Probing Cell Mechanics with Bead-Free Optical Tweezers in the Drosophila Embryo
Summary
We present a setup of optical tweezers coupled to a light sheet microscope, and its implementation to probe cell mechanics without beads in the Drosophila embryo.
Abstract
Morphogenesis requires coordination between genetic patterning and mechanical forces to robustly shape the cells and tissues. Hence, a challenge to understand morphogenetic processes is to directly measure cellular forces and mechanical properties in vivo during embryogenesis. Here, we present a setup of optical tweezers coupled to a light sheet microscope, which allows to directly apply forces on cell-cell contacts of the early Drosophila embryo, while imaging at a speed of several frames per second. This technique has the advantage that it does not require the injection of beads into the embryo, usually used as intermediate probes on which optical forces are exerted. We detail step by step the implementation of the setup, and propose tools to extract mechanical information from the experiments. By monitoring the displacements of cell-cell contacts in real time, one can perform tension measurements and investigate cell contacts' rheology.
Introduction
Embryonic development is a highly reproducible process during which the cells and tissues deform to shape the future animal. Such deformations have been shown to require the active generation of forces at the cell level1,2. To understand morphogenetic processes during which cells and tissues change their shape, it is therefore key to assess the mechanical properties of the cells involved, and to measure the forces within the tissue during the process3,4. Epithelial layers, especially in Drosophila, have been widely studied due to their quasi-2D geometry and to their easy manipulation.
A number of techniques have thus been developed by us and others to assess epithelial mechanics in vivo during development. We will give a quick overview of the three main techniques used in epithelial tissues. Laser ablation, a widely used method, allows to reveal the local mechanical stress at cell junctions5,6,7,8 or at larger scale9,10,11 by performing local cuts that disrupt the mechanical integrity of the target. The dynamics of the opening following the cut provides information both on the stress prior ablation, and on the rheology of the tissue12,13. A drawback of laser ablation is that it is invasive, as it requires the local disruption of the cell cortex. Hence, one can only perform a limited number of ablations if one wants to preserve tissue integrity. Another drawback is that ablations only provide relative estimates of tension at cell contacts, since the opening velocity is dependent on viscous friction, which is generally not known. Magnetic manipulation has also been developed and used in Drosophila, involving either the use of ferrofluids14 or ultramagnetic liposomes15. They can provide absolute measurements16,17, but are also invasive in the sense that they require the injection of probes at the desired location. This can be very tricky depending on the system, which is not always amenable to precise injections. A third technique, fully non-invasive, is force inference18,19,20. Force inference relies on the assumption of mechanical equilibrium at triple points (tricellular junctions, or vertices), and allows to infer tensions at all cell-cell contacts (and possibly, pressures in all cells) by solving an inverse problem. For tensions, each vertex provides two equations (X and Y). This yields a large system of linear equations which can be inverted under some conditions to assess tension at all cell contacts. While this method is very attractive, as it only requires a segmented image and no extra experiment or setup, its accuracy is yet to determine, and again it only provides relative values, unless an absolute calibration measurement is performed.
To overcome some of these limitations, we introduce in this article a setup of optical tweezers coupled to a light sheet microscope to apply controlled forces at the cell scale in the embryonic epithelium of Drosophila melanogaster. Optical tweezers have been used for numerous biological applications including the measurements on single proteins and manipulation of organelles and cells21. Here, we report applied forces in the range of a few dozen pN, which is small yet sufficient to induce local deformations of cell contacts and perform mechanical measurements in vivo. Typically, we use perpendicular deflection of cell contacts, monitored through the analysis of kymographs, to relate force to deformation. Importantly, our setup does not require the injection of beads at the desired location in the tissue, as optical tweezers are able to directly exert forces on cell-cell contacts. The coupling of the optical tweezers to a light sheet microscope allows one to perform rapid imaging (several frames per second), which is very appreciable for a mechanical analysis at short time scales, and with reduced phototoxicity, since the illumination of the sample is limited to the plane of imaging22.
Overall, optical tweezers are a non-invasive way to apply controlled forces at cell contacts in vivo in the Drosophila embryo, and to extract mechanical information such as stiffness and tension at cell contacts23, rheological properties24, and gradient or anisotropy of tension23.
Protocol
1. Setting-up the Light Sheet Microscope
- Refer to the description of the setup in previous publication25.
NOTE: The setup is composed of an upright microscope stage and a light sheet module producing a light sheet in the horizontal plane. A 10X excitation objective lens directs the light sheet into a glass cuvette (Figure 4). The detection objective lens has a high NA (1.1), which is necessary for efficient tweezing (see below).
2. Setting-up the Optical Tweezers Path
NOTE: Figure 1 gives a general scheme of the optical setup.
- Place the 1070 nm laser unit and fix the fiber on the optical table with a V-clamp mount. Ensure that the fiber collimator is parallel to the optical table (hence, horizontal) and that the height selected here will be the height for all the optical components aligned on the optical table.
NOTE: 100 mm can be a good choice for this height. - Turn the key of the infrared laser unit and press the "Select" button to turn on the alignment pointer. The light has to be horizontal outgoing of the fiber.
- Place the shutter in the optical path, and fix it to the optical table, so that the infrared laser beam passes through the aperture of the shutter. Ensure that the distance between the optical table and the center of the aperture of the shutter is the height chosen in the step 2.1.
- Turn the key of the shutter controller, select the manual mode with the arrows and press the "Enable" button to open the shutter.
- Place, align and fix the first galvanometer mirror.
NOTE: The two galvanometer mirrors have to be conjugated to the back aperture of the detection objective, so that when one of the mirrors is rotating, the laser beam does not go out of the back aperture of the objective. Note that the distance between the first galvanometer and the back aperture of the objective has to be calculated to evaluate the position of this galvanometer in relation to the objective (f1+f1+f2+f2+f3+f3+f4+f4 = 30+30+30+30+30+200+200+500+500 = 1550 mm; fi=focal length of lens n°i). - Turn on the galvanometer power supply. Ensure that the galvanometers are powered for the rest of the alignment protocol.
- Set the galvanometers to zero deflection (with NIMAX or optical tweezer software – see step 3 for galvanometer connections).
- Place, align and fix the relay telescope composed of the two lenses with a focal length of 30 mm.
- Place, align and fix the second galvanometer mirror. Note that the distance between the two galvanometers is 4f = 4 x 30 mm = 120 mm (f = focal length).
- Place and fix the telescope.
NOTE: The telescope is composed of two lenses that expand the beam to a width at least equal to the diameter of the back aperture of the objective lens. The lens with the smallest focal length should come first. - Place, align and fix the periscope bringing upwards the optical path closer to the entrance of the dichroic mirror rail.
- Fix the hot mirror in the rail, place the rail in the microscope. Ensure that the rail is sawed on the right side to allow the entrance of the infrared laser (Figure 2).
- Check that the laser light is going out of the microscope, first without the objective then with the objective. Correct the position of the laser before the objective with the bottom mirror of the periscope. Correct the position of the laser after the objective with the top mirror of the periscope.
- Put 1 µL of 500 nm fluorescent beads in the glass cuvette and add 10 mL of distilled water. Put the cuvette under the objective in the cuvette holder and adjust the focus of the objective (Z position) so that the objective touches the water surface.
- Start the acquisition homemade software controlling the AOTF, the camera and the piezoelectric stage. A free software such as micromanager can also be used. Turn on the live acquisition mode by pressing the "Live" push-button.
- Adjust the IR laser power to 1 W.
- Put on the goggles (and do not remove it before the end of the experiment) and switch on the laser. A bead should be trapped at the laser focus.
- If no bead is trapped at the laser focus, check if the red laser pointer (which is collinear with the IR laser) is coming out of the objective. If not, start again the alignment steps, in particular step 2.13. Or, increase the IR laser power.
- If the bead is trapped out of focus (of the imaging plane), gently move the position of the last lens of the last telescope (L4 in Figure 1) along the optical axis to bring the bead in focus with the imaging plane. If not enough, move the first lens of the last telescope (L3 in Figure 1) along the optical axis.
- If trapped bead is not centered in the image, use the 2 periscope mirrors to adjust the position of the trap. If the position of the bead is changed by changing the angle of the first mirror, also compensate the direction of the beam by changing the corresponding angle of the second mirror. Correct the X position only (1 screw for each mirror), then correct the Y position (1 screw for each mirror).
- If necessary, adjust the position of the 2nd telescope lens to observe the bead in focus.
- If necessary, adjust the angle of the periscope mirrors to have the bead centered in the image.
3. Interfacing the Instrument
NOTE: Figure 3 gives a general scheme of the National Instruments (NI) Card connections.
- Insert the output card (NI-9263) in the first slot of the chassis (cDAQ-9178). Note that other models of NI cards with at least 2 analog outputs and 3 analog inputs can be used.
- Insert the input card (NI-9215) in the second slot of the chassis.
- Connect the first galvanometer to the analog output AO0 and the analog input AI0 with BNC cables. Note that a T adapter can be used to connect 2 BNC cables to one. Refer to the galvanometer manual to localize the pins on the galvanometer driver board.
- In the same way, connect the second galvanometer to AO1 and AI1.
- Connect PFI1 to the trigger IN of the shutter (back of the controller).
- Connect PFI0 to the fire of the camera and to AI2.
- Connect AI3 to the trigger out of the shutter (back of the controller).
- Adjust the settings of the shutter controller. Use the arrows to change the parameter values and push the "Select" button to validate the choice and pass to the next parameter. Put the “time open sec” parameter to 000.000, “time closed sec” to 000.000, “mode” to SINGLE, and “trigger” to EXT. Allow the control of the shutter by pushing the "Enable" button.
- Open QtCreator (downloaded from https://www.qt.io/, free version).
NOTE: QtCreator is the Integrated Development Environment used to develop in C++ the Optical Tweezers software. Qt library is used here to create widgets. - Open the OT.pro file (provided in the Qt code files). This action will open the project. Change the name of the input and output in the AOGenerator.cpp file according to the NI card(s) used.
- Compile and run the OT software.
4. Calibration of the Optical Trap Position with Beads
- Recording of a calibration movie
- Put 1 µL of 500 nm red fluorescent beads in the glass cuvette, then fill the cuvette with 10 mL of distilled water. Figure 4 gives a view of the cuvette in the observation context.
- Fix the cuvette on the piezoelectric stage.
- Adjust the Z position of the cuvette so that the detection objective dips in the water.
- Turn on the infrared laser and set the power to 1 W.
- Turn on the image acquisition software.
- To acquire time lapse images, select the exposure time, the gain, the time between two images, the number of images to be acquired, and the laser power. Note that for optical tweezer calibration with beads, the illumination laser (561 nm) is not required. The 2-photon effect induced by the infrared laser is enough to excite the fluorescence of a trapped bead.
- Start the optical tweezers homemade software.
- Set the optical tweezers parameters to trace a circle. Set SamplingRate as 250 samp/s, the bufferSize as 1000, Waveform1 parameters (galvo 1) as Sinusoidal, nbPeriod 1, amplitude 0.5 V, phase 0.0 rad, Offset 0.0 V, Waveform 2 parameters (galvo 2) as Sinusoidal, nbPeriod 1, amplitude 0.5 V, phase 1.57 rad, Offset 0.0 V.
- Check the "AI Parameters" box, and the "Wait for it" … box.
- Start the image acquisition (500 or 1000 images with a high frame rate, such as 10 fps).
- Switch on the optical tweezers by pressing the "Generating!" push button.
- Allow the trapped bead to complete at least two full circles and stop the optical tweezers by un-pressing the "Generating!" button.
- Stop the image acquisition. Note that a tiff movie and a text file with galvanometer voltages are created in the default “C:/TEMP/” folder (unless a custom location is specified). Note that several calibration movies with associated data files can be created if needed.
- If the circle is not complete, the bead is lost during the circle trajectory. Maybe the speed is too high. Decrease it by decreasing the sampling rate in the OT software. Or, using the red laser pointer of the infrared laser, check if the laser is coming out of the objective during the whole circle trajectory. If not, the galvanometer mirrors may not be conjugated with the back focal plane of the objective lens. Correct the positions of the galvanometers in relation to the back focal plane of the objective, starting again the alignment steps from step 2.5.
- Set the amplitudes to zero and switch on the laser to trap a bead at a fix position. Place a landmark at the trap location.
- Position interpolation with image analysis
- Open Matlab, go to the calibration folder, and run the “position2tension” script (provided in the Matlab code files). This script computes the interpolation function translating galvanometer voltages to optical trap position.
- Choose the number of calibration movies will be used. Several movies can be selected with different trajectories, such as two perpendicular ellipses. Generally, a single calibration movie with a circular trajectory is enough.
- Provide the first and last image numbers in the dialog box (one dialog box per movie), sequences with a clear trajectory and good signal to noise ratio.
- Provide the path of the text file containing the voltages during the acquisition for each movie. Note that the script reads the file and calculates the mean voltages of the two galvanometers for each image.
- Provide the path of the corresponding calibration movie.
NOTE: The script detects the bead for each frame with a subpixel resolution by fitting a 2D Gaussian to the bead, using custom functions and functions extracted from MTT26. It displays a graph showing the bead trajectory. - Eliminate any poorly detected point (outliers) by clicking it.
- Check if the interpolation map for x and y laser positions computed and displayed by the script, is complete. If there is a lot of missing values (white regions in the map), repeat the operation with a new calibration movie, with a better signal to noise ratio, or/and with a slower speed of the galvanometers.
NOTE: These images and the interpolation values are automatically saved in the same folder as the image, with the name convertFct.mat.
5. Mounting Drosophila Embryos27
- Collect Drosophila embryos from a cage, incubated at 25 °C.
- Remove the yeast, passing the plate contents through a sieve (pore size should be about 100 µm).
- Wash the embryos with 100% bleach (2.6% sodium hypochlorite) for 45 s to remove the chorion.
- Wash the embryos with water for 2 min.
- Put the embryos on an agar pad with a brush.
- Select about 10 embryos according to the desired stage and align the embryos with a pike or a moist brush.
NOTE: Alignment should be done according to the region of interest (region of interest as close as possible to the detection objective). - Using a diamond-marking pen, cut a piece of a glass slide of 10 x 20 x 1 mm3.
- Add glue on one side of the cut piece, and let it dry for 20 s.
- Turn the cut piece over and place it on the line of embryos, to stick it at the edge of the slide.
- Install this preparation in the sample holder and place it in the cuvette. Figure 4 gives a view of the cuvette in observation context.
- Put the cuvette on the piezoelectric stage.
6. Trapping Experiment In Vivo
- Trapping of the cell contacts — oscillations (Figure 5)
- Find the location of interest in the tissue.
NOTE: Using flies in which cadherins are labeled can be helpful if one needs to work in the plane of adherens junctions. - Move the target junction’s midpoint on the trap position (landmark set at step 4.1.14) using the piezo stage.
- Set the trap parameters to achieve the oscillations perpendicular to the contact line. Set the phases as zero. Set the amplitudes in X and Y directions to have a movement perpendicular to the contact line. The amplitude should typically be 0.1 V.
- Start the acquisition (with a fast frame rate, such as 10 fps).
- Switch on the optical tweezers pressing the "Generating!" push button.
- When done, switch off the tweezers and stop image acquisition. A movie and a text file containing galvanometer voltages during acquisition should now appear in the specified folder.
- Trapping of the cell contacts — pull-release (Figure 6)
- Find the location of interest in the tissue.
- Move the target junction using the piezo stage so that its midpoint is typically 1 µm away from the trap position (landmark set at step 4.1.14).
- Set the amplitudes to 0 to have a steady trap.
- Start the image acquisition (with a fast frame rate, such as 10 fps).
- Switch on the optical tweezers pressing the "Generating!" push button.
- After the desired time, switch off the trap and wait for relaxation to occur.
- Stop the image acquisition. A movie and a text file containing galvanometer voltages during acquisition should now appear in the specified folder.
- Find the location of interest in the tissue.
- Semi-automatic kymograph generation and detection of the interface position.
- In the Matlab command window, load the interpolation map generated during the calibration procedure (4b): load('convertFct.mat'). This provides the fx and fy variables.
- In the Matlab command window, run autokymo(fx,fy), provided in the Matlab code files. The purpose of this function is to generate a kymograph along the laser trajectory, and to detect the position of the contact line with subpixel resolution for each frame.
- Select the prompted text file path containing the voltage values.
- Select the first and last frame numbers of the movie to be analyzed.
- Select the tiff movie file. Two new figures will be displayed: The first one is an image of the kymograph. The second one is a graph of the detection of the contact line and laser positions, versus the image number.
NOTE: The function saves the graph representing the position of the sample and the position of the laser versus the image number (matlab figure, jpg), the kymograph (tiff image) and the kymograph with the laser position (tiff image). For steady trap experiments, the kymograph has to be done manually, for instance using ImageJ. The detection of the interface from the kymograph can then be done using the same algorithm.
7. Mechanical Measurements
- Stiffness and tension measurements
- Perform an oscillation experiment (step 6.1) and the corresponding analysis (step 6.3) of the time lapse movie.
- Plot the interface position
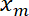 as a function of the trap position
as a function of the trap position 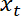 using Matlab plot function or any data plotting software.
using Matlab plot function or any data plotting software.
NOTE: This should be approximately linear. - Perform a linear fit, using Matlab “ezfit” free toolbox or any software allowing fits. The inverse of the fit’s slope provides the average ratio
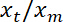 .
. - Perform estimates of the apparent bending stiffness of the interface, which, assuming small deformations and a quadratic potential for trapping, is given by
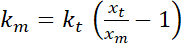 .
. 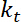 is the trap stiffness (see step 8 for trap stiffness calibration).
is the trap stiffness (see step 8 for trap stiffness calibration). - Perform estimates of tension, which can be defined as23:
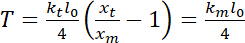 .
.
- Rheological measurements
- Perform pull release experiments (step 6.2) and the corresponding analysis (step 6.3)
- Fit the resulting curve with the custom rheological model.
8. Calibration of the Trap Stiffness
NOTE: The determination of the absolute forces requires the knowledge of the trap stiffness on interfaces. This can be accessed using beads through a two-step procedure.
- Determination of the cytosol viscosity
- Place the embryos in halocarbon oil and inject them using a microinjection setup with fluorescent polystyrene beads (1:1,000 stock dilution, 0.46 µm diameter)23,27.
- Place the injected embryos under the microscope.
- Track the movement of single beads found in the cytosol. Extract the mean square displacement of beads (use a large number of trajectories > 100 beads). Determine the bead diffusion constant from the slope of the mean square displacement. Relating the diffusion constant to viscosity by the Stokes-Einstein equation, deduce the effective viscosity of the cytosol. In the early embryo, a viscosity of approximately 3.5 Pa.s is reported23.
- Determination of trap stiffness on beads
- Trap single beads in the cytosol with optical tweezers.
- Move the bead in a stepwise fashion between two trap positions separated by 0.5 µm
- Determine the trap stiffness on beads, as the ratio between the drag coefficient and the bead relaxation time from one trap position to the other.
NOTE: The drag coefficient is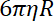 , where
, where 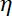 is the viscosity determined in (8.1) and
is the viscosity determined in (8.1) and 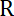 the bead radius. The relaxation time is obtained from exponential fits of the bead position, using Matlab or any software allowing fits. Typical values of the trap stiffness are 120 ±50 pN.µm-1 at 200 mW laser excitation.
the bead radius. The relaxation time is obtained from exponential fits of the bead position, using Matlab or any software allowing fits. Typical values of the trap stiffness are 120 ±50 pN.µm-1 at 200 mW laser excitation.
- Determination of trap stiffness on cell interface
- Focus the laser on a contact line, and deflect it (as detailed in step 6). Measure the deformation amplitude. This should be repeated on several contact lines to obtain a representative average.
- Compare the produced deformation with that induced by beads pushed against contact lines. Since the deformation is inversely proportional to the trap stiffness, deduce the trap stiffness on cell contacts.
NOTE: The trap stiffness on cell contacts was typically found 2- to 3-fold smaller than that on beads (0.46 µm diameter).
Representative Results
Figure 5 shows experimental data obtained by imposing a sinusoidal movement to the trap. The trap produces a deflection of the interface, as exemplified by the 3 snapshots showing 3 successive interface positions (Figure 5A)23. Recorded movies are used to generate a kymograph (Figure 5B) and are further analyzed to determine the position of the interface with subpixel resolution, using a Gaussian fit along the x direction for each frame. In the regime of small deformations, the trap and interface positions are proportional (Figure 5C). The amplitude of the interface deflection relative to that of the trap (Figure 5D) gives access to the interface tension or stiffness (see step 7.1).
Figure 6 shows experimental data obtained by performing pull release experiments. The optical trap is switched on approximately 1 µm away from the midpoint of the interface between two cells, which causes the interface to deflect towards the trap position (Figure 6A)24. The trap is then switched off after the desired amount of time. The position xm of the interface (Figure 6B) is obtained from kymographs (Figure 6C), again using Gaussian fits along the x direction for each frame. The resulting graph can be compared to hypothesized rheological models, for instance, a Maxwell model (Figure 6D).
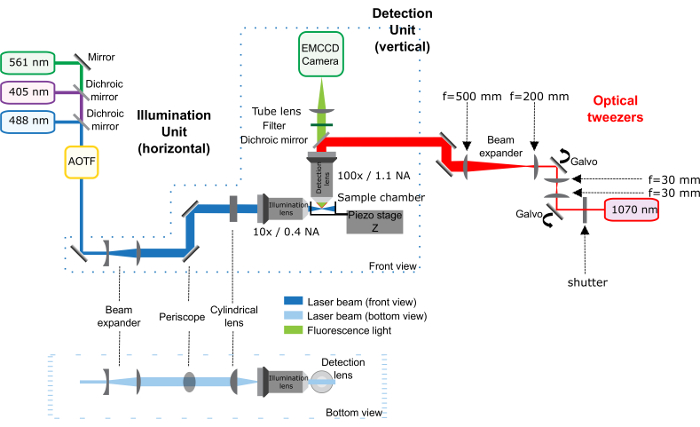
Figure 1: Schematic of the optical tweezers (red path) setup combined with the light sheet microscope. This figure has been modified from Bambardekar, K. et al.23.The light sheet microscope, composed by the illumination unit and the detection unit, is described previously25. The optical tweezers correspond to the red part of the scheme: The infrared laser passes through an optical shutter and 2 galvanometers which control the position of the trap in the sample. A 1-fold telescope is placed between the 2 galvanometers to keep the conjugation between them. Then, a telescope increases the size of the beam by 2.5-fold and the periscope brings it to the height of the microscope. The infrared laser enters the detection objective of the microscope thanks to a dichroic mirror.Important distances between optical elements are given directly in the figure. Distance between the last lens (focal length 500 mm) and the back aperture of the objective is 500 mm. Please click here to view a larger version of this figure.
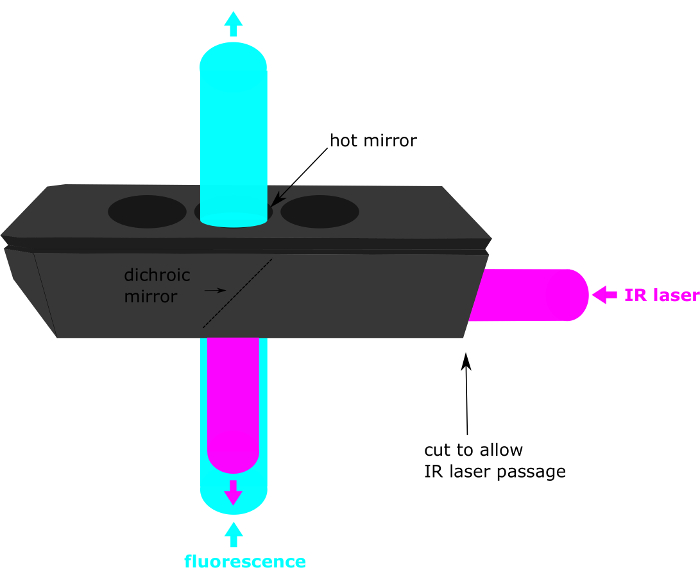
Figure 2: Homemade modified rail holding the dichroic mirror and the hot mirror. The dichroic mirror rail of the microscope has been cut on the left side to allow the entrance of the infrared laser. The dichroic mirror reflects the infrared light and transmits the visible light (fluorescence). Please click here to view a larger version of this figure.
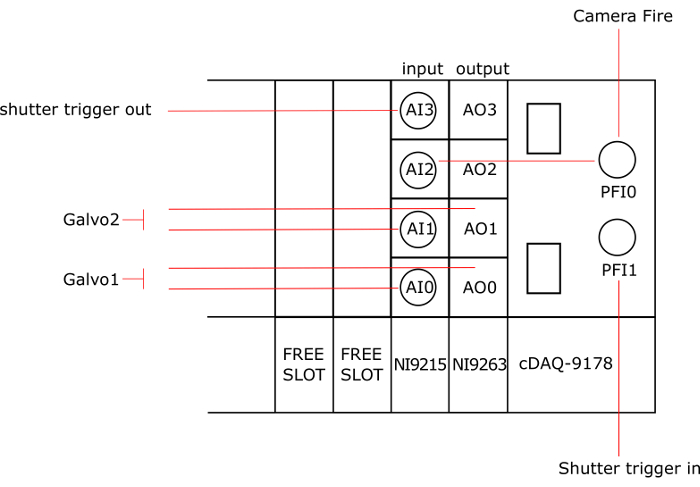
Figure 3: Connection of the instruments to the NI Cards. AO: analog output, AI: analog input, AO0 and AI0 are connected to galvo1, AO1 and AI1 are connected to galvo2, PFI0 is connected to the fire of the camera, to AI2 and PFI1 is connected to the trigger in of the shutter and AI3 is connected to the trigger out of the shutter. Please click here to view a larger version of this figure.
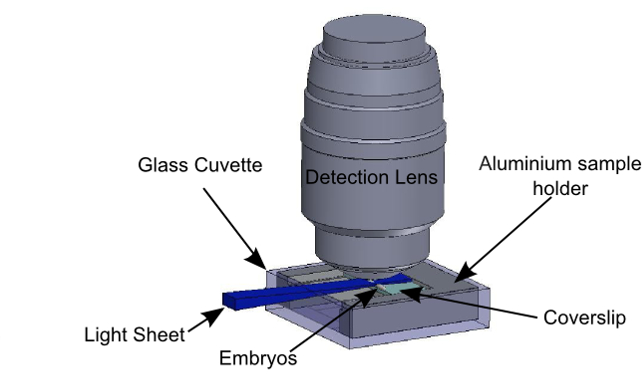
Figure 4: Sample holder in the observation context. This figure has been modified from Chardès, C., et al.25. The embryos are immobilized on the glass coverslip. The slide is held by the sample holder. The sample holder is inserted in the glass cuvette, held by the holder fixed to the piezoelectric stage. The light sheet is horizontal and illuminates the embryos from the side. The detection objective lens is vertical, above the sample, and dips into the cuvette.
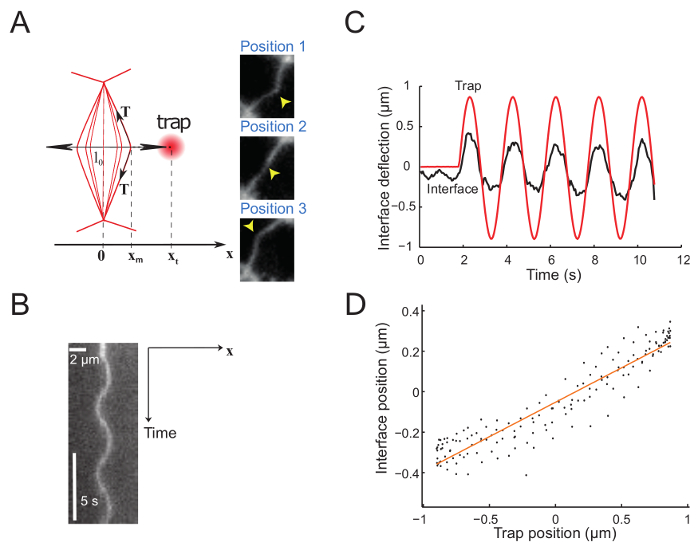
Figure 5: Interface deflection imposed by sinusoidal movement of the trap. This figure has been modified from Bambardekar, K. et al23. (A) The trap is moved sinusoidally perpendicular to the interface. The trap and interface positions are xt and xm, respectively. The right panels show three images of the interface at different positions. The laser trap position is marked by a yellow arrowhead. Interface are labelled with a membrane marker (GAP43::mcherry). (B) Kymograph along the axis defined by the direction of trap movement (perpendicular to the interface) (Period = 5 s). (C) Representative plot of deflection versus time showing both trap (red solid line) and interface positions (black solid line). (D) Interface position as a function of trap position during few cycles of laser oscillation (Amplitude = 0.5 µm, Period = 2 s). Please click here to view a larger version of this figure.
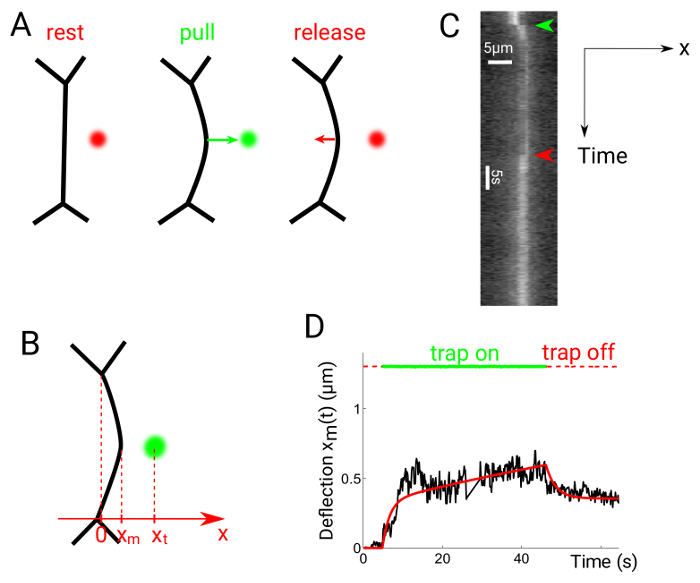
Figure 6: Interface deflection in pull-release experiments. This figure has been modified from Serge, A., et al.24. (A) The trap is switched on at a distance from the midpoint of the interface, then switched off again. (B) The trap and interface positions are xt and xm, respectively. Kymographs are generated along the x direction, perpendicular to the contact line's midpoint. (C) Kymograph of a pull-release experiment. Cell contacts are labelled with Utrophin::GFP. (D) Representative plot of deflection versus time showing both trap (dotted red/green lines) and interface position (black solid line). The solid red line shows a fit obtained using a Maxwell-like rheological model24. Please click here to view a larger version of this figure.
Supplementary Movie 1: Tweezing experiment. Pixel size = 194 nm, 10 fps, Trap oscillation period = 2 s, Labelling: Gap43::mCherry. Please click here to download this file.
Discussion
Optical tweezers allow to perform absolute mechanical measurements directly in the developing embryonic epithelium in a non-invasive manner. In that sense, it presents advantages over other methods such as laser ablation, which are invasive and provide relative measurements, magnetic forces, which require injections, or force inference, which relies on strong assumptions and also provide relative measurements.
The protocol includes a few critical steps. First, as the objective lens may show chromatic aberrations and that the laser trap pushes the object "downstream", it is important to check that the IR laser traps the bead in the imaging plane, and eventually correct for it (step 2.18). Second, the method relies on the measurements of the cell contacts' position. It is thus crucial to use a high-contrast fluorescent marker.
The quality of the objective lens and the laser beam are critical to effective trapping. The numerical aperture of the objective lens should exceed 1.0. If trapping is ineffective, make sure that the laser beam fills the rear aperture of the objective lens.
Our method comes with several limitations. First, it is not clear what exactly provides the support for optical trapping. Although a mismatch of refractive index is detected, its origin remains to be determined. Second, the calibration process, if one is interested in absolute measurements, can be a bit tedious, as it requires passive microrheology experiments, and calibration of the trap stiffness on beads. It is important to recognize that the calibration of the stiffness on cell contacts is subject to experimental uncertainty: it relies on measurements of the trap stiffness on beads, which can be measured only in the cytosol, but not near cell contacts. Third, it is unclear how versatile optical tweezers can be. Although they are able to deform the cells in the Drosophila embryo, other tissues might present higher tension or more difficult access (such as going through a cuticle) and therefore be less amenable to optical tweezing.
Optical forces are small (<few tens of pN) and may be thus insufficient to deform stiff or highly tensed structures. Magnetic tweezers on large particles would probably be more effective in this case.
We described here the coupling of optical tweezers to a light sheet microscope, but optical tweezers can be coupled to other types of microscopes, such as an epifluorescence or a confocal spinning disk microscope. The introduction of the IR trapping laser into the microscope depends on the microscope configuration. It mainly requires the possibility to add a dichroic mirror to combine the paths for imaging and optical manipulation. Most microscopy companies propose modular illumination systems, with a two-layer module, which allow this combination.
Several directions exist to improve or upgrade the technique. A possibility is to split the laser residence time among several positions or to use more advanced holographic techniques, to produce several traps. This could allow to create more complex force patterns on target cells or cell contacts. Another improvement could be to design a real-time feedback between the deflection caused and the position of the trap. This could allow proper creep experiments in which the force applied is maintained constant throughout the experiment.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by an FRM Equipe Grant FRM DEQ20130326509, Agence Nationale de la Recherche ANR-Blanc Grant, Morfor ANR-11-BSV5-0008 (to P.-F.L.). We acknowledge France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04-01, «Investments for the future»). We thank Brice Detailleur and Claude Moretti from the PICSL-FBI infrastructure for technical assistance.
Materials
| Ytterbium Fiber Laser LP, 10 W, CW | IPG Laser | YLM-10-LP-SC | including collimator LP : beam D=1.6 mm and red guide laser |
| Ø1/2" Optical Beam Shutter | Thorlabs | SH05 | |
| Small Beam Diameter Galvanometer Systems | Thorlabs | GVS001 | 1 for X displacement, 1 for Y displacement |
| 1D or 2D Galvo System Linear Power Supply | Thorlabs | GPS011 | galvanometers power supply |
| 2 lenses f = 30mm | Thorlabs | LB1757-B | relay telescope between 2 galva |
| Lens f = 200mm | Thorlabs | LB1945-B | 2.5X telescope |
| Lens f = 500mm | Thorlabs | LB1869-B | 2.5X telescope |
| Right-Angle Kinematic Elliptical Mirror Mount with Tapped Cage Rod Holes | Thorlabs | KCB1E | Periscope |
| Laser Safety Glasses, Light Green Lenses, 59% Visible Light Transmission, Universal Style | Thorlabs | LG1 | |
| 45° AOI, 50.0mm Diameter, Hot Mirror | Edmund Optics | #64-470 | |
| Multiphoton-Emitter HC 750/S | AHF | HC 750/SP | |
| CompactDAQ Chassis | National Instruments | cDAQ-9178 | |
| C Series Voltage Output Module | National Instruments | NI-9263 | Analog output module |
| C Series Voltage Input Module | National Instruments | NI-9215 | Analog input module |
| FluoSpheres Carboxylate-Modified Microspheres, 0.5 µm, red fluorescent (580/605), 2% solids | ThermoFisher Scientific | F8812 | calibration beads |
| C++ (Qt) home made optical tweezers software | developed by Olivier Blanc and Claire Chardès. Alternative solution: labview |
References
- Lecuit, T., Lenne, P. -. F., Munro, E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annual review of cell and developmental biology. 27 (1), 157-184 (2011).
- Heisenberg, C. -. P., Bellaïche, Y. Forces in tissue morphogenesis and patterning. Cell. 153 (5), 948-962 (2013).
- Sugimura, K., Lenne, P. -. F., Graner, F. Measuring forces and stresses in situ in living tissues. Development. 143 (2), 186-196 (2016).
- Campàs, O. A toolbox to explore the mechanics of living embryonic tissues. Seminars in cell & developmental biology. 55, 119-130 (2016).
- Kiehart, D. P., Galbraith, C. G., Edwards, K. A., Rickoll, W. L., Montague, R. A. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. Journal of Cell Biology. 149 (2), 471-490 (2000).
- Farhadifar, R., Roper, J. C., Aigouy, B., Eaton, S., Julicher, F. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Current Biology. 17 (24), 2095-2104 (2007).
- Rauzi, M., Verant, P., Lecuit, T., Lenne, P. F. Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nature Cell biology. 10 (12), 1401-1410 (2008).
- Ma, X., Lynch, H. E., Scully, P. C., Hutson, M. S. Probing embryonic tissue mechanics with laser hole drilling. Physical Biology. 6 (3), 036004 (2009).
- Hutson, M. S., Tokutake, Y., et al. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 300 (5616), 145-149 (2003).
- Bonnet, I., Marcq, P., Bosveld, F., Fetler, L., Bellaïche, Y., Graner, F. Mechanical state, material properties and continuous description of an epithelial tissue. Journal of the Royal Society, Interface / the Royal Society. 9 (75), 2614-2623 (2012).
- Etournay, R., Popović, M., et al. Interplay of cell dynamics and epithelial tension during morphogenesis of the Drosophila pupal wing. eLife. 4, e07090 (2015).
- Rauzi, M., Lenne, P. -. F. Cortical forces in cell shape changes and tissue morphogenesis. Current topics in developmental biology. 95, 93-144 (2011).
- Saha, A., Nishikawa, M., Behrndt, M., Heisenberg, C. -. P., Jülicher, F., Grill, S. W. Determining Physical Properties of the Cell Cortex. Biophysical journal. 110 (6), 1421-1429 (2016).
- Desprat, N., Supatto, W., Pouille, P. A., Beaurepaire, E., Farge, E. Tissue deformation modulates twist expression to determine anterior midgut differentiation in Drosophila embryos. Developmental cell. 15 (3), 470-477 (2008).
- Mitrossilis, D., Röper, J. -. C., et al. Mechanotransductive cascade of Myo-II-dependent mesoderm and endoderm invaginations in embryo gastrulation. Nature Communications. 8, 13883 (2017).
- Campàs, O., Mammoto, T., et al. Quantifying cell-generated mechanical forces within living embryonic tissues. Nature Methods. 11 (2), 183-189 (2013).
- Serwane, F., Mongera, A., et al. In vivo quantification of spatially varying mechanical properties in developing tissues. Nature Methods. 14 (2), 181-186 (2017).
- Chiou, K. K., Hufnagel, L., Shraiman, B. I. Mechanical stress inference for two dimensional cell arrays. PLoS computational biology. 8 (5), e1002512 (2012).
- Ishihara, S., Sugimura, K. Bayesian inference of force dynamics during morphogenesis. Journal of theoretical biology. 313, 201-211 (2012).
- Brodland, G. W., Veldhuis, J. H., Kim, S., Perrone, M., Mashburn, D., Hutson, M. S. CellFIT: a cellular force-inference toolkit using curvilinear cell boundaries. PLoS ONE. 9 (6), e99116 (2014).
- Svoboda, K., Block, S. M. Biological applications of optical forces. Annual Review of Biophysics and Biomolecular Structure. 23, 247-285 (1994).
- Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J., Stelzer, E. H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 305 (5686), 1007-1009 (2004).
- Bambardekar, K., Clément, R., Blanc, O., Chardès, C., Lenne, P. -. F. Direct laser manipulation reveals the mechanics of cell contacts in vivo. Proceedings of the National Academy of Sciences of the United States of America. , (2015).
- Clément, R., Dehapiot, B., Collinet, C., Lecuit, T., Lenne, P. -. F. Viscoelastic Dissipation Stabilizes Cell Shape Changes during Tissue Morphogenesis. Current biology: CB. 27 (20), (2017).
- Chardès, C., Ménélec, P., Bertrand, V., Lenne, P. -. F. Setting-up a simple light sheet microscope for in toto imaging of C. elegans development. Journal of visualized experiments. 87, e51342 (2014).
- Serge, A., Bertaux, N., Rigneault, H., Marguet, D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nature Methods. 5 (8), 687-694 (2008).
- Cavey, M., Lecuit, T. Imaging Cellular and Molecular Dynamics in Live Embryos Using Fluorescent Proteins. Drosophila. 420, 219-238 (2008).

