve to identify trajectories of cell maturation, which is of particular interest in tissues such as IMAT, in which the cellular origins of the mature adipocytes are still debated.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge Bryan Bergman, PhD at University of Colorado for providing the image of the IMAT biopsy in Figure 1C from the MoTrIMAT study (R01AG077956). We are grateful for the Study of Muscle, Mobility and Aging providing the IMAT sample from which data is shown in the representative results section. The National Institute on Aging (NIA) funded the Study of Muscle, Mobility and Aging (SOMMA; R01AG059416) and its ancillary studies SOMMA AT (R01AG066474) and SOMMA Knee OA (R01AG070647). Study infrastructure support was funded in part by NIA Claude D. Pepper Older American Independence Centers at University of Pittsburgh (P30AG024827) and Wake Forest University (P30AG021332) and the Clinical and Translational Science Institutes, funded by the National Center for Advancing Translational Science, at Wake Forest University (UL1 0TR001420).
Materials
| 0.2 µm corning syringe filters | Millipore Sigma | CLS431229 | |
| 1.7 mL DNA LoBind tubes | Eppendorf | 22431021 | low-bind tubes |
| 10% Tween 20 | Bio-Rad | 1662404 | |
| 100x protease inhibitor | Thermo Fisher Scientific | 78437 | |
| 10X Magnetic Separator | 10X Genomics | 230003 | |
| 10X Vortex Adapter | 10X Genomics | 330002 | |
| 15 mL canonical tubes | Sarstedt | 6,25,54,502 | |
| 2100 Bioanalyzer | Agilent | G2939BA | |
| 50 mL conical tubes | Sarstedt | 6,25,47,254 | |
| CellRanger | Genomics | N/A | |
| Chromium iX accesory kit | 10X Genomics | PN1000323 | |
| Chromium iX Controller | 10X Genomics | PN1000326 | |
| Chromium Next GEM Chip G Single Cell Kit | 10X Genomics | PN1000127 | |
| Chromium Next GEM Single Cell 3' Kit v 3.1 | 10X Genomics | PN1000269 | |
| Chromium Next GEM Single Cell 3' Gel Bead Kit v3.1 | 10X Genomics | PN1000129 | |
| Chromium Next GEM Single Cell GEM Kit v3.1 | 10X Genomics | PN1000130 | |
| Countess 3 Automated Cell Counter | Thermo Fisher Scientific | AMQAX2000 | Automated cell counter |
| Countess cell counting chamber slides | Thermo Fisher Scientific | C10228 | |
| DoubletFinder | R | N/A | |
| DPBS (no calcium, no magnesium) | Thermo Fisher Scientific | 14190144 | |
| DTT | Thermo Fisher Scientific | R0861 | |
| Dual Index Kit TT Set A, 96 rxns | 10X Genomics | PN1000215 | |
| Dynabeads MyOne SILANE | 10X Genomics | PN2000048 | |
| Falcon 100 µm Cell strainer | Corning Life Science | 352360 | |
| Falcon 40 µm Cell strainer | Corning Life Science | 352340 | |
| Glycerin (glycerol), 50% (v/v) Aqueous Solution | Ricca Chemical Company | 3290-32 | |
| KCL | Thermo Fisher Scientific | AM9640G | |
| Library Construction Kit v3.1 | 10X Genomics | PN1000196 | |
| MACS SmartStrainers (30µm) | Miltenyi Biotec | 130-098-458 | |
| Mastercycler Nexus Gradient Thermal cycler | Eppendorf | 6331000017 | |
| MgCl2 | Ambion | AM9530G | |
| Mortar and pestel | Health care logistics | 14075 | |
| NucBlue Live Ready Probes Reagent | Thermo Fisher Scientific | R37605 | |
| Nuclease Free Water (not DEPC treated) | Thermo Fisher Scientific | AM9930 | |
| Probumin Bovine Serum Albumin Fatty Acid Free, Powder | Sigma-Aldrich | 820024 | |
| Qiagen Buffer EB | Qiagen | 19086 | |
| Ribolock RNAse inhibitor | Thermo Fisher Scientific | EO0382 | |
| Seurat | R | N/A | |
| Sucrose | Sigma-Aldrich | S0389 | |
| SUPERasin 20 U/µL | Thermo Fisher Scientific | AM2695 | |
| ThermoMixer C | Eppendorf | 5382000015 | |
| Tissue homogenizer | Glass-Col | 099C K54 | |
| Tris buffer pH 8.0 | Thermo Fisher Scientific | AM9855G | |
| Triton X-100 | Thermo Fisher Scientific | AC327372500 | |
| UltraPure 0.5M EDTA pH 8.0 | Gibco | 15575020 |
References
- Goodpaster, B. H., Bergman, B. C., Brennan, A. M., Sparks, L. M. Intermuscular adipose tissue in metabolic disease. Nat Rev Endocrinol. 19 (5), 285-298 (2023).
- Sparks, L. M., Goodpaster, B. H., Bergman, B. C. The metabolic significance of intermuscular adipose tissue: Is IMAT a friend or a foe to metabolic health. Diabetes. 70 (11), 2457-2467 (2021).
- Gallagher, D., et al. Adipose tissue in muscle: A novel depot similar in size to visceral adipose tissue. Am J Clin Nutr. 81 (4), 903-910 (2005).
- Manini, T. M., et al. Reduced physical activity increases intermuscular adipose tissue in healthy young adults. Am J Clin Nutr. 85 (2), 377-384 (2007).
- Addison, O., Marcus, R. L., LaStayo, P. C., Ryan, A. S. Intermuscular fat: A review of the consequences and causes. Int J Endocrinol. 2014, 309570 (2014).
- Goodpaster, B. H., et al. Obesity, regional body fat distribution, and the metabolic syndrome in older men and women. Arch Intern Med. 165 (7), 777-783 (2005).
- Goodpaster, B. H., Thaete, F. L., Kelley, D. E. Thigh adipose tissue distribution is associated with insulin resistance in obesity and in type 2 diabetes mellitus. Am J Clin Nutr. 71 (4), 885-892 (2000).
- Goodpaster, B. H., et al. Association between regional adipose tissue distribution and both type 2 diabetes and impaired glucose tolerance in elderly men. Diabetes Care. 26 (2), 372-379 (2003).
- Sachs, S., et al. Intermuscular adipose tissue directly modulates skeletal muscle insulin sensitivity in humans. Am J Physiol Endocrinol Metab. 316 (5), E866-E879 (2019).
- Ford, H., Liu, Q., Fu, X., Strieder-Barboza, C. White adipose tissue heterogeneity in the single-cell era: From mice and humans to cattle. Biology (Basel). 12 (10), 1289 (2023).
- Wang, L., et al. Single-nucleus and bulk RNA sequencing reveal cellular and transcriptional mechanisms underlying lipid dynamics in high marbled pork. NPJ Sci Food. 7 (1), 23 (2023).
- Li, J., et al. Identification of diverse cell populations in skeletal muscles and biomarkers for intramuscular fat of chicken by single-cell RNA sequencing. BMC Genomics. 21 (1), 752 (2020).
- Lyu, P., Qi, Y., Tu, Z. J., Jiang, H. Single-cell RNA sequencing reveals heterogeneity of cultured bovine satellite cells. Front Genet. 12, 742077 (2021).
- Fitzgerald, G., et al. MME+ fibro-adipogenic progenitors are the dominant adipogenic population during fatty infiltration in human skeletal muscle. Commun Biol. 6 (1), 111 (2023).
- Cummings, S. R., et al. The study of muscle, mobility and aging (SOMMA): A unique cohort study about the cellular biology of aging and age-related loss of mobility. J Gerontol A Biol Sci Med Sci. 78 (11), 2083-2093 (2023).
- Whytock, K. L., et al. Isolation of nuclei from frozen human subcutaneous adipose tissue for full-length single-nuclei transcriptional profiling. STAR Protoc. 4 (1), 102054 (2023).
- 10x Genomics. Chromium Single Cell 3′ Reagent Kits User Guide (v3.1 Chemistry Dual Index), Document Number CG000315 RevE Available from: https://cdn.10xgenomics.com/image/upload/v1668017706/support-documents/CG000315_ChromiumNextGEMSingleCell3-_GeneExpression_v3.1_DualIndex__RevE.pdf (2022)
- Heumos, L., et al. Best practices for single-cell analysis across modalities. Nat Rev Genet. 24 (1), 550-572 (2023).
- Hao, Y., et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. 42 (2), 293-304 (2023).
- McGinnis, C. S., Murrow, L. M., Gartner, Z. J. DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8 (4), 329-337 (2019).
- Yang, S., et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol. 21 (2), 57 (2020).
- Common considerations for quality control filters for single cell RNA-seq data. 10X Genomics Available from: https://www.10xgenomics.com/analysis-guides/common-considerations-for-quality-control-filters-for-single-cell-rna-seq-data (2022)
- Luecken, M. D., Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 15 (6), e8746 (2019).
- Emont, M. P., et al. A single-cell atlas of human and mouse white adipose tissue. Nature. 603 (7903), 926-933 (2022).
- Hildreth, A. D., et al. Single-cell sequencing of human white adipose tissue identifies new cell states in health and obesity. Nat Immunol. 22 (5), 639-653 (2021).
- Whytock, K. L., et al. Single cell full-length transcriptome of human subcutaneous adipose tissue reveals unique and heterogeneous cell populations. iScience. 25 (8), 104772 (2022).
- Probst, V., et al. Benchmarking full-length transcript single cell mRNA sequencing protocols. BMC Genomics. 23 (1), 860 (2022).
- CG000148 Rev A Technical Note – Resolving cell types as a function of read depth and cell number. Technical note. 10X Genomics Available from: https://assets.ctfassets.net/an68im79xiti/6gDArDPBTOg4IIkYEO2Sis/803be2286bb (2018)
- Gupta, A., et al. Characterization of transcript enrichment and detection bias in single-nucleus RNA-seq for mapping of distinct human adipocyte lineages. Genome Res. 32 (2), 242-257 (2022).
- Bakken, T. E., et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 13 (12), e0209648 (2018).
- Wu, H., Kirita, Y., Donnelly, E. L., Humphreys, B. D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol. 30 (1), 23-32 (2019).
- Kim, N., Kang, H., Jo, A., Yoo, S. -. A., Lee, H. -. O. Perspectives on single-nucleus RNA sequencing in different cell types and tissues. J Pathol Transl Med. 57 (1), 52-59 (2023).
- Avila Cobos, F., Alquicira-Hernandez, J., Powell, J. E., Mestdagh, P., De Preter, K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun. 11 (1), 5650 (2020).

