Confocal Live Imaging of Shoot Apical Meristems from Different Plant Species
Summary
This protocol presents how to live image and analyze the shoot apical meristems from different plant species using laser scanning confocal microscopy.
Abstract
The shoot apical meristem (SAM) functions as a conserved stem cell reservoir and it generates almost all aboveground tissues during the postembryonic development. The activity and morphology of SAMs determine important agronomic traits, such as shoot architecture, size and number of reproductive organs, and most importantly, grain yield. Here, we provide a detailed protocol for analyzing both the surface morphology and the internal cellular structure of the living SAMs from different species through laser scanning confocal microscope. The whole procedure from the sample preparation to the acquisition of high resolution three-dimensional (3D) images can be accomplished within as short as 20 minutes. We demonstrate that this protocol is highly efficient for studying not only the inflorescence SAMs of the model species but also the vegetative meristems from different crops, providing a simple but powerful tool to study the organization and development of meristems across different plant species.
Introduction
The plant meristem contains a pool of undifferentiated stem cells and continuously sustains the plant organ growth and development1. During the postembryonic development, almost all aboveground tissues of a plant are derived from the shoot apical meristem (SAM). In crops, the activity and size of the SAM and its derived floral meristems are tightly associated with many agronomic traits such as shoot architecture, fruit production, and seed yield. For example, in tomato, an enlarged SAM causes an increase in the shoot and inflorescence branching, and thus results in generating extra flower and fruit organs2. In maize, an increase in SAM size leads to a higher seed number and total yield3,4. In soybean, the meristem indeterminacy is also closely associated with the shoot architecture and yield5.
The morphology and anatomy of SAMs can be characterized by several different methods, including histological sectioning/staining and scanning electron microscopy (SEM)6, both of which have greatly advanced the meristem research through providing either the sectional view or a three-dimensional (3D) surface view of SAMs. However, both methods are time consuming, involving several experimental steps from the sample preparation to the data acquisition, and these methods mainly depend on fixed samples. Recent advances in laser scanning confocal microscopy technique have overcome these limitations and provide us with a powerful tool to investigate the cellular structure and developmental process of plant tissues and organs7,8. Through optical rather than physical tissue sectioning, confocal microscopy allows the collection of a series of z-stack images and the subsequent 3D reconstruction of the sample through image analysis software.
Here, we describe an efficient procedure for investigating both inside and surface structures of the living SAMs from different plant species using laser scanning confocal microscopy, which potentially allows researchers to accomplish all the experimental process within as short as 20 minutes. Different from other published methods for live confocal imaging of Arabidopsis inflorescence SAMs9,10,11,12,13,14,15 and Arabidopsis flowers12,13, here we demonstrate that this protocol is highly efficient for studying not only the inflorescence meristems of the model species but also the vegetative shoot apical meristems from different crops, such as tomato and soybean. This method does not rely on transgenic fluorescent markers, and potentially can be applied to study the shoot meristems from many different species and cultivars. In addition, we also introduce the simple image processing steps for viewing and analyzing different SAMs in a 3D view. Taken together, this simple method will facilitate researchers better understanding both the structure and developmental process of the meristems from both model organisms and crops.
Protocol
1. Media and imaging dishes preparation
- MS plates: Add 0.5x Murashige & Skoog MS medium, 1% agar into deionized water and then adjust pH to 5.8 using potassium hydroxide solution (Optional: Add 1% sucrose for the long-term plant growing). Autoclave and pour plates.
- Imaging dishes: Fill plastic Petri dishes (6 cm wide, 1.5 cm depth) to 0.5-0.8 cm with 1.5% molten agarose.
2. Plant growth
- Arabidopsis growth
- Sow sterilized seeds on MS plates and place plates under 4 °C for two days. Then, move MS plates to a short day (8 h light/ 16 h dark), at 22 °C for two weeks.
- Transplant seedlings to soil and grow them in a short day (8 h light/ 16 h dark) at 22 °C for four weeks.
- Transfer plants to continuous light, at 22 °C to induce the transition from the vegetative to the reproductive stage and for imaging the inflorescence SAMs.
- Tomato and soybean growth
- Incubate the seeds covered with wet filter paper under 28 °C till they germinate.
- Transplant seedlings to soil and grow them in a long day (16 h light/ 8 h dark) at 25 °C for one week or longer for imaging the vegetative SAMs.
3. Dissection of the shoot apex
- Dissection of the inflorescence shoot apex
- Cut the inflorescence shoot apex together with 1-2 cm main stem from the bolted Arabidopsis plants with a razor blade. Hold the basal part of the main stem and remove as many older flower organs as possible from the main stem by dissecting out the peduncles with jewelry forceps.
CAUTION: Avoid cutting fingers when using a razor blade. Dispose of the used razor blades to a proper sharps’ container. - Hold the attached stem of the shoot apex with jewelry forceps or fingers in the field of the stereomicroscope, continue removing the rest of flowers till nearly the whole SAM can be viewed from the eyepieces. Remove peduncles clearly at the junction of the main stem.
- Cut the inflorescence shoot apex together with 1-2 cm main stem from the bolted Arabidopsis plants with a razor blade. Hold the basal part of the main stem and remove as many older flower organs as possible from the main stem by dissecting out the peduncles with jewelry forceps.
- Dissection of the vegetative shoot apex
- To view the vegetative SAMs from either tomato or soybean, dissect out the cotyledons, leaves, and roots from the plants.
- Hold the hypocotyls of the plants under the stereomicroscope and further dissect out the leaf primordia covering the vegetative SAMs using jewelry forceps.
4. Staining
- To directly visualize cells in SAMs, use freshly prepared propidium iodide (PI) solution to stain cell walls. Dissolve PI powder in sterile, deionized water, make 1mL PI staining solution at the concentration of 1 mg/mL and store the PI solution in microcentrifuge tube covered with aluminum foil.
- Pipette 50 µL PI solution in a clean and empty Petri dish and dip the whole dissected shoot apex into dye for 2 min. Rinse the stained shoot apex twice in sterile, deionized water. During the staining process, immerse the whole inflorescence SAM or vegetative SAM into the PI solution to achieve the uniform staining.
5. Image collection
NOTE: For this method, all the SAMs are imaged using an upright confocal microscope and with a 20x water-dipping lens. As described in other protocols9,12,13,15, it is also feasible to image SAMs using an inverted microscope. In addition, the live imaging can be achieved using different brands of confocal microscopes, with the same sample preparation steps. In this study, the imaging steps are described in detail as an example.
- Pierce a hole at the center of an imaging dish using forceps and stick the stained shoot apex upright in the medium.
- Fill the imaging dish with sterile, deionized water to completely immerse the sample. Viewing from the stereo microscope, pipette up and down to remove air bubbles trapped around the meristem. Then, adjust the angle of the stem in the agar to make sure that the SAM is fully visible from directly above.
- Place the imaging dish on the sample stage of the confocal microscope. Lower water-dipping lens and raise the microscope sample stage to let the tip of lens dip into the water.
- Open the confocal microscope software and locate the SAM in the brightfield in the eyepieces. Move the SAM sample right below the objective lens through adjusting the XY controller, then focus on the SAM from eyepieces through cautiously adjusting the Z controller.
- Operate the acquisition function in the confocal microscope software (see Table of Materials), start the Live mode to view the sample from the computer screen, and set up all parameters for the laser scanning experiment.
- When adjusting the parameters, use Range Indicator function to define whether the signal is saturated or not.
- Optionally, apply the Reuse function to reload all the parameter settings from the selected confocal file.
NOTE: Suggested imaging parameters: Laser line (excitation): 515 nm or 561 nm; Emission 570-650 nm; pinhole: 1 airy unit (AU); Gain: 600-750, scan mode: Frame; Frame size: either 512 X 512 or 1024 X 1024; scanning speed: from 7 to Max; Scanning direction: bi-direction; Averaging number: 2-4; Averaging method: mean; Bit depth: 16 Bit; Scanning Interval: 0.5-1 µM. Further, optimize all these parameters based on the nature of different plant samples and the specific imaging needs.
6. Image processing
- For visualizing optical orthogonal and transverse section views, use the same commercial software for the imaging acquisition. Open the original confocal file, click ortho menu, select ortho. Then select either x position, y position and z position of the image, and save the images as the tiff files.
- Optionally select 3D distance function to define the physical distance between two points that have been selected from the stack of the confocal images.
- Alternatively, use Fiji/Image J, the open resource image processing package to visualize the orthogonal and transverse section views.
- Open the original confocal file with Fiji, click image menu, select Stacks and then select Orthogonal views.
- Select XY, YZ, and XZ planes in the middle position and save as Tiff format images.
- For visualizing a 3D transparent projection, use the same software.
- Open the original confocal file, click 3D menu, and select Transparent to generate a 3D projection view.
- Optionally click 3D menu, select Appearance, and then select Transparency to adjust three parameters of the projection including Threshold, Ramp and Maximum for the transparency of the 3D image.
- Click 3D menu, select Appearance, and select Light to adjust the brightness of the 3D image.
- Export the projected images and save them as the Tiff files.
- For visualizing a 3D maximum intensity projection, open the confocal files with the same software, and click the 3D menu.
- Select 3D menu and select Maximum.
- Alternatively, use Fiji/Image J to visualize a 3D maximum intensity projection.
- Open the original confocal file with Fiji, click Image menu and select stacks.
- Select 3D project and save as Tiff format images.
- For visualizing the depth coding view of the 3D images, use the same software.
- Click 3D menu and select Appearance.
- Select Special and select Depth Coding.
- Alternatively, use Fiji/Image J to visualize the depth coding view.
- Open the confocal files, click image menu, select Hyperstack.
- Select Temporal-Color code and save as Tiff format images.
NOTE: The depth coding z-stack also can be achieved through the plugin Z Code Stack for Fiji.
- For visualizing the 3D rotation view as shown (Movie 1 and Movie 2), use the same software (See the Table of Materials).
- Open the confocal files, click 3D menu, and select Series.
- Select Render series, and select one of the four options including Turn around x, Turn around y, Start and end, and Position list.
- Save the render series as the AVI files.
- Alternatively, use Fiji/Image J to visualize the 3D rotation view.
- Open the original confocal file with Fiji, click Image menu, and select Stacks.
- Select 3D project and save as AVI format videos.
Representative Results
To evaluate the efficiency of our protocol and to explore the morphology of the meristems from different species, we have performed the confocal live imaging experiments on the inflorescence meristem from Arabidopsis and the vegetative meristems from both tomato and soybean. In this study, Arabidopsis ecotype Landsberg erecta, tomato cultivar Micro-Tom and soybean cultivar Williams 82 have been used as examples.
Seen from the orthogonal section through the middle of the Arabidopsis SAM, it is clear that PI was able to stain the horizontal walls from almost all the cells at the multiple cell layers (Figure 1A). Shown from one transverse section through the corpus of the inflorescence SAM, the cells from the XY plane are also clearly imaged (Figure 1B). In the 3D projection view, the inflorescence meristem forms a dome like structure and is surrounded by the developing flower primordia, where the cells are also stained and imaged (Figure 1 C-D) (Movie 1).
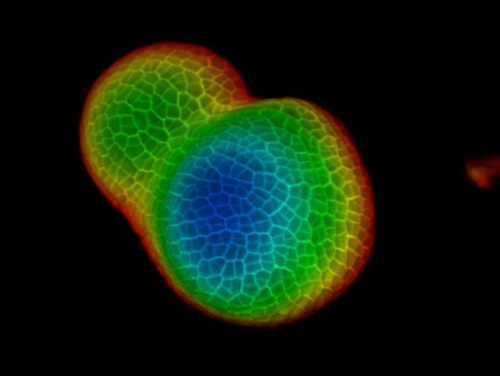
Seen from the orthogonal section through the middle of the tomato SAM, it is clear that PI was able to stain the horizontal walls from cells at the multiple cell layers, although the PI signal from the deep interior region is slightly lower. From one transverse section through the deep layers of the vegetative SAM, the cells from the XY planes are also clearly imaged, and the boundary formed between the vegetative meristem and leaf primordia is also imaged (Figure 2B). The 3D project view can further provide a comprehensive view of the shape and organization of the vegetative meristem from tomato (Figure 2C-D) (Movie 2).
In the orthogonal view through the middle of the soybean SAM, we can see the dome-like vegetative meristem and its derived new leaf primordia (Figure 3A). In the 3D projection view, both the soybean vegetative meristem and the tomato vegetative meristem form the dome like structure, however, the shape of the soybean vegetative meristem is different from that of the tomato meristem, and the organization and patterns of the leaf primordia surrounding these two SAMs are distinct (Figure 3B-C).
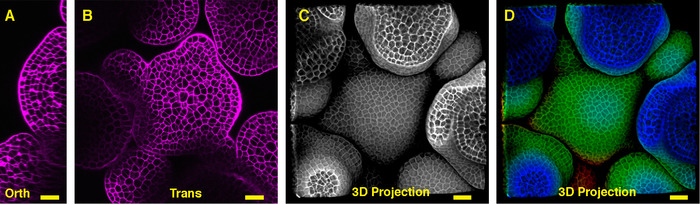
Figure 1: Live imaging and analyzing the inflorescence SAM of Arabidopsis. A and B. optical orthogonal (Orth) and transverse (Trans) section views of middle plane of the same SAM, PI (propidium iodide) stain (purple). C. a 3D projection of the same SAM, PI stain (gray). D. depth color coding of the 3D projection, with blue indicating the top surface layer and red representing the deepest layer. Cell walls were stained with PI. Scale bars: 20 µm (A, B); 50 µm (C, D). Please click here to view a larger version of this figure.
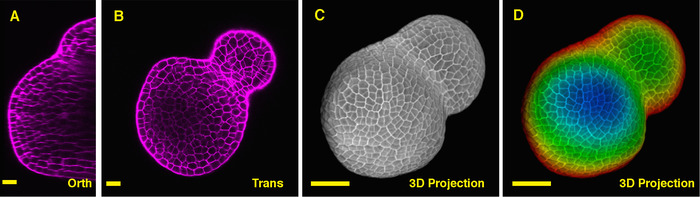
Figure 2: Live imaging and analyzing the vegetative SAM of tomato. A and B. optical orthogonal (Orth) and transverse (Trans) section views of middle plane of the same SAM. C. a 3D projection of the same SAM. D. depth color coding of the 3D projection, with blue indicating the top surface layer and red representing the deepest layer. Cell walls were stained with PI. Scale bars: 20 µm (A, B); 50 µm (C, D). Please click here to view a larger version of this figure.
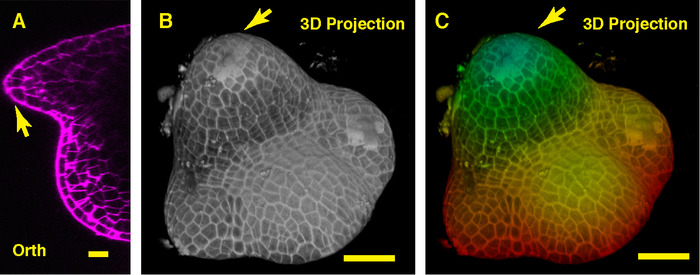
Figure 3: Live imaging and analyzing the vegetative SAM of soybean. A. an optical orthogonal (Orth) section view of the middle plane of the vegetative SAM. B. a 3D projection of the same SAM. C. depth color coding of the 3D projection, with blue indicating the top surface layer and red representing the deepest layer. Cell walls were stained with PI. Scale bars: 20 µm (A); 50 µm (B, C). Arrow: leaf primordium. Please click here to view a larger version of this figure.
Video 1: 3D rotation of the Arabidopsis inflorescence meristem in Figure 1. Please click here to view this video. (Right-click to download.)
Video 2: 3D rotation of the tomato vegetative meristem in Figure 2. Please click here to view this video. (Right-click to download.)
Discussion
Here, we describe a simple imaging method that can be applied to the study of shoot apical meristems from different plants with minor modification, opening a new avenue to study the meristem regulation at both vegetative and reproductive stages in model plants and crops. In contrast to the SEM and histological staining methods, this protocol can help reveal both surface view and internal cellular structures of the SAMs, without the need for labor-intensive sample fixation and/or tissue sectioning steps. This protocol is also compatible to the established imaging method for the fluorescence based reporters in the SAMs, potentially providing a good cellular resolution which allows us to obtain a 3D view of expression patterns of key genes and proteins in the SAM16,17,18,19. In addition, the associated image processing procedure described here will be able to greatly help researchers analyze and compare the SAMs from different species in 3D, advancing both evolutionary-developmental biology studies and agricultural research.
There are a few critical steps in this simple protocol. First, sample preparation and dissection. A plant SAM is usually hidden by developing primordia and young organs at the shoot tip, and it cannot be directly imaged under a confocal microscope. To image the vegetative SAM, it is necessary to remove all the leaves and older leaf primordia that cover on top of the SAM using fine jewelry forceps. To image the inflorescence SAM, it is necessary to carefully remove all the young flowers to expose the SAM, also using fine jewelry forceps.
Second, the live cell staining using the PI solution. In this protocol, we use freshly prepared PI solution (1 mg/mL) to stain cell walls to directly visualize live SAMs, which is fast, efficient, and easy to perform. Although the PI solution can be stored at 4 °C for several weeks, a freshly prepared solution usually gives the best staining result for the shoot apical meristems. Although PI mainly does not cross the membrane of living cells and it stains intact cells with the clear cellular outline, it can easily penetrate the damaged/dead cells and strongly stain the nuclei and other internal membrane systems in those damaged areas, potentially affecting the quality of the confocal images. Thus, it will be essential to avoid any physical damages while preparing the SAM samples for the staining and live imaging. On the other hand, PI at a high concentration shows a toxic effect to plant cells, and thus, FM4-64 can be used as a substitute for PI to stain the living SAM cells9. However, FM4-64 labels plasma membrane, which may potentially be taken up into cell interior through endocytosis, making it challenging to stain the samples with high cellular resolution.
Third, imaging acquisition. 1). High numerical apertures (NA), the water-dipping lens is critical for the live imaging of the SAMs. Water has a higher index of refraction than air and the high NA of the objective lens helps collect signal photons scattered through deep cell layers of SAMs. 2). The settings of laser power (watts or watts per surface) can be highly variable between different confocal microscopes. Higher laser power may allow better signal collection but leads to more photodamage/ photobleaching on the samples. There are trade-offs between resolution, bleaching and imaging time. In general, selecting a larger frame size leads to better XY resolution but more imaging time, and selecting a smaller scanning interval leads to a better Z resolution but also more imaging time. Longer imaging time potentially causes more photodamage/ photobleaching
With this method, sometimes it may not be easy to observe cellular details in deeper tissue layers, likely due to the limitation of the confocal detection and the inefficient PI staining in internal tissues. Based on our experiences, increasing the concentration of PI solution and/or the staining time can help get a better stain. Alternatively, the modifications to the staining procedure can be made. For example, the modified pseudo-Schiff propidium iodide (mPS-PI) method works well for the fixed tissues20. And it is still necessary to find alternative dyes with better staining for the living tissues in future. In addition, it is also interesting to test whether the method described here can be generally applied to the study of SAMs from all the other flowering plants, considering the fact that some plant species have special cellular contents or different cell wall compositions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge Purdue Bindley Bioscience Center Imaging Facility for accessing the laser scanning confocal microscope and for the technical support, and the authors appreciate the help from Andy Schaber in the Purdue Bindley Imaging Facility. This activity was funded by Purdue University as part of AgSEED Crossroads funding to support Indiana’s Agriculture and Rural Development.
Materials
| Agar Phyto | Dot Scientific Inc. | DSA20300-1000 | |
| Agarose | Dot Scientific Inc. | AGLE-500 | |
| Forceps | ROBOZ | RS-4955 | Dumont #5SF Super Fine Forceps Inox Tip Size .025 X .005mm, for dissecting shoot apices. |
| LSM 880 Upright Confocal Microscope | Zeiss | ||
| Murashige & Skoog MS medium | Dot Scientific Inc. | DSM10200-50 | |
| Plan APO 20x/1.1 water dipping lens | Zeiss | ||
| Plastic petri dishes 100 mm X 15 mm | CELLTREAT Scientific Products | 229694 | Use as making MS plates |
| Plastic petri dishes60 mm X 15 | CELLTREAT Scientific Products | 229665 | Use as imaging dishes |
| Propagation Mix | Sungro Horticulture | ||
| Propidium iodide | Acros Organics | 440300250 | 1 mg/mL solution in water, to stain the cell walls |
| Razor blade | PERSONNA | 62-0179 | For cutting shoot apex from plants |
| Stereomicroscope | Nikon | SMZ1000 | |
| Tissue | VWR | 82003-820 | |
| Zen black | Zeiss | Image acquisition software |
References
- Meyerowitz, E. M. Genetic control of cell division patterns in developing plants. Cell. 88 (3), 299-308 (1997).
- Xu, C., et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nature Genetics. 47 (7), 784-792 (2015).
- Bommert, P., Nagasawa, N. S., Jackson, D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nature Genetics. 45 (3), 334-337 (2013).
- Je, B. I., et al. Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nature Genetics. 48 (7), 785-791 (2016).
- Ping, J., et al. Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell. 26 (7), 2831-2842 (2014).
- Vaughan, J. G., Jones, F. R. Structure of the angiosperm inflorescence apex. Nature. 171, 751 (1953).
- Sijacic, P., Liu, Z. Novel insights from live-imaging in shoot meristem development. Journal of Integrative Plant Biology. 52 (4), 393-399 (2010).
- Tax, F. E., Durbak, A. Meristems in the movies: live imaging as a tool for decoding intercellular signaling in shoot apical meristems. Plant Cell. 18 (6), 1331 (2006).
- Grandjean, O., et al. In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell. 16 (1), 74-87 (2004).
- Heisler, M. G., Ohno, C. Live-imaging of the Arabidopsis inflorescence meristem. Methods in Molecular Biology. 1110, 431-440 (2014).
- Tobin, C. J., Meyerowitz, E. M. Real-time lineage analysis to study cell division orientation in the Arabidopsis shoot meristem. Methods in Molecular Biology. 1370, 147-167 (2016).
- Prunet, N. Live confocal Imaging of developing Arabidopsis flowers. Journal of Visualized Experiments. (122), e55156 (2017).
- Prunet, N., et al. Live confocal imaging of Arabidopsis flower buds. Developmental Biology. 419, 114-120 (2016).
- Reddy, G. V., Heisler, M. G., Ehrhardt, D. W., Meyerowitz, E. M. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 131, 4225-4237 (2004).
- Nimchuk, Z. L., Perdue, T. D. Live Imaging of Shoot Meristems on an Inverted Confocal Microscope Using an Objective Lens Inverter Attachment. Frontiers in Plant Science. 8, 773 (2017).
- Zhou, Y., et al. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science. 361 (6401), 502-506 (2018).
- Zhou, Y., et al. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature. 517 (7534), 377-380 (2015).
- Nimchuk, Z. L., Zhou, Y., Tarr, P. T., Peterson, B. A., Meyerowitz, E. M. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development. 142 (6), 1043 (2015).
- Li, W., et al. LEAFY Controls Auxin Response Pathways in Floral Primordium Formation. Science Signaling. 6 (270), ra23 (2013).
- Truernit, E., et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell. 20 (6), 1494-1503 (2008).

