Visualizing Yeast Organelles with Fluorescent Protein Markers
Summary
Here we describe the use of a set of fluorescent protein-based organelle markers in live-cell imaging of the budding yeast, Saccharomyces cerevisiae.
Abstract
The budding yeast, Saccharomyces cerevisiae, is a classic model system in studying organelle function and dynamics. In our previous works, we have constructed fluorescent protein-based markers for major organelles and endomembrane structures, including the nucleus, endoplasmic reticulum (ER), Golgi apparatus, endosomes, vacuoles, mitochondria, peroxisomes, lipid droplets, and autophagosomes. The protocol presented here describes the procedures for using these markers in yeast, including DNA preparation for yeast transformation, selection and evaluation of transformants, fluorescent microscopic observation, and the expected outcomes. The text is geared toward researchers who are entering the field of yeast organelle study from other backgrounds. Essential steps are covered, as well as technical notes about microscope hardware considerations and several common pitfalls. It provides a starting point for people to observe yeast subcellular entities by live-cell fluorescent microscopy. These tools and methods can be used to identify protein subcellular localization and track organelles of interest in time-lapse imaging.
Introduction
Subcellular compartmentalization into membrane-bound organelles is a common principle in the organization of eukaryotic cells. Each organelle fulfills specific functions. Like in many other aspects of eukaryotic biology, the budding yeast, Saccharomyces cerevisiae, has been a classic model system in elucidating the basic principles of organelle organization and dynamics. Examples include the seminal discoveries in the protein secretion pathway, the peroxisomal protein import pathway, and the autophagy pathway1,2,3.
In typical nutrient-rich conditions, fast-growing yeast cells contain endoplasmic reticulum (ER), early Golgi, late Golgi/early endosomes, late endosomes, vacuoles, and mitochondria. Some peroxisomes, lipid droplets, and autophagosomes (even fewer than the first two, mainly of the Cvt vesicle type, which are present in nutrient-rich conditions4) are also present, but not as prominent as it would be under specific culturing conditions (lipid-rich media, starvation media, etc.). Compared to other common eukaryotic models, yeast cells are quite small; the diameter of a typical yeast cell is around 5 µm, compared to tens of micrometers for most animal and plant cells. As a result, in the same imaging field that normally contains a single adherent animal cell, one normally sees tens of yeast cells at various cell cycle stages. Besides the size difference, yeast organelle morphology also has some peculiar features. At the ultrastructural level, yeast ER is composed of sheets and tubules, like in other systems. Under fluorescent microscopy, yeast ER manifests as two rings with some interconnecting structures in between. The inner ring is the nuclear ER, which is continuous with the nuclear envelope, and the outer ring is the peripheral ER, which is a tubular network lying beneath the plasma membrane5. Similar to plant cells but different from animal cells, a hybrid organelle, the late Golgi/early endosome, sits at the intersection between the secretory pathway and endocytic pathway6,7. Morphologically, yeast Golgi apparatuses are dispersed in the cytoplasm. Vacuoles are functionally analogous to lysosomes in animal cells. They often occupy large portions of the cytoplasm and undergo frequent fission and fusion. Besides the use of fluorescent colocalization markers, the vacuolar membrane can be distinguished from the nuclear ER by at least two criteria: The vacuolar membrane is generally more rounded than the nuclear ER, and the concaving appearance of the vacuole in DIC is also more pronounced than that of the nucleus.
Routinely, we use a set of fluorescent protein-based markers to visualize the aforementioned organelles in live yeast cells (Table 1). The fidelity and functionality of these organelle markers have been experimentally verified7,8. These marker constructs are intended to introduce fluorescent protein chimera cassettes into the yeast genome. As outlined below, in preparation for yeast transformation, linear DNA fragments are generated either by enzymatic digestion or PCR amplification7,8. The linear DNA fragments get integrated into the genome via homologous recombination. For plasmids described in this protocol, three types of design are employed. In the first type, covering the majority of the plasmids, it is often possible to obtain transformants carrying multiple copies of the construct. This is usually undesired because it introduces expressional and possibly functional variations across transformants. Single-copy transformants need to be identified through imaging as described in this protocol, by immunoblotting or by carefully designed PCR tests. In the second type, covering GFP-Sed5, GFP-Pep12, and GFP-Atg8, only single-copy integration is produced in haploid yeast cells. Both the first type and the second type keep the endogenous copy of the marker gene intact in the genome. A third type of plasmid design, covering Sec7-2GFP and Vph1-2GFP, is intended to introduce C-terminal knock-ins, leading to the chimeras being the sole copy of the corresponding marker gene.
Here we describe the procedure to utilize these organelle markers, provide exemplary microscopy images, and discuss precautions geared toward researchers new to yeast organelle imaging.
Protocol
1. Yeast strain construction
- Obtain marker plasmids and a suitable yeast strain.
NOTE: The plasmids are available from Addgene. This protocol utilizes TN124 (MATa ura3 trp1 pho8Δ60 pho13Δ::LEU2), BY4741(MATa leu2Δ0 ura3Δ his3Δ1 met15Δ0), and DJ03 (BY4741 trp1Δ::MET15) as examples. One important consideration for strain choice, other than the nature of the scientific question, is the compatibility of selection markers. The organelle marker plasmids described here use URA3 and TRP1 as the selection markers. Therefore, the genotype of the recipient strain needs to be ura3 and trp1. Otherwise, one needs to modify the strain or the plasmids. - Prepare yeast media according to the following recipes.
NOTE: SMD (synthetic minimal dextrose; 2% glucose, 0.67% yeast nitrogen base (YNB) without amino acids, 30 mg/L adenine, 30 mg/L lysine, 30 mg/L methionine, 20 mg/L histidine, 20 mg/L uracil, 50 mg/L tryptophan, 50 mg/L leucine). SMD+CA (SMD with the addition of 0.5% casamino acids). YPD (Yeast extract peptone dextrose; 1% yeast extract, 2% peptone, 2% glucose). SD-N (synthetic dextrose without nitrogen; 0.17% YNB without amino acids and ammonium sulfate, 2% glucose). Please refer to the Discussion section for considerations on the choice of the medium. - Prior to the transformation step, generate linearized DNA fragments by enzymatic digestion or PCR amplification of an organelle marker plasmid.
NOTE: Table 1 lists the restriction enzyme sites and PCR primers to generate linear fragments from the organelle marker plasmids. - Culture yeast cells in liquid YPD medium and transform yeast with linear DNA fragment.
NOTE: Yeast transformation can be performed using the conventional LiAc-based method9 or other methods of choice. It is advisable to include appropriate controls for transformation, in particular, a negative control with no plasmid or plasmid-derived DNA. - Incubate on an appropriate selection plate (e.g., for URA3 selection, use SMD-Ura medium) at 30 °C for 2-3 days.
NOTE: It takes about 2 days for single colonies to show up. - To verify fluorescent chimera expression and integration copy number, pick eight colonies from the selection plate, re-streak on fresh selection plates, and incubate at 30 °C.
2. Fluorescence microscopy: general procedures and single time-point imaging
- Culture yeast cells overnight in SMD liquid medium at 30 °C with shaking.
- The next morning, measure the optical density of yeast culture at 600 nm (hereafter referred to as OD600) in a spectrophotometer or plate reader.
NOTE: With some practice on inoculating yeast, one can usually ensure that OD600 of all samples is lower than 2 at this stage. If it ends up higher, it is advisable to dilute more in the next step and allow more time for yeast cells to recover. - Dilute yeast culture to approximately 0.2 OD600 using a fresh medium.
- Continue culturing till OD600 reaches approximately 0.8-1.0.
NOTE: The doubling time of yeast is about 1.5-2 h. - Put a cover glass on top of tissue paper on a flat surface, spread 5 µL of 1 mg/mL concanavalin A on the top side of the cover glass, and wait for 5 min (Figure 1A)10.
NOTE: This preparation step can be done ahead of time. - Transfer 100 µL of yeast culture to the top side of the cover glass, and wait for 5 min.
- Combine cover glass with a supporting glass slide, with the yeast cells sandwiched in-between; press with appropriate force to secure the attachment.
NOTE: It takes some practice to find the right pressure, so that yeast cells form an immobile single layer but are not crushed (Figure 1B). The liquid medium will be pushed out and absorbed by the underlying tissue paper during this step. - Mount the sample slide on the microscope stage. Locate and focus on a patch of yeast cells using differential interference contrast (DIC) or phase contrast (PC) illumination.
NOTE: Do not use fluorescence to locate cells; otherwise, the signal may get bleached prior to data collection. - Manually configure three sets of parameters to collect image z-stacks: z-sectioning, imaging channels, and exposure parameters.
NOTE: See Supplementary Figure 1 for GUI examples of parameter settings in one commercial software. The exact look differs across software platforms, but the options are more or less the same.- Z-sectioning: Collect slices at 0.5 µm stepping for 15 slices (covering 7 µm depth, generally sufficient for normal haploid yeast cells).
- Imaging channels: Select DIC or PC for cell contours and appropriate fluorescence channels as needed.
- Excitation light intensity and exposure time: Set the excitation light intensity and exposure time as appropriate for the sample.
NOTE: As a starting point, use 100% for excitation light intensity and 100 ms for exposure time; decrease light intensity if photobleaching becomes obvious with the progression of z-stack collection or if the signal exceeds camera recording capacity (i.e., signal saturation); increase the light intensity if the signal-to-noise ratio is low.
- Write down the imaging settings and use the same settings for all samples to be compared.
NOTE: Do not use the "auto" setting for data collection and image visualization. It is important to record the settings in a lab note so that the exact same parameters can be re-applied in future experimental repeats. Some microscope controlling software applications have the ability to save and re-apply settings; even so, it is advisable to have the settings recorded independently because software profiles may get deleted or modified unintentionally by co-workers. - Save the data in 16-bit multichannel stack format.
NOTE: Do not save as 8-bit RGB pictures (or 24 bit, if counting all three colors). See the image visualization section (Section 4) for limitations of the 8-bit format. - Move to a completely different area for the next image stack collection.
NOTE: Adjacent areas may have experienced photobleaching and phototoxicity. As a result, the image stack collected from these areas may be misleading.
3. Time-lapse imaging
NOTE: The procedure of time-lapse imaging differs from the one for single time-point imaging in two areas: sample preparation and imaging parameters.
- Sample preparation
- Coat a 35 mm glass-bottom dish with 1 mg/mL concanavalin A and wait 5 min or more.
- Add 1.5 mL of yeast liquid culture to the dish, and wait for 5 min to allow the yeast cells to settle to the glass surface.
- Using a pipet, aspirate the liquid medium from the edge of the dish, then gently rinse the patch of yeast sediment with about 1 mL of fresh medium to remove insecurely attached cells.
- Repeat the rinsing 2-3 times. Aspirate with a pipet, then gently add 2 mL of fresh culture medium.
- Imaging parameters
- Z-sectioning: Collect slices at 0.5 µm stepping for 15 slices.
- Imaging channels: Select as needed.
- Excitation light intensity and exposure time: Use the minimal necessary to discern the subcellular structure under investigation.
NOTE: For time-lapse imaging, photobleaching and phototoxicity from repeated illumination limit the total number of time points that can be collected. Therefore, excitation intensity and exposure time are generally set to low values so that more time points can be imaged. - Imaging interval: Set the timing intervals appropriate for the biological process being investigated.
NOTE: For example, the formation of an autophagosome under starvation conditions takes about 5-10 min; an interval of 1 min or less is preferred to track its dynamics. Under nutrient-rich conditions, the yeast cell cycle occurs on the hour scale; a longer interval, such as 10-20 min, can be utilized.
- If proper hardware is available, maintain the dish temperature at 30 °C.
NOTE: Most biological processes in yeast cells can also be imaged at room temperature (RT), except at a slower pace in general.
4. Visualization of image stacks and assessment of integration copy number
- Install Fiji or ImageJ software for image visualization and analysis11,12.
NOTE: Fiji is a tool collection based on ImageJ, suitable for general purpose biological image data visualization and processing. For image processing listed in this protocol, ImageJ without additional plugins is sufficient. - Pick appropriate parameters for image display and comparison.
- In Fiji, open a couple of z-stack images.
- Drag the z-position in each stack to the mid-section (or other positions if the structure under investigation is better visualized there).
- Go to Image > Adjust > Brightness/Contrast and click on Reset.
- In the Brightness/Contrast (B&C) window, use the scroll bars to change two parameters, Minimum and Maximum values, to discern the structure under investigation clearly.
NOTE: Generally, the Minimum value is set to be close to the average value in empty areas, and the Maximum is set to be close to the maximum in the image. Those values can be inferred from the displayed histogram (Figure 2A). If the imaging workstation is color-calibrated, the minimum value can be set slightly higher to hide more background signals. - In the Brightness/Contrast (B&C) window, click on Set and select Propagate to all other Open Images checkbox in the popup window Set Display Range.
NOTE: This operation applies the same brightness/contrast settings (including the minimum & maximum values) to all opened images so that the images can be cross-compared. - Look across different images and scroll through the z-stack in each. If the images look good with a proper dark background and little overblown saturations, write down the Minimum and Maximum settings.
- For multichannel images, pick image display settings for each channel:
- Go to Image > Color > Channels Tool, select the Greyscale mode in the popup window Channels.
- Go through each channel, pick the proper Brightness/Contrast for that channel, and write down the settings.
- With the Brightness/Contrast adjusted, go back to the Channels window and change to Composite mode for simultaneous visualization of multiple channels.
NOTE: The pseudo color for each channel can be manually changed in Channels to suit personal preference. The checkboxes are for showing/hiding particular channels.
- If needed, use the checkboxes in the Channels window to show or hide particular channels.
- If desired, generate stack projections by going to Image > Stacks > Z Project.
NOTE: Several projection modes are available. Max intensity is a good choice for quick assessment. The nature of the research should be considered to determine if a particular projection mode or individual slices should be used for quantification. Projection can also be limited to select z-slices to reduce the interference of out-of-focus light. - Screen for single-copy-integration transformants.
- Once the same Brightness/Contrast settings are applied across all opened images, compare the image intensities across samples to infer plasmid integration number.
NOTE: Images should be acquired by following the general procedure for single time point imaging, beginning with overnight culturing in a liquid medium. Multi-copy integrations (See colony 2 & 3 in Figure 2B for examples) look substantially brighter than single-copy ones (colony 1 in Figure 2B). In contrast, single-copy ones look similar to each other (Figure 2B). - Examine images from eight transformants for each strain, ensuring to use the same Brightness/Contrast settings.
- Re-streak and save three or more single-copy transformants for subsequent study.
- Once the same Brightness/Contrast settings are applied across all opened images, compare the image intensities across samples to infer plasmid integration number.
- Export images for presentation.
- Go to Image > Duplicate to make a copy of the image of interest, and give it a name of choice.
- Go to Image > Type > 8-bit to convert the duplicate copy from 16-bit to 8-bit and save it as needed.
NOTE: Be aware that once this is applied, if the Brightness/Contrast settings are not written down previously, there is no easy way of retrieving that information, and thus no easy way of re-applying the same setting in subsequent image visualizations. This is also the reason why the previous duplication operation is recommended. - To split a z stack into a collection of individual images, go to Image > Stacks > Stack to Images and save as needed.
NOTE: Images can also be cropped to a smaller area in Fiji.
Representative Results
Organelle morphology and dynamics are subject to change as yeast cells respond to external and internal signals. Here, we provide representative images of yeast organelles in the mid-log phase (Figure 3A,B). As mentioned previously, several organelles have their distinct morphological features, thus are easy to recognize without extensive comparison with other organelle markers. These include ER, mitochondria, and vacuoles. Note that in some laboratory strains, including the ones we are showing here, the fission-fusion balance of vacuoles is skewed to the extent that many log-phase cells contain a single vacuole. Golgi apparatuses, endosomes, lipid droplets, and peroxisomes are generally too small for normal light microscopy to resolve their contours. So, they all appear as cytoplasmic dots. Ascertaining their identities requires cross-comparison with other organelle markers, which we have performed for the present marker set in log-phase cells7.
As presented in the Introduction, most of our marker constructs have the potential to be integrated multiple times in the genome. To avoid unnecessary complications, transformants with single-copy integration can be screened when verifying their marker expression by fluorescent microscopy (Figure 2B). The fluorescence signal in transformants carrying multiple copies is much brighter than those with a single copy.
Preparation of microscope slides containing live yeast cells is a simple procedure that can be done quickly to screen through tens of samples in a single experiment. However, it takes some practice to find the "right" pressure that secures a single layer of cells on slides without crushing them (Figure 1B). Too little force, cells stay in multiple layers and likely float around; too much force, the cells are essentially mechanically lysed. With experience, one can also spot dead cells and old cells, which should rarely appear in wild-type log-phase populations.
Time-lapse imaging is a powerful technique for investigating membrane dynamics. Here we demonstrate the frequent formation and consumption of autophagosomes in starved yeast cells labeled with GFP-Atg8 (Figure 4). There are claims in the literature that a yeast cell only has a single subcellular site that generates autophagosomes. It sounds correct if one only checks single-time-point snapshots since, on average, there is about one dot per cell. However, one can see easily in time-lapse imaging that in a single yeast cell, different instances of autophagosome biogenesis can occur independently at different subcellular locations over time.
Besides tracking the dynamics of each organelle, another common use of organelle markers is to determine the subcellular localization of a protein of interest. In practice, proteins in cells are often localized to more than one organelle. Here, we use Sft1 as an example; Sft1 is a v-SNARE functioning in intracellular transport13. GFP-tagged Sft1 manifests as multiple cytoplasmic dots (Figure 5). By cross-comparison with co-expressed red organelle makers, one can see that Sft1 partially colocalizes with early Golgi and late Golgi/early endosome markers, which provide important clues for understanding Sft1 function.
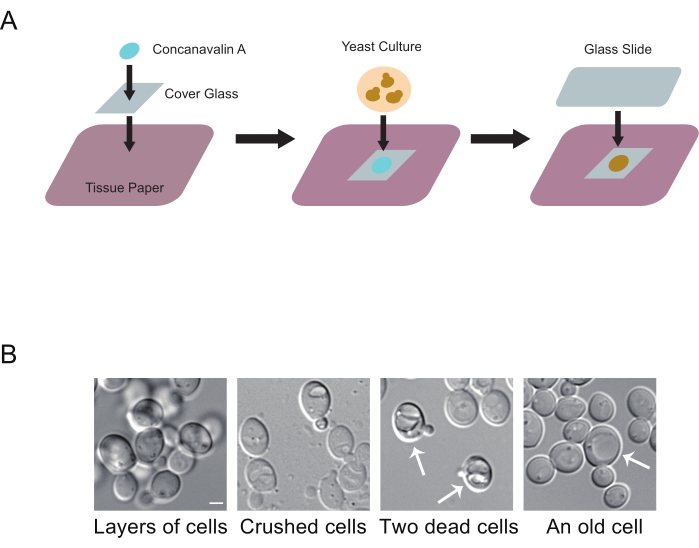
Figure 1: Sample slide preparation. (A) Carton illustration of the preparation. (1) Place a cover glass on tissue paper, spread the concanavalin A solution to the top side of the cover glass, and wait for 5 min or longer. (2) Add ~100 µL of yeast culture medium to the top side of the cover glass, and wait for 5 min for the cells to settle. (3) Cover the cover glass with a glass slide and press to combine it with the cover glass. Excess liquid is absorbed by the underlying tissue paper. (B) Appearance of yeast cells in undesirable situations. From left to right: formation of layers of cells when insufficient force is applied in slide preparation; crushed yeast cells when excessive force is applied in slide preparation; two potentially dead cells, marked by arrows; an old cell with a very large vacuole, marked by arrows. Scale bar: 2 µm. Please click here to view a larger version of this figure.
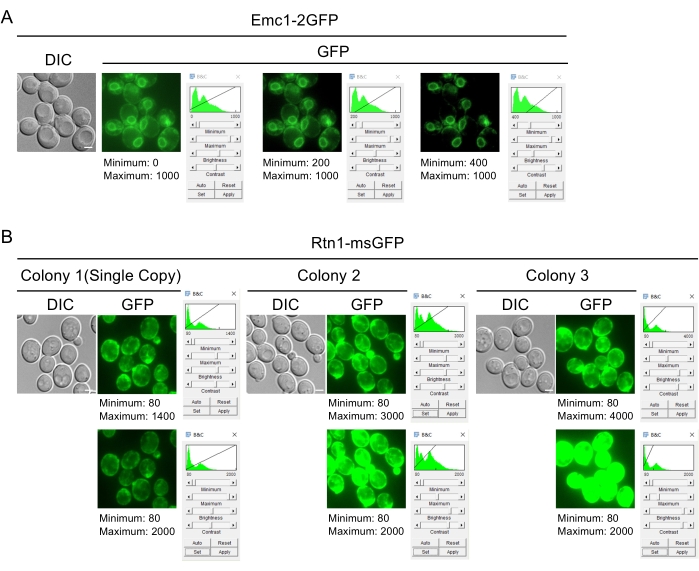
Figure 2: Adjusting image display and identifying single-copy integration transformants. (A) Effect of adjusting minimum display value. The visual appearance of a 16-bit image in ImageJ/Fiji depends on the user-determined display range. Here, the effect of adjusting the minimum value on the appearance of the endoplasmic reticulum is shown. The value can be set in the Brightness and Contrast (B&C) window. The visual appearance of the same 16-bit image varies as this setting is changed. On the left, with a value of 0, the contrast is low, with the background signal visible in areas devoid of cells. On the right, a value of 400 is substantially higher than the background; as a result, the peripheral endoplasmic reticulum (i.e., the network beneath the plasma membrane) becomes obscured. The histogram in the Brightness and Contrast (B&C) window provides information on the distribution of signal intensity across the image, which can be used as a guide in picking the appropriate display range. Scale bar: 2 µm. (B) Inferring construct integration number among transformants by comparing the fluorescent signal intensity. Note that the same exposure parameters should be employed when capturing the images. In this example, images from three different transformants are being compared. Colony 1 is a single-copy-integration transformant; colony 2 and colony 3 are multi-copy-integration transformants. In the top row, different minimum and maximum thresholds were applied to each image (80-1400, 80-3000, 80-4000, respectively), with higher maximum values applied to brighter samples. In this view, similar subcellular distribution patterns can be seen in all three samples. In the bottom row, the same minimum and maximum thresholds (80-2000) were applied to all three images. In this view, higher signal levels in colony 2 and colony 3 are apparent, reflecting the fact that they carry multiple copies of the Rtn1-msGFP construct in their genomes. Scale bar: 2 µm. Please click here to view a larger version of this figure.
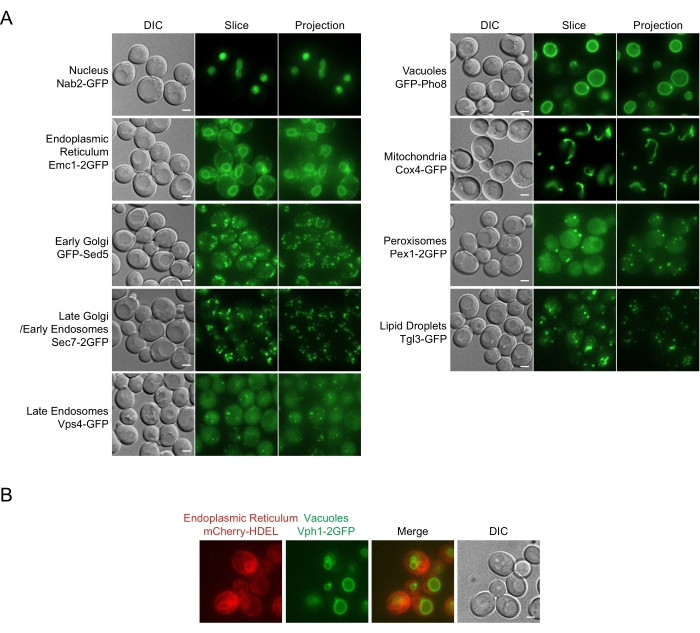
Figure 3: Typical morphology of major yeast organelles. (A) Representative snapshot images. The fluorescent protein constructs and the organelles they represent are indicated to the left of the images. DIC: differential interference contrast, a single slice. Slice: A single slice in the z-stack of the fluorescent channel. Projection: Max intensity projection of the fluorescent channel. Note that here the mapping of 16-bit to 8-bit was done differently for slices and projections to enhance the visibility of organelles in slices. Scale bar: 2 µm. (B) Morphological differences between nuclear endoplasmic reticulum/nuclear envelope and vacuole. Both the nuclear endoplasmic reticulum and vacuoles manifest as circular structures when imaged in the center of the cell. Compared with the vacuoles, the nuclear endoplasmic reticulum is generally less rounded, and the corresponding area in DIC is also less pronounced. Scale bar: 2 µm. Please click here to view a larger version of this figure.
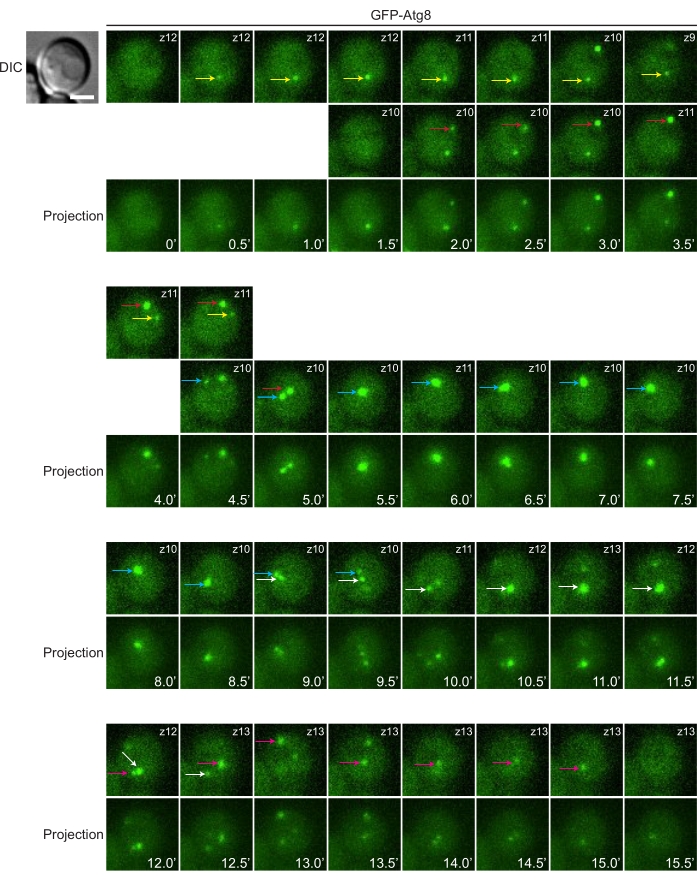
Figure 4: Following the dynamics of autophagosomes by time-lapse imaging. Yeast cells expressing GFP-Atg8 were grown to mid-log phase in YPD medium, then shifted to SD-N medium for starvation. After 45 min of the medium switch, yeast cells were mounted on glass-bottom dishes and incubated with SD-N for time-lapse imaging. Image stacks were collected at 30 s intervals. In this example, five rounds of autophagosome formation and disappearance can be seen. The dots, representing Atg8-positive autophagic structures, were manually tracked over time, and labeled with arrows of different color (i.e., arrows of the same color denote the same dot being followed). Note that at any particular time point, different dots (if there is more than one) may appear in different z-positions. Individual slices containing the tracked dots in focus are shown in the top one or two rows, with the z-position indicated in the top right corner (i.e., z10 denotes the 10th slice, and z12 denotes the 12th slice). The use of max-intensity projection is a convenient approach to glance through all structures. However, the contour of each individual structure is not as clear as in single slices. Max intensity projections are shown in the row below the slices. Scale bar: 2 µm. Please click here to view a larger version of this figure.
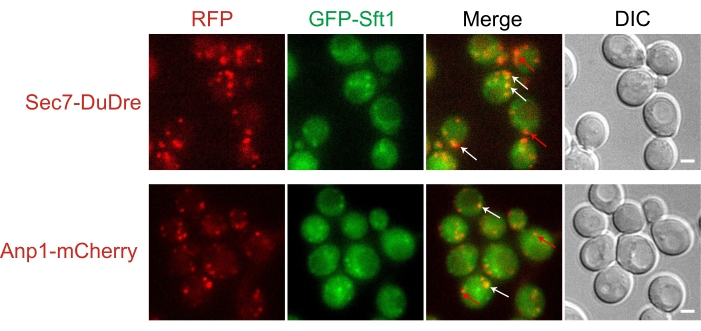
Figure 5: Using organelle markers to determine protein subcellular localization. Representative slices of yeast cells co-expressing GFP-Sft1 with either Sec2-DuDre (late Golgi/early endosome marker) or Anp1-mCherry (early Golgi marker). Sft1 displayed partial colocalization with both organelle markers. White arrows, the incidence of colocalization; red arrows, the incidence of no colocalization. Scale bar: 2 µm. Please click here to view a larger version of this figure.
Supplementary Figure 1: Screenshots showing the selection of imaging parameters in VisiView software. (A) Setting excitation light intensity. As a starting point, set 100% for all channels. See text for general considerations in adjusting excitation parameters. (B) Selecting the light channels to be imaged. This example demonstrated the selection of two fluorescence channels: GFP and mCherry. In VisiView, each channel needs to be picked from a drop-down menu. In many other software applications, all channels can be configured in a spreadsheet-like single graphical interface. Note that in this particular software, there is a checkbox Same Exposure/Gain for all Wavelengths, which needs to be unchecked to allow unique exposure duration for each channel. (C) Setting the number of slices and stepping for collecting z-stacks. 3.5 µm in each direction, covering 7 µm of depth at 0.5 µm steps. (D) Picking the order of channel switching and z-stack progression in multichannel imaging. In most microscope controlling software, one can choose to finish all channels at a z-position before moving on to the next slice, or finish a complete z-stack in each channel before moving to the next channel. This choice influences both time consumption in image acquisition and accuracy of colocalization for moving objects. (E) Setting the number of time points and intervals for time-lapse imaging. Please click here to download this File.
Table 1: List of organelle marker plasmids. Please click here to download this Table.
Discussion
The protocol described here provides a simple start for people entering from other research fields to explore imaging yeast organelles. Before moving on to specific topics, we would like to emphasize one more time that one needs to refrain from excessive use of automatic features in imaging software. Microscopy images are not just pretty pictures, they are scientific data, and therefore their acquisition and interpretation should be treated accordingly. It is especially important that image collection parameters be selected conscientiously, and the concepts of 16-bit versus 8-bit images be familiarized.
This protocol list four different yeast media: SMD, SMD+CA, YPD, and SD-N. SMD stands for “synthetic minimal dextrose”. The advantage of SMD is that it produces low autofluorescence. Thus, SMD is the preferred medium for fluorescent imaging. SMD+CA contains casamino acids, which provide richer nutrition than SMD, with the limitation that it is no longer purely synthetic. Being nutrient-rich, YPD is good as a general medium for the culturing of yeast cells, but it is less ideal for imaging because of the high autofluorescence it brings. SD-N is a nitrogen starvation medium which can be used to induce autophagy and the proliferation of lipid droplets. SD-N is also synthetic and generates low autofluorescence.
Note that the amino acid and nucleobase supplement in the SMD recipe used in this protocol is suitable for many common laboratory strains, but not for all. SMD needs to provide essential nutrients, the scope of which is strain genotype-dependent. When selecting for transformants with an auxotrophic gene allele complemented, the corresponding nutrient ingredient is omitted to create the drop-out medium (i.e., uracil is left out to select for Ura+ cells). For more information, please refer to other publications on this topic14. Also, note that for media sterilization, amino acid and nucleobase supplements shall not be autoclaved. It needs to be filtered. Glucose turns dark when autoclaved for a long time, which should also be avoided.
When studying organelle dynamics and intracellular protein trafficking, it is important to avoid overexpressing fluorescent protein constructs. Overexpression tends to disturb protein complex stoichiometry and introduce unexpected gain-of-function consequences, which may either obscure or eliminate the original subcellular distribution pattern under investigation.
With this consideration in mind, we recommend using a good inverted wide-field fluorescence microscope for observing proteins expressed at their endogenous levels. The working environment needs to be kept dim with all lighting in the ceiling being switched off. Avoid using point-scanning laser confocal microscopes. At the time of writing, most point-scanning laser confocal microscopes are not as sensitive in detecting low fluorescent signals as high-end wide-field fluorescence microscopes. Within available wide-field fluorescence microscopes, pick one with a good CMOS camera and a high numerical aperture objective. Objectives with high magnification (like 100x) are preferred, mainly because yeast cells are small. Ideally, the pixel pitch of the camera should satisfy Nyquist sampling frequency when paired with the objective of choice. If the microscope is instead equipped with a camera of larger pixel pitch for the detection of extremely low signals, spatial detail recovery by computational processing is worth trying15. The consumables in this microscopy procedure are generic in nature. One potential issue is that some suppliers’ cover glass is not cleaned to a high standard, therefore may produce fluorescence that interferes with sample observation. Switching to a different supplier is a simple solution.
It is essential that yeast cells are maintained in a healthy condition for imaging. Yeast on agar plates can survive for months at 4 °C. However, if an old plate is used to inoculate a liquid culture, ugly senescent cells will frequently appear in the field of view. This is avoided by the use of a freshly streaked plate.
If one cannot detect the fluorescent signal of a chimera, check its expression by immunoblotting. If the construct is expressed and the molecular weight looks reasonable, then it is most likely a hardware configuration issue. Consult with a microscope specialist. If no expression is detected, the problem is likely an error in strain construction. Verify the plasmid is correct and used in the intended way (i.e., enzyme digested or PCR amplified).
When yeast cells are attached to glass surfaces for microscopic observation, even if a liquid medium is present and temperature is maintained, the environment is not identical to that in a test tube with shaking. Access to air is limited, and the mechanical clue is also different. There is a chance that these differences may alter the biological process under investigation16. In such rare situations, one needs to find more ingenious ways to keep the particular process going.
Acknowledgements
The authors would like to thank members of the Xie lab for their generous help in manuscript preparation. This work was supported by National Natural Science Foundation of China (grant 91957104), Shanghai Municipal Education Commission (grant 2017-01-07-00-02-E00035), and Shanghai Municipal Science and Technology Commission (grant 22ZR1433800).
Materials
| Adenine | Sangon Biotech | A600013 | |
| Casaminoacid | Sangon Biotech | A603060 | |
| Concanavalin A from canavalia ensiformis (Jack bean) | Sigma Aldrich | L7647 | |
| D-Glucose | Sangon Biotech | A501991 | |
| Fiji | https://fiji.sc/ | ||
| Glass-bottom petri dish | NEST | 706001 | Φ35 mm |
| ImajeJ | https://imagej.net/ | ||
| Inverted florescence microscope | Olympus | IX83 equipped with UPLXAPO 100X oil immersion objective, Lumencor Spectra X light source, and Hamamatsu Orca Flash4.0 LT camera. | |
| L-Histidine | Sangon Biotech | A604351 | |
| L-Leucine | Sangon Biotech | A100811 | |
| L-Lysine | Sangon Biotech | A602759 | |
| L-Methionine | Sangon Biotech | A100801 | |
| L-Tryptophan | Sangon Biotech | A601911 | |
| Microscope cover glass | CITOTEST | 10222222C | 22 mm x 22 mm, 0.16–0.19 mm |
| Microscope slides | CITOTEST | 1A5101 | 25 mm x 75 mm, 1–1.2 mm |
| Peptone | Sangon Biotech | A505247 | |
| Uracil | Sangon Biotech | A610564 | |
| Visiview | Visitron System GmbH | https://www.visitron.de/products/visiviewr-software.html | |
| Yeast extract | Sangon Biotech | A100850 | |
| Yeast nitrogen base without amino acids | Sangon Biotech | A610507 | |
| YNB without amino acids and ammonium sulfate | Sangon Biotech | A600505 |
References
- Levine, B., Klionsky, D. J. Autophagy wins the 2016 Nobel prize in physiology or medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proceedings of the National Academy of Sciences of the United States of America. 114 (2), 201-205 (2017).
- Spang, A. Anniversary of the discovery of sec mutants by Novick and Schekman. Molecular Biology of the Cell. 26 (10), 1783-1785 (2015).
- Walter, T., Erdmann, R. Current advances in protein import into peroxisomes. The Protein Journal. 38 (3), 351-362 (2019).
- Farre, J. C., Subramani, S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nature Reviews. Molecular Cell Biology. 17 (9), 537-552 (2016).
- West, M., Zurek, N., Hoenger, A., Voeltz, G. K. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. Journal of Cell Biology. 193 (2), 333-346 (2011).
- Day, K. J., Casler, J. C., Glick, B. S. Budding yeast has a minimal endomembrane system. Developmental Cell. 44 (1), 56-72 (2018).
- Zhu, J., et al. A validated set of fluorescent-protein-based markers for major organelles in yeast (Saccharomyces cerevisiae). mBio. 10 (5), 19 (2019).
- Li, D., et al. A fluorescent tool set for yeast Atg proteins. Autophagy. 11 (6), 954-960 (2015).
- Gietz, R. D., Woods, R. A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods in Molecular Biology. 313, 107-120 (2006).
- Caloca, B., et al. Comparison of concanavalin A and poly-l-lysine as cell adhesives for routine yeast microscopy applications. Yeast. , (2021).
- Rueden, C. T., et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 18 (1), 529 (2017).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Banfield, D. K., Lewis, M. J., Pelham, H. R. B. A SNARE-like protein required for traffic through the Golgi complex. Nature (London). 375 (6534), 806-809 (1995).
- Curran, B. P., Bugeja, V. Basic investigations in Saccharomyces cerevisiae. Methods in Molecular Biology. 1163, 1-14 (2014).
- Zhao, W., et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy. Nature Biotechnology. , (2021).
- He, C. W., et al. Membrane recruitment of Atg8 by Hfl1 facilitates turnover of vacuolar membrane proteins in yeast cells approaching stationary phase. BMC Biology. 19 (1), 117 (2021).

.
