환경 검체에서 박테리오파지 검출
English
Share
Overview
출처: 이안 페퍼 박사와 찰스 게르바 박사의 연구소 – 애리조나 대학교
데모 저자: 알렉스 와시미
바이러스는 진핵생물과 대핵생물을 모두 감염시키는 생물학 실체의 유일한 그룹입니다. 그(것)들은 신진 대사 능력이 없는 의무 기생충이고, 복제하기 위하여, 호스트 물질 대사에 의지하여 호스트 세포 안쪽에 자기 조립하는 바이러스성 부분을 생성합니다.
바이러스는 초미세이며, 전자 현미경의 더 큰 해상도로만 볼 수있는 가벼운 현미경으로 볼 수 없을 정도로 작습니다. 바이러스 성 입자는 단백질 하위 단위 또는 카포머로 구성된 캡슐화로 알려진 단백질 코트로 둘러싸인 DNA 또는 RNA 인 핵산 게놈으로 구성됩니다. 좀 더 복잡한 바이러스에서 캡시드는 추가 지질 봉투로 둘러싸여 있으며 일부는 스파이크 모양의 표면 부속물이나 꼬리를 가지고 있습니다.
인간과 동물의 장관을 감염시키는 바이러스는 장 바이러스로 알려져 있습니다. 배설물에서 배설되어 국내 폐수에서 분리될 수 있습니다. 박테리아를 감염시키는 바이러스는 박테리오파주로 알려져 있으며, 대장균 균을 감염시키는 바이러스는 대장균(도1)이라고합니다. 대장균 박테리아의 파지는 대장균 박테리아가 발견되는 곳이면 어디에서나 발견됩니다.
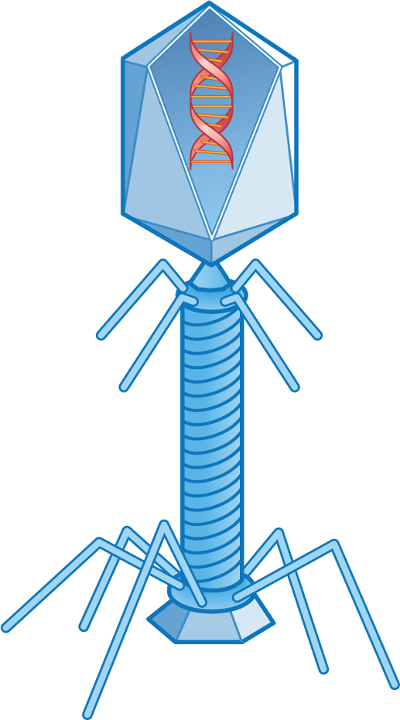
그림 1. 대장균 T2.
Principles
Procedure
Results
Dilution of sewage sample = 10-1
Number of plaques obtained = 9
Therefore, phage concentration in sewage sample
= 10 x 9 ÷ 1 mL
= 90 plaque-forming units / mL
Raw sewage typically contains 103 – 104 coliphage per mL, with a range of 102 – 108 per mL.
Applications and Summary
There are many potential applications of coliphages as environmental indicators. These include their use as indicators of sewage contamination, efficiency of water and wastewater treatment, and survival of enteric viruses and bacteria in the environment. The use of bacteriophages as indicators of the presence and behavior of enteric bacteria and animal viruses has always been attractive because of the ease of detection and low cost associated with phage assays. In addition, they can be quantified in environmental samples within 24 h as compared to days or weeks for enteric viruses.
1E. coli strain ATCC 15597 usually will produce the greatest number of plaques from sewage samples. It should be grown overnight in a 250-mL Erlenmeyer flask containing 100 mL of nutrient or trypticase soy broth and incubated under shaking conditions at 35 °C. 3 h before the phage assay inoculate one mL of this culture into a fresh flask containing 100 mL of nutrient or trypticase soy broth and place in a shaking water bath at 35-37 °C. This will ensure that the bacteria are in the log phase of growth.
Transcript
Viruses are infectious biological particles that are responsible for many diseases, cold and flu, to hepatitis and HIV.
Viruses are biological particles consisting of either a DNA or RNA genome, wrapped inside a protein coat known as a capsid, sometimes with an additional lipid envelope. Viruses have no metabolic or reproductive ability on their own, and must invade living cells and hijack their cellular machinery in order to make more copies of themselves.
Both prokaryotic cells, such as bacteria, and eukaryotic cells, such as those of humans, can be infected by specific classes of viruses. Specifically, bacteriophages are viruses that infect bacteria. For example, coliphages are those phages that infect E. coli, a common gut bacterium, some strains of which can cause food poisoning, and which is an indication of fecal contamination of the water supply.
While phages themselves are not generally known to be pathogenic in humans, there is evidence that they are reliable surrogate indicators for disease-causing enteroviruses that are also fecally-transmitted but difficult to assay. The availability of relatively quick and inexpensive methods to enumerate bacteriophages makes them an attractive tool for the assessment of fecal contamination in environmental samples.
This video will introduce the principles behind phage enumeration; demonstrate a protocol for quantifying phages, known as the plaque assay; and finally, explore several environmental science applications for the detection and counting of phages and other viruses.
Bacteriophages, like all viruses, must parasitize living cells, in this case bacteria, in order to reproduce.
Phages do so by landing and attaching to the bacterial cell surface and injecting their genetic materials into the cell. Once inside the cell, the viral genome is replicated and the protein components of the viral capsid are produced, both using the host cell’s biochemical machinery. Once the phage particles are assembled, they are released from the bacteria, often by lysing the host and bursting out, killing the host cell in the process.
A widely used method for determining phage concentration in a sample takes advantage of this lytic activity. In this technique, the phage from the sample is mixed with bacteria and soft agar. This mixture is poured onto Petri dishes with regular agar as a substrate, and the top layer forms an overlay.
The bacteria are at such a high concentration that they form a continuous lawn. The bacteria should be obtained from culture that is in the log phase of growth, to ensure that every bacterium that the phages infect is alive and allows the phage to produce progeny.
When a phage particle infects and lyses a bacterium, the phage progeny will spread to nearby bacterial cells and continue the infection. The soft agar restricts the diffusion of the phage particles. Eventually, an area of clearing, known as a plaque, will be formed.
If the phage is diluted to a low enough concentration, then discrete, individual plaques can be observed on the bacterial lawn. These can be counted and used to calculate the number of plaque-forming units, or PFUs, of phage per mL of the original sample.
Now that you understand how phages infect bacteria and how this activity can be used to measure phage concentration, let’s go through a protocol for using a plaque assay to enumerate phages in environmental water samples.
One day before performing the assay, inoculate a colony of E. coli strain ATCC 15597 into 100 mL of trypticase soy broth in a 250-mL Erlenmeyer flask. Incubate it with shaking at 35 °C overnight.
3 h before the assay, inoculate 1 mL of the overnight E. coli culture into a fresh 100 mL of broth. Place this new culture into a shaking water bath at a temperature between 35 and 37 °C. This ensures that the bacteria are in the log phase of growth when the assay begins.
To start the plaque assay, make 10- and 100-fold serial dilutions of the water sample, using 9 mL of Tris buffer.
Using a steam bath, melt three 5-mL tubes of 0.7% soft top agar, either trypticase soy or nutrient agar. Once melted, place in a water bath at 45 to 48 °C for at least 15 min for the agar’s temperature to drop to 45 °C.
To the first tube of soft agar, add 1 mL of the previously prepared log-phase E. coli culture and 1 mL of the undiluted water sample. Remove the tube from the water bath and gently rock between your hands for 2-3 s to mix the suspension.
Pour the soft agar over a previously prepared Petri dish with trypticase soy or nutrient agar. Quickly rotate the plate to spread, making sure that it covers the entire surface.
Repeat the inoculation and plating for the other two tubes, using 1 mL of each of the diluted samples.
Once the top agar has solidified, invert the dishes and incubate them at 37 °C for 48 h.
After the incubation, count the number of plagues on each plate. From the count, calculate the phage concentration in the original sample
For example, if 9 plaques were obtained from the 10-fold dilution plate, then there are 9 times 10 divided by 1 mL, or 90 PFUs/mL of coliphage in the original water sample.
Now that you have seen how phage plaque assays are performed, let’s look at how plaque assays can be used to enumerate phage and other types of viruses from a variety of sources.
Plaque assay-based methods can be used to isolate bacteriophage from different environmental samples such as soil. In this example, researchers first collected phage from soil by filtration. The phage was then used to infect the common soil bacteria Arthrobacter in a plaque assay. Phages were picked from individual plaques and streaked onto new agar plates, and then overlaid with bacteria-containing top agar. Phage concentration decreases along the length of the streak, so that discrete plaques, likely formed by a single type of phage, might be obtained. These individual plaques could then be picked to further analyze the phages within.
In addition to bacteriophages, plaque assays can also be performed with other viruses, including those, such as influenza, that infect mammals. To do this, mammalian cells are first grown up as monolayers in tissue culture dishes. Media containing the viruses are then added to the cells to allow infection to occur, before the cells are overlaid with an immobilizing medium such as the gel-like agarose. After an incubation period that could last up to two weeks, the infected cells are fixed and stained to allow the plaques to be visualized and counted.
Finally, in addition to samples collected from the environment, plaque assays are useful for detecting and enumerating viruses in tissue samples from infected individuals. Here, researchers obtained and homogenized lung tissues from mice infected with gamma-herpesviruses. This virus-containing homogenate was then used to infect mammalian cell culture. The number of plaques could then be counted to provide an estimate for the viral titer in the infected lung tissues.
You have just watched JoVE’s video on the detection of bacteriophages in environmental samples. You should now understand the basic biology of phages, how to perform a plaque assay to quantitate phages in an environmental sample, and how plaque assays can be used to study phages and other viruses in environmental or clinical samples. As always, thanks for watching!



