Selected Reaction Monitoring Mass Spectrometry for Absolute Protein Quantification
Summary
This protocol describes how to perform absolute quantification assays of target proteins within complex biological samples using selected reaction monitoring. It was used to accurately quantify proteins of the mouse macrophage chemotaxis signaling pathway. Target peptide selection, assay development, and qualitative and quantitative assays are described in detail.
Abstract
Absolute quantification of target proteins within complex biological samples is critical to a wide range of research and clinical applications. This protocol provides step-by-step instructions for the development and application of quantitative assays using selected reaction monitoring (SRM) mass spectrometry (MS). First, likely quantotypic target peptides are identified based on numerous criteria. This includes identifying proteotypic peptides, avoiding sites of posttranslational modification, and analyzing the uniqueness of the target peptide to the target protein. Next, crude external peptide standards are synthesized and used to develop SRM assays, and the resulting assays are used to perform qualitative analyses of the biological samples. Finally, purified, quantified, heavy isotope labeled internal peptide standards are prepared and used to perform isotope dilution series SRM assays. Analysis of all of the resulting MS data is presented. This protocol was used to accurately assay the absolute abundance of proteins of the chemotaxis signaling pathway within RAW 264.7 cells (a mouse monocyte/macrophage cell line). The quantification of Gi2 (a heterotrimeric G-protein α-subunit) is described in detail.
Introduction
Proteomic experiments that use mass spectrometry (MS) can be designed to use either non-targeted (shotgun) or targeted methods. Discovery proteomics generally relies on bottom-up shotgun MS, either by using a traditional data-dependent acquisition mode, or by using one of the recently developed data-independent techniques (e.g., MSE, SWATH)1,2. Shotgun proteomics is a powerful tool for high-throughput peptide identification and relative quantification, but it is generally unsuitable for absolute quantification or for targeting small, defined sets (~tens) of proteins. The MS method most often used for targeted proteomics is selected reaction monitoring (SRM) because of its high sensitivity, speed, and dynamic range3-5. Alternatives to SRM include parallel reaction monitoring, which takes advantage of high-resolution, full MS scanning6.
SRM is usually performed using a nano-flow reversed-phase high-performance liquid chromatography (nano-RP-LC) instrument coupled to a nano-electrospray ionization (nano-ESI) ion source attached to a triple quadrupole mass spectrometer (QqQ-MS). In a typical experiment, sample proteins are proteolytically digested, and the resulting peptides are chromatographically separated, desorbed, and ionized. The resulting precursor ions are m/z filtered by the first quadrupole (Q1) and fragmented in the second quadrupole (q2) by colliding them with a collision gas. The resulting fragment ions are m/z-filtered in the third quadrupole (Q3) and quantified by a dynode. Each precursor and fragment ion pair is referred to as a transition, and each transition is monitored for a specified time period (the dwell time; typically 2-50 msec). During LC-SRM, the QqQ-MS cycles through a predefined list of transitions (the duty cycle is typically ≤3 sec), and a chromatogram of each transition is produced.
Alternative strategies for protein quantification typically use immunoassays such as dot blots, Western blots, ELISAs, antibody microarrays, reverse phase protein microarrays, microfluidic immunoassays, digital ELISAs, and microsphere-based immunoassays7. The best immunoassays can be significantly more sensitive than LC-SRM, and sample throughput of immunoassays can be significantly higher than that of LC-SRM5. However, developing immunoassays can be expensive and/or time consuming, and the resulting assays can be vulnerable to cross-reactivity and/or interference, incompatible with cell/tissue lysis/homogenization methods, and/or not amenable to multiplexing5,8. Some of these issues can be addressed by coupling antibody- and MS-based techniques. For example, target proteins can be enriched using immunoprecipitation prior to proteolysis and LC-SRM9-12. Alternatively, the SISCAPA technique employs immunoprecipitation subsequent to proteolysis at the peptide level13,14. In addition to immunoenrichment strategies, immunodepletion of high abundance proteins can be employed to increase LC-SRM sensitivity by reducing interference by coeluting analytes15,16.
MS-based protein quantification can be divided into relative and absolute quantification, and also into label-free and stable isotope labeling (e.g., metabolic labeling, chemical labeling, and heavy-labeled protein and peptide internal standards). Label-free techniques can be useful for relative protein quantification, but are unsuitable for accurate absolute quantification. By comparison, labeling techniques have reduced error associated with sample preparation and MS variance, and are often used for relative protein quantification17. For example, stable isotope labeled proteome (SILAP) standards prepared using a cultured human cell line enabled relative quantification of potential biomarkers via LC-SRM of human serum18. Accurate absolute protein quantification by MS requires that purified, quantified, isotopically-labeled protein or peptide internal standards be spiked-into biological samples prior to MS. The incorporation of heavy isotope labeled internal standards into an LC-SRM workflow enables absolute quantification that has been shown to be highly reproducible and transferable between laboratories16,19.
Stable isotope labeled internal standards for absolute protein quantification by MS include peptide standards prepared using solid phase synthesis20, proteins composed of concatenated protease-cleavable peptide standards21, and full-length protein standards22. Target protein covalent modification and incomplete sample preparation (i.e., incomplete sample lysis and homogenization, and incomplete protein solubilization, denaturation, alkylation, and proteolysis) can undermine accurate quantification. Internal protein standards are the least likely to be affected by most of these potential problems, but they are usually the most difficult to prepare. An alternative is to analyze each target protein using multiple internal peptide standards which are designed to include amino- and carboxy-terminal native flanking residues. Regardless of which type of internal standard is employed, it should be spiked-into the biological samples at as early a point during sample preparation as possible. Also, multiple sample preparation techniques (e.g., different denaturation conditions) should be tested. The usage of multiple orthogonal experimental techniques (experimental cross-validation) is a viable strategy for overcoming most potential quantification challenges23-25.
LC-SRM quantification of proteins is a highly flexible technique that has been used in a wide variety of applications. Notably, it has been used to study peptide and protein biomarkers within clinical samples such as serum, core biopsies, and fine needle aspirates5. LC-SRM has also been used to measure the stoichiometry of protein complexes5,26, to detect botulinum neurotoxins27, to quantify protein phosphorylation dynamics within signaling pathways5, and to quantify changes in protein conformation28.
Our laboratory is using LC-SRM to quantify the signaling proteins that mediate macrophage chemotaxis to support the development of chemotaxis pathway simulations. The overall scheme of the protocol (Figure 1) begins with ranking the tentative target peptides. Subsequently, crude external peptide standards are synthesized and used to develop LC-SRM assays for qualitative analyses of biological samples. If the biological sample-derived target peptide is detected, purified heavy-labeled internal peptide standards are prepared for quantitative LC-SRM. This protocol can be used to accurately quantify proteins from a wide variety of biological samples, and to support investigations of a wide variety of protein targets.
Protocol
NOTE: This method has been previously described56.
1. Peptide Target Selection
- Compile a list of the target proteins, and include a small number of housekeeping proteins for normalization across biological samples, and also include an internal protein standard (e.g., firefly luciferase). Digest the target proteins into tryptic peptides in silico using a software tool such as Protein Digestion Simulator29,30.
- Require that the peptides are fully tryptic and contain no missing trypsin cleavage sites. Avoid peptides with neighboring trypsin cleavage sites to avoid potentially incomplete trypsin digestion31.
- Require that the length of each peptide is 5-20 residues. Consider longer peptides, but note that they are generally more expensive to synthesize.
- Avoid a peptide if it corresponds to a natural genetic variant (e.g., a single-nucleotide polymorphism). Also confirm that the trypsin sites are unaffected (Step 1.2).
- Avoid a peptide if it corresponds to a site of posttranslational modification (PTM). Similarly, avoid a peptide if it would be prone to chemical modification during LC-MS sample preparation. Specifically, avoid peptides prone to cysteine and methionine oxidation, asparagine and glutamine deamidation, and amino-terminal glutamine formation of pyroglutamate. Confirm that the trypsin sites are unaffected (Step 1.2).
- Furthermore, avoid peptides that originate from protein amino- and carboxy-termini because protein termini are prone to posttranslational modification (amino-terminal methionine loss and acetylation, and carboxy-terminal amidation).
- Require that each target peptide is unique to each target protein. If this is not possible, require that the peptide is unique to a set of isoforms/homologues. Treat peptides differing only by leucine/isoleucine substitution as if they were identical when determining peptide uniqueness because these perform nearly identically during LC-SRM.
- If a comprehensive transcriptomic analysis of all of the biological samples has been performed (e.g., RNA-seq), determine peptide uniqueness using in silico translations of the transcript sequences32 instead of using the entire proteome of the species.
- Require that the target peptides are proteotypic. Identify proteotypic peptides using shotgun mass spectrometry1,2 of the biological samples being studied. Alternatively, retrieve peptide identifications of proteotypic peptides from online proteomics databases such as the NIST mass spectrometry libraries33 (http://peptide.nist.gov/), The Global Proteome Machine34 (X! HUNTER spectral libraries from http://www.thegpm.org/HUNTER/index.html, and GPMDB peptide identifications from http://gpmdb.thegpm.org/), PeptideAtlas35 (http://www.peptideatlas.org/), and the PRIDE database36 (http://www.ebi.ac.uk/pride/archive/).
- If quantitative transcript-level analyses of the biological samples have been performed (e.g., RNA-seq, qPCR), then avoid targeting peptides that correspond to the relatively low abundance transcripts.
- If needed, select target peptides that could be used for LC-SRM assays of target protein orthologs (e.g., to assay both the human and the mouse forms of a target protein).
- Select at least two target peptides per target protein (if possible).
2. Preparation of Peptide Standards
NOTE: This section of the protocol describes the preparation of a set of twenty lyophilized peptide standards (each being 1 nmol in quantity) for downstream analyses. For a different number of peptides, or for different peptide quantities, it will need to be adjusted accordingly.
- Prepare lyophilized peptide standards (≥1 nmol) using a peptide synthesis technology such as SPOT synthesis20,37.
- Ensure that cysteine residues of external peptide standards are carbamidomethylated (the chemical structure formed by iodoacetamide alkylation). In contrast, ensure that cysteine residues of internal peptide standards are unmodified (these will be alkylated during sample preparation, described below).
- Design internal peptide standards so that they contain amino- and carboxy-terminal native flanking residues (to control for trypsin digestion efficiency; each is typically three to six residues long).
- Design internal peptide standards so that they are stable isotope labeled. Consider the natural isotopic profile of the target peptide and the mass measurement accuracy of the MS when selecting a peptide labeling strategy. Do not use deuterated peptide standards because peptide deuteration causes reversed-phase LC retention times to shift38.
Note: Typically, tryptic internal peptide standards are synthesized using ~98% pure [13C6, 15N4]Arg and [13C6,15N2]Lys incorporated at the carboxy-termini. - Purify internal peptide standards using HPLC39, and accurately quantify them40.
- Add 100 µl of 0.1% v/v formic acid, 20% v/v acetonitrile (ACN) to each 1 nmol lyophilized peptide standard to produce a peptide concentration of 10 µM (use care as the lyophilized peptide can have a very low-density and can be lost easily). Vortex the samples for 2 min and bath sonicate them for 5 min to ensure that peptide dissolution is complete.
- Pool the dissolved peptides, and concentrate the resulting mixture in a vacuum concentrator to a final volume of 80 µl. Add 20 µl of ACN to dissolve any precipitated peptide.
Note: The sample now contains 10 µM of each peptide and 20% v/v ACN. - For internal peptide standards, prepare a dilution series for quantitative LC-SRM:
- Prepare 100 µl of a 1 µM dilution: combine 90 µl of 20% v/v ACN and 10 µl of the 10 µM peptide mixture.
- Prepare 100 µl of a 100 nM dilution: combine 90 µl of 20% v/v ACN and 10 µl of the 1 µM peptide mixture.
- For external peptide standards, prepare 100 µl of a 1 µM dilution for LC-MS by combining 90 µl of 0.1% v/v formic acid and 10 µl of the 10 µM peptide mixture.
Note: The sample now contains 1 µM of each peptide, 0.1% v/v formic acid, and 2% v/v ACN, and is ready to be used for LC-SRM assay development.
3. LC-SRM Assay Development
- Analyze the mixtures of the external peptide standards (roughly 1-10 pmol of each peptide per injection) by shotgun MS using a nano-flow HPLC system coupled to a triple quadrupole mass spectrometer (LC-QqQ-MS(/MS)).
- Use an LC-MS system equipped with a capillary column packed with C-18 media (≤5 µm diameter, ~200 Å pores, length ≥10 cm, i.d. = 50-100 µm) and a nano-ESI tip (typically produced using a laser-puller). Ensure that each LC-MS run includes a ~60 min linear gradient (typically, 0-40% Solvent B), a column regeneration step (~80% Solvent B), and a column re-equilibration step (~0% Solvent B) (Solvent A = 0.1% v/v formic acid in H2O, Solvent B = 0.1% v/v formic acid in ACN, flow rate = 200-800 nl/min, ESI voltage = 1,800 V, Q3 isolation width = 0.7 m/z, q2 argon pressure = 1.5 mTorr).
- For each precursor ion scan, dynamically select the top ~10 most intense precursor ions for tandem mass spectrometry (MS/MS). Run each sample in duplicate so that two different collision energy ramps are used: one designed to optimally fragment +2 precursor ions, and the other for +3 precursor ions41.
- Perform the LC-QqQ-MS(/MS) analyses using Q3 precursor ion scanning (Q1 precursor ion scanning can cause tailing of the precursor ion peaks resulting from low-energy argon collisions in q2).
- Make sure that the LC-MS system is performing adequately by analyzing QC samples and technical replicates. Prevent sample carryover by using freshly made LC columns and by running multiple blanks between samples.
- Analyze the resulting shotgun LC-QqQ-MS(/MS) data using database searching against the sequences of the external peptide standards42. If appropriate, discard ambiguous peptide identifications using the peptide identification confidence scores (e.g., expectation values) or by using statistical modeling42. Regardless, manually review all of the peptide identifications43 to ensure that they are all unambiguous.
- Use the shotgun MS peptide identifications to construct a spectrum library using a software program such as Skyline.
- Prepare an LC-SRM transition list using the 3-10 most intense transitions per precursor ion (+2 and +3 precursor ions; +1 and +2 fragment ions; y-, b-, and a-ions that are ≥2 residues long).
- Discard a transition if the precursor and fragment ion m/z values overlap within the mass measurement accuracy of the QqQ-MS (precursor ion fragmentation is sometimes incomplete; remember to consider the monoisotopic and the heavy natural isotope forms, as well as the unlabeled and heavy-labeled forms).
- Similarly, discard a transition if the fragment ion m/z overlaps that of a different fragment ion from the same precursor ion.
- Use the resulting transition lists to perform LC-SRM analyses of the mixtures of the external peptide standards (perform mass spectrometry as described in Step 3.1; Q1 and Q3 isolation width = 0.7 m/z; dwell time = 2-50 msec).
- Manually review the resulting LC-SRM data and delete any poorly performing assays (the analysis of LC-SRM data is described in Section 5).
4. LC-SRM Assays of Biological Samples
- Prepare the dishes of cultured cells44. If multiple experimental conditions are being compared, block and randomize the samples to reduce any possible systematic biases45.
- For each dish of cells, aspirate the cell culture medium to waste and suspend the cells in a serum free buffer44. If necessary, wash the cells in a serum free buffer to remove any remaining extracellular protein.
- For each sample, count the number of viable and total cells by using a viability stain (e.g., trypan blue) and a hemocytometer or an automated cell counter44.
- Pellet the cells by centrifugation, and aspirate the supernatants to waste44 (a typical cell pellet volume is ~30 µl).
- Add 400 µl of Urea Lysis Buffer or Surfactant Lysis Buffer to each cell pellet (100 mM HEPES∙NaOH pH 8, 10 µM bestatin hydrochloride, 10 µM pepstatin A, and either 8 M urea or 0.1% w/v surfactant (respectively); freshly prepared; use an acid-labile MS-compatible surfactant such as RapiGest SF or PPS Silent Surfactant). If using other protease inhibitors, ensure that they do not inhibit trypsin.
- Mix each sample using gentle pipetting, and transfer each to a 2 ml tube (with a screw cap with an O-ring) containing ~100 µl of 0.1 mm zirconia/silica beads (note that the beads can cause snap-cap tubes to leak). Lyse the cells by vortexing the samples for 5 min at full speed (this lyses the cells by bead beating).
- Alternatively, lyse the cells using a different mechanical method (e.g., using a French press or homogenization tips).
Note: Avoid centrifuging cell lysates because this might pellet precipitated protein that could otherwise be tryptically digested and detected by LC-MS.
- Alternatively, lyse the cells using a different mechanical method (e.g., using a French press or homogenization tips).
- For the surfactant denatured samples, incubate them at 90 °C for 10 min to assist sample homogenization and protein denaturation.
- Bath sonicate the samples for 10 min at room temperature to assist homogenization and protein denaturation.
Note: The lysates and lysis buffer can be stored at -80 °C (the lysis buffer will be needed for Steps 4.9 and 4.13). - Perform a protein concentration assay46 (e.g., a bicinchoninic acid assay) of the lysates (and of the lysis buffer as a control experiment).
- For qualitative analyses (i.e., no internal peptide standards will be spiked-into the samples), pipette 200 µg (protein mass) of cell lysate into a fresh 1.5 ml microcentrifuge tube. For quantitative analyses, prepare four such samples for a stable isotope dilution series.
- For each sample, add an internal protein standard (e.g., add ~5 pmol of ~98% pure firefly luciferase).
- For quantitative analyses, add the stable isotope dilution series of the equimolar mixture of the internal peptide standards (e.g., 0, 0.2, 2, 20 pmol of each peptide) to the samples.
- In parallel, prepare control samples using the internal peptide standards alone (i.e., no cell lysate) (e.g., 0, 0.2, 2, 20 pmol of each peptide).
- Also prepare control samples using the external and internal peptide standards (for example: 2 pmol of each external peptide standard, and 0, 0.2, 2, and 20 pmol of each internal peptide standard).
Note: Be sure that the peptide standards are free of formic acid as it may interfere with the trypsin digestion.
- Add lysis buffer so that all of the samples are identical volumes. Note: A typical sample volume is 70 µl, so this volume will be used for this protocol.
- Reduce protein cysteine residues by adding 0.7 µl of 1 M DTT (freshly prepared) to each sample (the final DTT concentration is 10 mM) and incubating the samples at 60 °C for 30 min.
- Alkylate protein cystines by adding 7 µl of buffered iodoacetamide (500 mM iodoacetamide, 1 M HEPES∙NaOH pH 8; freshly prepared) to each sample (the final iodoacetamide concentration is 50 mM) and incubating the samples at room temperature for 20 min in darkness (iodoacetamide is light sensitive). If necessary, quench the remaining iodoacetamide by adding DTT to a final concentration of 50 mM.
- For each of the samples denatured using urea, add 482 µl of 100 mM HEPES∙NaOH pH 8 so that the final urea concentration is 1 M (trypsin is significantly inhibited at >1 M urea47).
- Tryptically digest the proteins into peptides.
- For each sample that contains cell lysate, add 8 µl of 0.5 µg/µl sequencing grade modified trypsin so that the final trypsin concentration is 1:50 (protein w:w) trypsin:sample, and incubate the sample at 37 °C for 18 hr.
- For each sample that does not contain cell lysate, add 0.5 µL of 0.5 µg/µl sequencing grade modified trypsin, and incubate the sample at 37 °C for 2 hr (each of these samples are for control experiments, and typically contain only 10 ng-10 µg of peptide + protein mass).
- For each of the urea denatured samples, add 440 µl of 2% v/v formic acid (the final formic acid concentration is 1% v/v). Confirm that these samples are pH ~3.
- For each of the surfactant denatured samples, add 914 µl of 0.5% v/v TFA (the final TFA concentration is 0.5% v/v). Confirm that these samples are pH ~1.5. Incubate these samples at 37 °C for 60 min to hydrolyze the surfactant.
- Microcentrifuge all of the samples at 21,000 × g for 20 min at room temperature to pellet the surfactant tail group and any other precipitates that would clog a C-18 SPE cartridge.
- Solid phase extract each of the supernatants using a disposable C-18 SPE cartridge(Buffer A = 0.1% v/v formic acid; Buffer B = 0.1% v/v formic acid, 80% v/v ACN; ~100 mg C-18 resin per cartridge). Force the mobile phases through the C-18 SPE cartridge using a flat-surfaced rubber bulb, using an extraction manifold, or by centrifuging the cartridge in a 15 ml centrifuge tube at 10 × g for 5 min.
- Wet the column by applying 1 ml of Buffer B, equilibrate it by applying 1 ml of Buffer A twice, apply the sample, and wash the cartridge by applying 1 ml of Buffer A twice. Elute the peptides by applying 1 ml of Buffer B slowly (~2 min).
- Concentrate each eluate in a vacuum concentrator to a final volume of 100 µl to evaporate away ACN. Add 200 µl of H2O to each sample, and concentrate each in a vacuum concentrator to a final volume of 98 µl to evaporate away any possible remaining ACN.
- Add 2 µL of 5% v/v formic acid in ACN to each sample (the final sample concentrations are 0.1% v/v formic acid, 2% v/v ACN).
Note: The samples can be stored at -80 °C. - Analyze the samples using LC-SRM (as in Step 3.4). For qualitative LC-SRM analyses, run the biological samples and the external peptide standards, and analyze all of the resulting data together. For quantitative LC-SRM analyses, run the isotope dilution series of each biological sample, and also run the samples consisting of the peptide standards alone, and analyze all of the resulting data together.
5. LC-SRM Data Analysis
NOTE: Peptide identification and quantification can be highly simplified and partially automated using software such as Skyline, but it is still strongly recommended that all data annotation be manually reviewed. Also, it is best to exclude protein level information during manual annotation of LC-SRM data to prevent bias.
- For each peptide identification, confirm that the shape of each transition elution profile is roughly Gaussian. For each transition, confirm that the elution profile is the product of multiple signals measured by the MS detector (i.e., is not the product of one or two random spikes of MS noise).
- For each peptide identification, ensure that the entire peptide elution profile is selected. Avoid manually adjusting the elution profile boundaries of subcomponents of the peptide identification (the heavy-labeled peptide form, the unlabeled form, individual precursor ions, and individual transitions).
- Confirm that each transition elution profile has a signal-to-noise ratio (s/n) ≥3. Note: “Noise” refers to random MS noise, not to reproducible signal from coeluting analytes. Chromatogram smoothing functions can greatly assist the analysis of noisy data.
- For each peptide identification, confirm that all of the transitions have nearly identical elution profiles (not just equal peak apex times; the elution profiles may have different amplitudes).
- For LC-SRM analyses of a large quantity of the peptide standards (typically ≥100 fmol of each peptide), confirm that the corresponding peptide elution profiles are very intense (typically s/n ≥100), and that the peptide identifications are unambiguous. Note: These highly confident identifications will be critical for the identification of the corresponding biological sample-derived peptides.
- For each peptide identification, confirm that the relative transition intensities match those of the confidently identified peptide standard. If necessary, discard noisy, relatively low-intensity transition elution profiles, but confirm that none of the relatively intense transitions are missing. Disregard relatively intense transition elution profiles that are significantly and unambiguously affected by a coeluting contaminant.
- For each peptide identification, estimate the probability that it was produced randomly by background signal (random MS noise plus reproducible signal from coeluting analytes).
- Estimate this probability manually by examining the entire LC-SRM chromatogram and estimating the uniqueness of each peptide identification. Produce a set of confident peptide identifications that has an estimated false discovery rate (FDR) of 5%. Use stricter criteria (e.g., FDR ≤1%) if a higher confidence set of identifications is required (use of looser criteria is not recommended).
- Alternatively, use the mProphet algorithm48 or the AuDIT algorithm49, but manually review the results.
- For each peptide identification, confirm that the observed elution time is consistent with the hydrophobicity of the peptide (Skyline is designed to perform this calculation).
- For each peptide, confirm that its elution time is consistent across the LC runs.
NOTE: LC-SRM analyses sometimes produce elution profiles that are systematically skewed between runs. Systematic shifts are to be expected after instrument alterations such as replacing a column. Also, early eluting peptides can be prone to shifting (especially if the column is loaded close to capacity), and late eluting peptides can also be prone to shifting. Peptide identification is now complete. - To improve peptide quantification, reinspect each peptide identification and discard a transition elution profile if it is especially noisy compared to the other transition elution profiles of the peptide identification. Discard a transition elution profile if its relative transition intensity is wrong (compared to the peptide standard).
- Inspect the boundaries of each precursor ion and transition elution profile. If necessary, carefully adjust the elution profile boundaries to crop away background signal (or to improve background signal estimation).
- For each internal peptide standard, check if it is contaminated with a detectable amount of light peptide (determined by the LC-SRM analyses of the internal peptide standards alone, described above). Subsequently, discard any light peptide elution profiles that were significantly compromised by light-peptide contamination within the internal peptide standards.
- Quantify each transition elution profile by calculating its LC peak area. If appropriate, deduct the estimated background signal. Quantify each peptide elution profile by summing the corresponding transition quantification values (hereafter, this is referred to as the “Sum_Peak_Area” value). For each biological sample peptide Sum_Peak_Area value, calculate the biological sample peptide molar abundance using data from LC-SRM of internal or external peptide standards (use only the transitions that were successfully quantified by both the biological sample peptide LC-SRM and that of the peptide standard).
- Quantification using external peptide standards (not recommended):
- Plot the external peptide standard Sum_Peak_Area values versus the external peptide standard molar abundance values. Produce a standard curve by fitting a linear regression to the data (typically, only the linear component of the dynamic range should be used; any nonlinear component should be used with extreme caution). For each biological sample peptide Sum_Peak_Area value, use the standard curve to calculate the peptide molar abundance.
Note: This quantification method is not recommended because it requires that the sample preparation and LC-SRM be demonstrability robust because the biological samples and external peptide standards are prepared and analyzed by LC-SRM separately. Also, it does not account for matrix effects (effects due to components of a sample other than the analyte).
- Plot the external peptide standard Sum_Peak_Area values versus the external peptide standard molar abundance values. Produce a standard curve by fitting a linear regression to the data (typically, only the linear component of the dynamic range should be used; any nonlinear component should be used with extreme caution). For each biological sample peptide Sum_Peak_Area value, use the standard curve to calculate the peptide molar abundance.
- Quantification using internal peptide standards (recommended):
- For each biological sample peptide Sum_Peak_Area value and corresponding heavy-labeled internal peptide standard Sum_Peak_Area value, calculate the light/heavy ratio. Use this ratio as a measure of the corresponding light/heavy peptide molar abundance ratio to calculate the molar abundance of the biological sample peptide. Specifically, the biological sample peptide molar abundance equals the internal peptide standard molar abundance multiplied by the light/heavy Sum_Peak_Area ratio.
- For each biological replicate and target peptide, identify the linear range of the LC-SRM quantification by creating a standard curve using a linear regression. Specifically, plot the internal peptide standard molar abundance values versus the heavy/light Sum_Peak_Area ratio values (note that the biological sample mass is constant across the dataset). Require that each biological sample peptide Sum_Peak_Area value is within the linear range (typically, only the linear component of the dynamic range should be used; any nonlinear component should be used with extreme caution). Likewise, perform this step for each target peptide transition.
Note: Analyte quantification by LC-QqQ-SRM should be linear over a wide dynamic range (~10,000). At the low end, background signal (random MS noise plus reproducible signal from coeluting analytes) will reduce quantification accuracy and precision. At the high end, detector saturation will cause nonlinearity (ultimately, this can cause elution profiles to be flat starting at a specific signal intensity).
- Quantification using external peptide standards (not recommended):
- If appropriate, perform a global normalization across the samples, such as a central tendency normalization50, to correct for slight differences in sample quantity prior to the spiking-in of the internal peptide standards. If necessary, use only the housekeeping proteins from Step 1.1.
- If necessary, impute missing quantification values51,52.
Note: It is improper to simply replace missing values with half the lower limit of quantification (LLOQ) or half the limit of detection (LOD) because doing so would artificially reduce quantification variance, and could result in a statistical test (e.g., an ANOVA) producing a false positive result (a type I error). However, ignoring missing values can also be problematic. For example, if half the true abundance values are below the LOD, and the other half are above the LLOQ, then the mean observed abundance would overestimate the mean true abundance. - Ensure that the assays satisfy the broadly accepted guidelines for LC-SRM23-25. Notably, require that the coefficient of variation of the quantification values is typically ≤25% for clinical assays, and ≤35% for non-clinical assays25. For LC-SRM assays related to healthcare or veterinary products or services that are to be considered for regulatory approval, require that the assays satisfy the stricter precision requirements for such assays: “The precision determined at each concentration level should not exceed 15% of the coefficient of variation (CV) except for the LLOQ, where it should not exceed 20% of the CV”23,24.
Representative Results
The development of predictive computational models of signal transduction pathways is one of the fundamental goals of systems biology53. Unfortunately, even for signaling pathways that have been studied extensively and have a high clinical significance, it is still not generally possible to quantitatively predict pathway behavior in response to perturbations (e.g., this is true for the MAPK/ERK pathway54). Recently, an investigation employed targeted proteomics, transcriptomics, and computational modeling and simulation to study the mouse macrophage chemotaxis signaling pathway56. The focus of the investigation was sphingosine-1-phosphate mediated chemotaxis of RAW 264.7 cells (a mouse monocyte/macrophage cell line). To facilitate pathway modeling, LC-SRM assays were developed and performed to measure the absolute abundance of the chemotaxis pathway proteins within RAW 264.7 cells. The resulting abundance values were used as parameters of the pathway model.
The overall experimental scheme (Figure 1) began with a list of target proteins, which included Gi2, a heterotrimeric G-protein α-subunit. The success of the overall protocol is highly dependent on the selection of peptide targets that are proteotypic and quantotypic, such as YDEAASYIQSK. Mixtures of external peptide standards were prepared and analyzed by shotgun LC-QqQ-MS(/MS). The resulting YDEAASYIQSK tandem mass spectrum was composed of several fragment ions, and it had a low background (Figure 2). The spectra were used to compose LC-SRM target lists containing the top ten most intense fragment ions per precursor ion.
A mixture of 409 external peptide standards (“JPT409”) was analyzed in triplicate by qualitative LC-SRM (data not shown). This sample included three Gi2 peptides, and the identification of all three was judged to be confident. Subsequently, RAW 264.7 samples (biological replicates “RAW1” and “RAW2”) and the JPT409 sample were each analyzed in duplicate (two LC-SRM technical replicates) by qualitative LC-SRM (these six LC-SRM analyses were performed as similarly as possible). The YDEAASYIQSK peptide was confidently identified in all six analyses (Figures 3A–C). The YDEAASYIQSK transition intensity patterns were consistent across the six LC-SRM analyses, and these were roughly similar to the shotgun LC-MS(/MS) pattern (the “Library” replicate of Figure 3C).
The pattern of transition intensities alone is not always sufficient for the confident identification of a peptide. The peptide hydrophobicity and measured LC retention time must be consistent. Also, the external peptide standards and the corresponding biological sample peptides must have approximately equal retention times. The hydrophobicity (estimated using the SSRCalc version 3.0 100 Å algorithm55) and observed retention time of YDEAASYIQSK were found to be consistent (Figure 4A). Also, the YDEAASYIQSK retention time was measured using both the RAW 264.7 analyses and the analyses of the external peptide standards, and all of the retention time values were approximately equal (Figure 4B).
Most of the qualitative LC-SRM analyses of the biological samples were successful, so corresponding heavy-labeled, purified, quantified internal peptide standards were prepared and spiked-into RAW 264.7 cell lysates. Isotope dilution series LC-SRM was used to analyze samples consisting of the internal peptide standards alone (sample “R0”), the internal and the external peptide standards (sample “R1”), and six RAW 264.7 biological replicates (samples “R2”-“R7”). Two different protein denaturants were used to test for any possible protein solubilization, denaturation, alkylation, and/or digestion issues that might undermine accurate target protein quantification (samples R2-R4 used urea, and samples R5-R7 used RapiGest SF). The light and heavy forms of YDEAASYIQSK contained nearly identical transition intensity patterns and elution profiles (Figures 5A–C). LC-SRM summed peak area ratios were used as a measure of relative peptide abundance, and the YDEAASYIQSK ratios were used to calculate Gi2 abundance values in units of copies per RAW 264.7 cell. In parallel, a second Gi2 internal peptide standard (IAQSDYIPTQQDVLR) was used to perform quantitative Gi2 LC-SRM assays of the same RAW 264.7 samples, and the two assays produced highly similar Gi2 abundance measurements across all of the biological replicates (Figure 5D). The agreement of the two Gi2 assays is strong evidence that both peptides are quantotypic, and that all of the quantitative Gi2 LC-SRM assays were accurate and precise.
Overall, 35 proteins were quantified using 58 internal peptide standards (Table 1). Notably, the LC-SRM assays of the internal protein standard (firefly luciferase; Step 4.11) were accurate and precise. The comprehensive set of Skyline data from this investigation is available at the Panorama online LC-SRM database (in the Manes_RAW_Chemotaxis folder at https://panoramaweb.org/labkey/project/NIH_NitaLazar/begin.view).
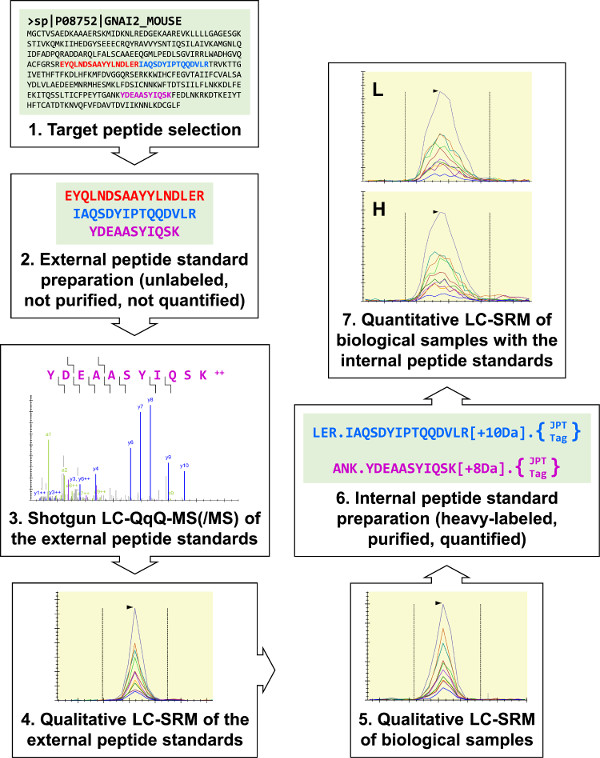
Figure 1: Overview of the protocol. Three tentative target peptides for Gi2 (a heterotrimeric G-protein α-subunit) were selected for external peptide standard synthesis. These were analyzed by shotgun LC-MS(/MS) to develop three Gi2 LC-SRM assays, and these assays were used to perform qualitative analyses of biological samples. All three Gi2 target peptides were identified, and two were selected for internal peptide standard preparation. Trypsin-cleavable “JPT-Tags” were used to quantify the internal peptide standards using UV spectrophotometry. The internal peptide standards were used to perform quantitative Gi2 LC-SRM assays of RAW 264.7 samples. Please click here to view a larger version of this figure.
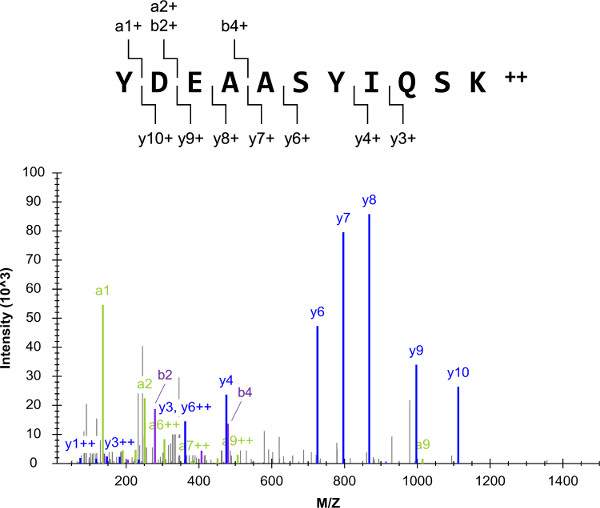
Figure 2: Shotgun LC-MS(/MS). The depicted mass spectrum originated from the analyses of one of the Gi2 external peptide standards (YDEAASYIQSK). For this precursor ion, the top ten most intense transitions were selected for LC-SRM (excluding the a11+ fragment ion due to its short length). Please click here to view a larger version of this figure.
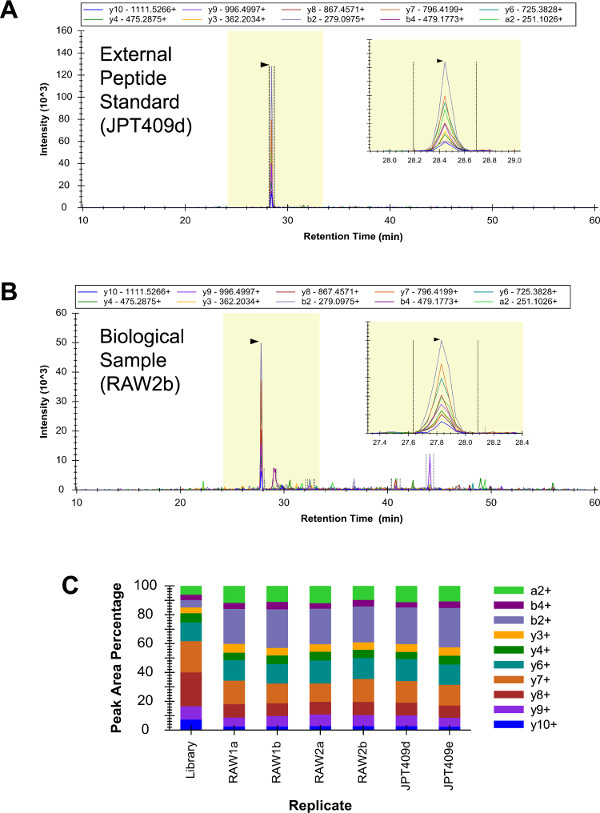
Figure 3: Qualitative LC-SRM of Gi2. The YDEAASYIQSK chromatogram from the JPT409 analyses contained a very low background and the peptide was confidently identified (A). The corresponding RAW 264.7 analyses resulted in more background signal, but the peptide identification was still unambiguous (B). The relative transition intensity patterns were consistent across all six LC-SRM analyses (two technical replicates of three independent samples), and these were roughly similar to the corresponding shotgun LC-MS(/MS) pattern (the “Library” replicate) (C). Please click here to view a larger version of this figure.
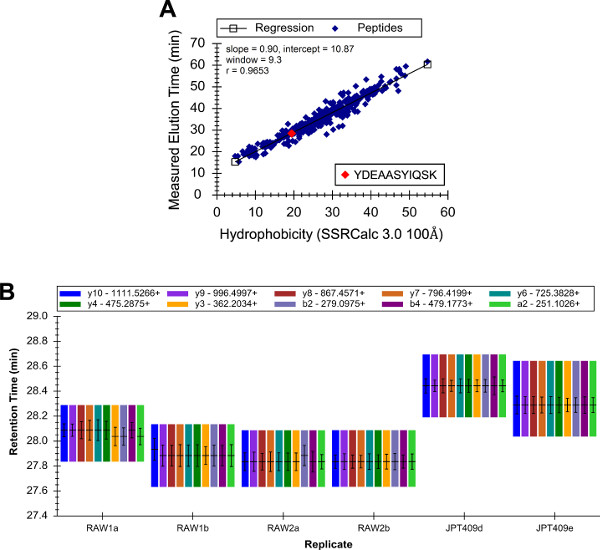
Figure 4: Peptide retention time prediction and variation across runs. A linear regression was calculated using the estimated hydrophobicity and measured qualitative LC-SRM elution time of all of the target peptides, and the predicted and measured elution times of YDEAASYIQSK were consistent (A). Additionally, the consistency of the observed elution time of each peptide across the LC-SRM analyses was determined. The elution times of YDEAASYIQSK spanned a range of ~40 sec, which was consistent with the precision of the LC-SRM instrument that was used (values are the peak apex time +/− the full width at half-max, and also the full width at the base of the peak) (B). Please click here to view a larger version of this figure.
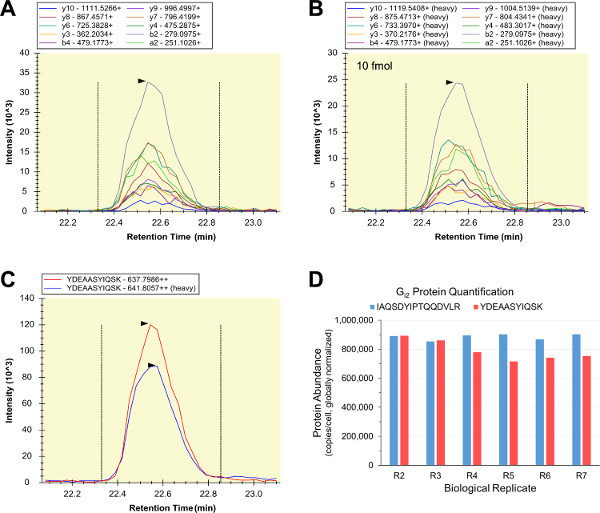
Figure 5: Quantitative LC-SRM of Gi2. The transition chromatograms of the biological sample peptide (A) and the internal peptide standard (B) had consistent relative transition intensities, and were summed (C) (depicted is the “R2” analysis for YDEAASYIQSK using 10 fmol internal peptide standard). The light and heavy elution profiles were consistent (C), and the ratio of the area under these curves was used as a measure of the light/heavy peptide abundance. A second Gi2 internal peptide standard (IAQSDYIPTQQDVLR) was used for quantitative LC-SRM in parallel, and the resulting Gi2 abundance values were consistent across both the six biological replicates and the two target peptides (overall, n = 12 and CV = 8.38%) (D). Please click here to view a larger version of this figure.
| UniProt Accession | Target Protein | Target Peptide | Abundance (fmol/μg) | Abundance (copies/cell, normalized) | CV |
| P08659 | Luciferase | (UV at 280 nm) | 23.824 | n/a | n/a |
| P08659 | Luciferase | VVDLDTGK | 24.103 | n/a | 7% |
| P08659 | Luciferase | VVPFFEAK | 24.717 | n/a | 4% |
| P60710 | Actin, cytoplasmic 1 | IWHHTFYNELR | 760.448 | 61,762,598 | 3% |
| P16858 | GAPDH | LISWYDNEYGYSNR | 357.803 | 28,906,524 | 14% |
| P06151 | Lactate dehydrogenase | LLIVSNPVDILTYVAWK | 129.623 | 10,633,145 | 30% |
| P99024 | Tubulin β 5 | ALTVPELTQQVFDAK | 78.765 | 6,398,971 | 3% |
| P20152 | Vimentin | SLYSSSPGGAYVTR | 121.100 | 9,807,488 | 10% |
| P60766 | CDC42 | DDPSTIEK | 86.647 | 6,957,317 | 27% |
| P60766 | CDC42 | QKPITPETAEK | 26.475 | 2,153,669 | 6% |
| Q8C3J5 | DOCK2 | ETLYETIIGYFDK | 1.459 | 118,539 | 15% |
| Q8C3J5 | DOCK2 | ISSSPTHSLYVFVR | 1.795 | 143,440 | 33% |
| Q8BPU7 | ELMO1 | ALTTKPSSLDQFK | 2.302 | 186,754 | 7% |
| Q8BPU7 | ELMO1 | SAIDISILQR | 1.244 | 99,728 | 25% |
| Q8BGM0 | FGR | GAYSLSIR | 1.099 | 89,473 | 17% |
| Q8BGM0 | FGR | WTAPEAALFGR | 0.319 | 24,766 | 6% |
| P27601 | Gα 13 | GIHEYDFEIK | 0.843 | 68,467 | 15% |
| P27601 | Gα 13 | VFLQYLPAIR | 1.295 | 106,176 | 23% |
| P08752 | Gα(i) 2 | IAQSDYIPTQQDVLR | 10.900 | 885,701 | 2% |
| P08752 | Gα(i) 2 | YDEAASYIQSK | 9.774 | 790,616 | 9% |
| Q9DC51 | Gα(k) | EYQLNDSASYYLNDLDR | 6.574 | 537,862 | 15% |
| Q9DC51 | Gα(k) | ISQTNYIPTQQDVLR | 3.504 | 285,784 | 10% |
| P62874 | Gβ 1 | AGVLAGHDNR | 17.078 | 1,385,578 | 4% |
| Q9CXP8 | Gγ 10 | DALLLGVPAGSNPFR | 2.167 | 179,178 | 36% |
| P63213 | Gγ 2 | EDPLLTPVPASENPFR | 3.284 | 266,360 | 8% |
| Q80SZ7 | Gγ 5 | VSQAAADLK | 6.087 | 493,512 | 6% |
| P08103 | HCK | GPVYVPDPTSSSK | 1.143 | 93,044 | 12% |
| P08103 | HCK | IIEDNEYTAR | 0.944 | 77,147 | 19% |
| P43406 | Integrin α V | AGTQLLAGLR | 0.276 | 22,264 | 32% |
| P43406 | Integrin α V | SHQWFGASVR | 0.443 | 34,235 | 5% |
| P25911 | LYN | VIEDNEYTAR | 1.461 | 119,377 | 13% |
| Q5SW28 | PI3K regulatory 5 | AGFPGILDTASPGK | 0.301 | 24,331 | 11% |
| Q8K3B3 | PI3K regulatory α | LYEEYTR | 0.472 | 38,592 | 16% |
| Q8K3B3 | PI3K regulatory α | TWNVGSSNR | 0.524 | 42,697 | 12% |
| Q8K3B3 | PI3K regulatory α | VLSEIFSPVLFR | 0.440 | 35,902 | 20% |
| Q5U3K7 | PI3K regulatory β | DTPDGTFLVR | 0.188 | 15,262 | 30% |
| Q5U3K7 | PI3K regulatory β | IAEIHESR | 0.282 | 22,561 | 13% |
| Q0VGQ5 | PI3K α | LINLTDILK | 0.102 | 8,494 | 38% |
| Q8CI98 | PI3K δ | HEVQEHFPEALAR | 0.178 | 14,566 | 22% |
| Q8CI98 | PI3K δ | ITEEEQLQLR | 0.481 | 38,709 | 24% |
| Q9ES52 | PIP3 5-phosphatase 1 | IVVLAKPEHENR | 0.486 | 39,194 | 19% |
| Q9ES52 | PIP3 5-phosphatase 1 | LSQLTSLLSSIEDK | 2.056 | 167,123 | 7% |
| Q69ZK0 | PIP3-dependent Rac GEF 1 | DSVLSYTSVR | 0.647 | 52,712 | 32% |
| Q69ZK0 | PIP3-dependent Rac GEF 1 | NQLLLALLK | 0.354 | 27,425 | 5% |
| Q9CQE5 | RGS 10 | ASSQVNVEGQSR | 2.460 | 199,782 | 4% |
| Q9CQE5 | RGS 10 | WASSLENLLEDPEGVQR | 2.647 | 206,005 | 11% |
| Q9CX84 | RGS 19 | AEANQHVVDEK | 0.495 | 39,753 | 21% |
| Q9CX84 | RGS 19 | LIYEDYVSILSPK | 0.846 | 68,481 | 22% |
| B9EKC3 | Rho GAP 5 | DGLAQELANEIR | 0.448 | 34,653 | 10% |
| Q99PT1 | Rho GDI 1 | SIQEIQELDK | 3.156 | 267,063 | 16% |
| Q99PT1 | Rho GDI 1 | VAVSADPNVPNVIVTR | 37.077 | 3,006,168 | 5% |
| Q61599 | Rho GDI 2 | LNYKPPPQK | 37.975 | 3,086,596 | 3% |
| Q61599 | Rho GDI 2 | YVQHTYR | 21.436 | 1,711,908 | 47% |
| Q61210 | Rho GEF 1 | FDGAEGSWFQK | 2.149 | 176,139 | 47% |
| Q61210 | Rho GEF 1 | SGLELEPEEPPGWR | 2.911 | 236,483 | 8% |
| Q9QUI0 | RhoA | QVELALWDTAGQEDYDR | 43.500 | 3,532,438 | 8% |
| P70336 | ROCK2 | GAFGEVQLVR | 0.527 | 42,800 | 23% |
| P70336 | ROCK2 | IYESIEEAK | 1.011 | 83,794 | 33% |
| P70336 | ROCK2 | LEGWLSLPVR | 0.573 | 48,259 | 22% |
| Q8R0X7 | S1P lyase 1 | AGYPLEKPFDFR | 1.787 | 147,043 | 27% |
| Q8R0X7 | S1P lyase 1 | TPEIVAPESAHAAFDK | 3.562 | 291,791 | 18% |
Table 1: Quantitative LC-SRM of RAW 264.7 cell proteins. Thirty-five RAW 264.7 cell proteins were quantified using fifty-eight internal peptide standards and six biological replicates. Five of the target proteins were housekeeping proteins (actin, GAPDH, lactate dehydrogenase, tubulin, and vimentin), and were quantified to enable normalization across biological samples (Step 1.1). In addition, an internal protein standard was spiked-into each cell lysate and quantified by LC-SRM (4.765 pmol of firefly luciferase per 200 µg sample; 98% pure by SDS-PAGE; quantified spectrophotometrically at 280 nm; Step 4.11). The CV values were calculated across the six biological replicates using the globally normalized abundance values (except for luciferase; Step 5.15).
Discussion
Absolute protein quantification is essential for a very diverse range of biomedical applications such as biomarker validation and signal transduction pathway modeling. Recently, targeted proteomics using LC-SRM has benefited from improvements to numerous technologies including peptide standard preparation, HPLC, QqQ-MS, and LC-SRM data analysis. Consequently, it has become a powerful alternative to immunoassays. Immunoassays can be extremely sensitive and high-throughput, but developing a robust immunoassay can be extremely challenging because immunoassays can be vulnerable to cross-reactivity and/or interference, incompatible with cell/tissue lysis/homogenization methods, and/or not amenable to multiplexing5,8. For example, the most comprehensive test for cross-reactivity is to perform the immunoassay using samples that originated from gene knockouts, which can be challenging to prepare.
This protocol describes target peptide selection, LC-SRM assay development, qualitative and quantitative LC-SRM, and LC-SRM data analysis. It was used to measure the absolute abundance of 36 chemotaxis pathway proteins within RAW 264.7 cells56, but its applicability extends far beyond this specific application. Though it was designed to quantify proteins within cell pellets, it could be adjusted for the analysis of other biological samples (e.g., biofluids) and other SRM targets (e.g., phosphopeptides). For example, changing the homogenization and/or protein digestion protocol may significantly improve solubilization, denaturation, alkylation, digestion, and quantification of especially difficult target proteins (e.g., membrane proteins), or might enable the analysis of especially challenging samples (e.g., samples containing <100 µg protein mass).
The initial selection of target peptides is critical but can be time consuming. For hundreds of target proteins, an ad hoc score could be used for each criteria, and the tentative target peptides can be ranked based on the sum of the scores, as has been done previously56. Alternatively, this analysis can be automated using PeptidePicker, a web interface that greatly simplifies target peptide selection30 (http://mrmpeptidepicker.proteincentre.com/).
After the target peptides have been selected and the LC-SRM assays have been developed, it is critical that qualitative LC-SRM assays of the biological samples be performed because even a highly proteotypic and quantotypic peptide will not be detectable if the protein is expressed at levels below the sensitivity threshold of the instrument, or if the background interference is especially problematic. A second dimension of separation (e.g., strong cation exchange HPLC, high-pH reversed-phase HPLC, and gel-free electrophoresis) can increase proteomic depth, but will require significantly more instrument time and data analysis. Alternatively, an enrichment strategy (e.g., peptide- and protein-level immunoenrichment and cell fractionation) can improve proteomic depth, and immunodepletion of highly abundant proteins can be used to reduce interference by coeluting analytes.
LC-SRM of ~tens of samples for ~hundreds or ~thousands of transitions typically requires extensive instrument time. Though it was not used for this study, LC-SRM scheduling (measuring transitions during pre-specified elution time windows) enables the analysis of more transitions per run. Also, scheduling reduces the SRM duty cycle during relatively vacant periods of the LC gradient, and this can result in improved peptide identification and quantification. However, scheduling requires that the peptide elution time be confidently determined from a qualitative LC-SRM analysis. Subtle changes to the sample preparation, the LC-MS instrument, or the LC-MS method can cause scheduling to crop or even entirely miss target peptides. For example, the biological sample YDEAASYIQSK elution profiles were slightly shifted relative to those of external peptide standard (Figure 4B), possibly due to matrix effects.
In summary, a step-by-step protocol is presented for the development and application of LC-SRM for absolute protein quantification. Absolute quantitation of proteins by LC-SRM has already been demonstrated to be reproducible between laboratories16,19. Proteomics technologies, including sample preparation (e.g., automation), liquid chromatography, mass spectrometry, and data analysis, are improving rapidly, and is enabling LC-SRM to become practical for large-scale research and clinical applications. The specificity, sensitivity, accuracy, reproducibly, and high-throughput of quantitative LC-SRM makes it a powerful tool for basic research and biomedicine.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases.
Materials
| Acetonitrile (ACN), LC-MS grade | Fisher | A955-1 | |
| BCA (bicinchoninic acid) protein assay kit | Fisher | 23235 | |
| Beads for bead beating, zirconia-silica, 0.1mm | BioSpec Products | 11079101z | |
| Bestatin hydrochloride | Sigma | B8385-10MG | |
| Cell culture DMEM (with glucose, without L-glutamine) | Lonza | 12-614F | |
| Cell culture EDTA, 500mM, pH8 | Gibco | 15575 | |
| Cell culture fetal bovine serum (FBS) | Atlanta Biologicals | S11550 | |
| Cell culture L-glutamine | Sigma | G8540-25G | |
| Cell culture phosphate buffered saline (PBS) pH 7.4 | Gibco | 10010-049 | |
| Cell culture Trypan Blue viability stain, 0.4% w/v | Lonza | 17-942E | |
| Cellometer Auto T4 cell counter | Nexcelom Bioscience | Cellometer Auto T4 | |
| Cellometer Auto T4 disposable counting chambers | Nexcelom Bioscience | CHT4-SD100-014 | |
| Dithiothreitol (DTT) | Sigma | D5545-5G | |
| Formic acid, LC-MS grade, ampules | Fisher | A117-10X1AMP | |
| Hemocytometer, Neubauer-improved, 0.1mm deep | Marienfeld-Superior | 0640030 | |
| HEPES, 1M, pH 7.2 | Mediatech | 25-060-CI | |
| Hydrochloric acid, 37% w/w | VWR | BDH3028-2.5LG | |
| Iodoacetamide | Sigma | I1149-5G | |
| Laser Based Micropipette Puller | Sutter Instrument Co. | P-2000 | |
| LC Magic C18AQ, 5µm, 200Å, loose media | Michrom Bioresources | PM5/61200/00 | |
| LC Halo ES-C18, 2.7µm, 160Å, loose media | Michrom Bioresources | PM3/93100/00 | |
| LC coated silica capillary, 50µm id | Polymicro Technologies | 1068150017 | |
| LC vial, autosampler, 12x32mm polypropylene | SUN SRI | 200-268 | |
| LC vial screw cap, autosampler, pre-slit PTFE/silicone | SUN SRI | 500-061 | |
| Luciferase, from Photinus pyralis | Sigma | L9506-1MG | |
| Pepstatin A | EMD Millipore | 516481-25MG | |
| pH strips colorpHast (pH 0.0-6.0) | EMD Chemicals | 9586-1 | |
| PhosStop phosphatase inhibitor cocktail | Roche | 04906837001 | |
| RapiGest SF | Waters | 186001861 | |
| Sep-Pak SPE, C18 1ml 100mg cartridge | Waters | WAT023590 | |
| Sep-Pak SPE, extraction manifold, 20 position | Waters | WAT200609 | |
| Sep-Pak SPE, flat-surfaced rubber bulb | Fisher | 03-448-25 | |
| Sodium hydroxide (NaOH) | Fisher | S318-500 | |
| SpeedVac vacuum concentrator | Fisher | SPD111V | |
| Trifluoroacetic acid (TFA), LC-MS grade | Fisher | A116-50 | |
| Trypsin, sequencing grade, modified | Promega | V5113 | |
| Tube decapper for Micronic tubes | USA Scientific | 1765-4000 | |
| Tubes, 2ml microcentrifuge, o-ring screw-cap, sterile | Sarstedt | 72.694.006 | |
| Urea | Sigma | U0631-500g | |
| Water, LC-MS grade | Fisher | W6-1 |
References
- Cox, J., Mann, M. Quantitative high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 80, 273-299 (2011).
- Zhang, Y., Fonslow, B. R., Shan, B., Baek, M. C., Yates, J. R. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 113, 2343-2394 (2013).
- Boja, E. S., Rodriguez, H. Mass spectrometry-based targeted quantitative proteomics: achieving sensitive and reproducible detection of proteins. Proteomics. 12, 1093-1110 (2012).
- Gillette, M. A., Carr, S. A. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Methods. 10, 28-34 (2013).
- Picotti, P., Aebersold, R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 9, 555-566 (2012).
- Lesur, A., Domon, B. Advances in high-resolution accurate mass spectrometry application to targeted proteomics. Proteomics. , (2015).
- Wild, D. . The immunoassay handbook : theory and applications of ligand binding ELISA., and related techniques. , (2013).
- Sturgeon, C. M., Viljoen, A. Analytical error and interference in immunoassay: minimizing risk. Ann Clin Biochem. 48, 418-432 (2011).
- Adrait, A., et al. Development of a Protein Standard Absolute Quantification (PSAQ) assay for the quantification of Staphylococcus aureus enterotoxin A in serum. J Proteomics. 75, 3041-3049 (2012).
- Lin, D., Alborn, W. E., Slebos, R. J., Liebler, D. C. Comparison of protein immunoprecipitation-multiple reaction monitoring with ELISA for assay of biomarker candidates in plasma. J Proteome Res. 12, 5996-6003 (2013).
- Weiss, F., et al. Catch and measure-mass spectrometry-based immunoassays in biomarker research. Biochim Biophys Acta. 1844, 927-932 (2014).
- Yassine, H., et al. Mass spectrometric immunoassay and MRM as targeted MS-based quantitative approaches in biomarker development: potential applications to cardiovascular disease and diabetes. Proteomics Clin Appl. 7, 528-540 (2013).
- Zhao, L., et al. Quantification of proteins using peptide immunoaffinity enrichment coupled with mass spectrometry. J Vis Exp. , (2011).
- Becker, J. O., Hoofnagle, A. N. Replacing immunoassays with tryptic digestion-peptide immunoaffinity enrichment and LC-MS/MS. 4, 281-290 (2012).
- Wasinger, V. C., Zeng, M., Yau, Y. Current status and advances in quantitative proteomic mass spectrometry. Int J Proteomics. 2013, 180605 (2013).
- Abbatiello, S. E., et al. Large-scale inter-laboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol Cell Proteomics. , (2015).
- Rodriguez-Suarez, E., Whetton, A. D. The application of quantification techniques in proteomics for biomedical research. Mass Spectrom Rev. 32, 1-26 (2013).
- Wehr, A. Y., Hwang, W. T., Blair, I. A., Yu, K. H. Relative quantification of serum proteins from pancreatic ductal adenocarcinoma patients by stable isotope dilution liquid chromatography-mass spectrometry. J Proteome Res. 11, 1749-1758 (2012).
- Kennedy, J. J., et al. Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Nat Methods. 11, 149-155 (2014).
- Jensen, K. J., Shelton, P. T., Pedersen, S. L. . Peptide synthesis and applications. , (2013).
- Pratt, J. M., et al. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat Protoc. 1, 1029-1043 (2006).
- Brun, V., et al. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics. 6, 2139-2149 (2007).
- . . Guidance for Industry: Bioanalytical Method Validation. , (2001).
- . . Guidance for Industry: Bioanalytical Method Validation. , (2013).
- Carr, S. A., et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics. 13, 907-917 (2014).
- Ori, A., Andres-Pons, A., Beck, M. The use of targeted proteomics to determine the stoichiometry of large macromolecular assemblies. Methods Cell Biol. 122, 117-146 (2014).
- Rosen, O., Feldberg, L., Gura, S., Zichel, R. A new peptide substrate for enhanced botulinum neurotoxin type B detection by endopeptidase-liquid chromatography-tandem mass spectrometry/multiple reaction monitoring assay. Anal Biochem. , (2015).
- Feng, Y., et al. Global analysis of protein structural changes in complex proteomes. Nat Biotechnol. 32, 1036-1044 (2014).
- MacLean, B., et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26, 966-968 (2010).
- Mohammed, Y., et al. PeptidePicker: a scientific workflow with web interface for selecting appropriate peptides for targeted proteomics experiments. J Proteomics. 106, 151-161 (2014).
- Rodriguez, J., Gupta, N., Smith, R. D., Pevzner, P. A. Does trypsin cut before proline. J Proteome Res. 7, 300-305 (2008).
- Min, X. J., Butler, G., Storms, R., Tsang, A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33, W677-W680 (2005).
- Lam, H., et al. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 5, 873-875 (2008).
- Craig, R., Cortens, J. P., Beavis, R. C. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 3, 1234-1242 (2004).
- Desiere, F., et al. The PeptideAtlas project. Nucleic Acids Res. 34, D655-D658 (2006).
- Vizcaino, J. A., et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063-D1069 (2013).
- Frank, R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports–principles and applications. J Immunol Methods. 267, 13-26 (2002).
- Ong, S. E., Kratchmarova, I., Mann, M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J Proteome Res. 2, 173-181 (2003).
- Mant, C. T., et al. HPLC analysis and purification of peptides. Methods Mol Biol. 386, 3-55 (2007).
- Alterman, M. A., Hunziker, P. . Amino acid analysis : methods and protocols. , (2012).
- Maclean, B., et al. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal Chem. 82, 10116-10124 (2010).
- Nesvizhskii, A. I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 73, 2092-2123 (2010).
- Tabb, D. L., Friedman, D. B., Ham, A. J. Verification of automated peptide identifications from proteomic tandem mass spectra. Nat Protoc. 1, 2213-2222 (2006).
- Freshney, R. I. . Culture of animal cells : a manual of basic technique and specialized applications. , (2010).
- Oberg, A. L., Vitek, O. Statistical design of quantitative mass spectrometry-based proteomic experiments. J Proteome Res. 8, 2144-2156 (2009).
- Noble, J. E., Bailey, M. J. Quantitation of protein. Methods Enzymol. 463, 73-95 (2009).
- Kiser, J. Z., Post, M., Wang, B., Miyagi, M. Streptomyces erythraeus trypsin for proteomics applications. J Proteome Res. 8, 1810-1817 (2009).
- Reiter, L., et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 8, 430-435 (2011).
- Abbatiello, S. E., Mani, D. R., Keshishian, H., Carr, S. A. Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin Chem. 56, 291-305 (2010).
- Callister, S. J., et al. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J Proteome Res. 5, 277-286 (2006).
- Karpievitch, Y. V., Dabney, A. R., Smith, R. D. Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinformatics. 13, S5 (2012).
- Oh, S., Kang, D. D., Brock, G. N., Tseng, G. C. Biological impact of missing-value imputation on downstream analyses of gene expression profiles. Bioinformatics. 27, 78-86 (2011).
- Germain, R. N., Meier-Schellersheim, M., Nita-Lazar, A., Fraser, I. D. Systems biology in immunology: a computational modeling perspective. Annu Rev Immunol. 29, 527-585 (2011).
- Futran, A. S., Link, A. J., Seger, R., Shvartsman, S. Y. ERK as a model for systems biology of enzyme kinetics in cells. Curr Biol. 23, R972-R979 (2013).
- Krokhin, O. V. Sequence-specific retention calculator. Algorithm for peptide retention prediction in ion-pair RP-HPLC: application to 300- and 100-A pore size C18 sorbents. Anal Chem. 78, 7785-7795 (2006).
- Manes, N. P., Angermann, B. R., Koppenol-Raab, M., An, E., Sjoelund, V. H., Sun, J., Ishii, M., Germain, R. N., Meier-Schellersheim, M., Nita-Lazar, A. Targeted Proteomics-Driven Computational Modeling of Macrophage S1P Chemosensing. . Mol Cell Proteomics. , .

