Performing Spectroscopy on Plasmonic Nanoparticles with Transmission-Based Nomarski-Type Differential Interference Contrast Microscopy
Summary
The goal of this protocol is to detail a proven approach for the preparation of plasmonic nanoparticle samples and for performing single particle spectroscopy on them with differential interference contrast (DIC) microscopy.
Abstract
Differential interference contrast (DIC) microscopy is a powerful imaging tool that is most commonly employed for imaging microscale objects using visible-range light. The purpose of this protocol is to detail a proven method for preparing plasmonic nanoparticle samples and performing single particle spectroscopy on them with DIC microscopy. Several important steps must be followed carefully in order to perform repeatable spectroscopy experiments. First, landmarks can be etched into the sample substrate, which aids in locating the sample surface and in tracking the region of interest during experiments. Next, the substrate must be properly cleaned of debris and contaminants that can otherwise hinder or obscure examination of the sample. Once a sample is properly prepared, the optical path of the microscope must be aligned, using Kohler Illumination. With a standard Nomarski style DIC microscope, rotation of the sample may be necessary, particularly when the plasmonic nanoparticles exhibit orientation-dependent optical properties. Because DIC microscopy has two inherent orthogonal polarization fields, the wavelength-dependent DIC contrast pattern reveals the orientation of rod-shaped plasmonic nanoparticles. Finally, data acquisition and data analyses must be carefully performed. It is common to represent DIC-based spectroscopy data as a contrast value, but it is also possible to present it as intensity data. In this demonstration of DIC for single particle spectroscopy, the focus is on spherical and rod-shaped gold nanoparticles.
Introduction
Since the 1980s, differential interference contrast (DIC) microscopy has largely been viewed as an important imaging method reserved for microscale objects within the biological sciences. However, during its development in the 1950s and 1960s, it was intended as a technique for materials science1. With the recent advancements in the material sciences related to plasmonic nanoparticles, an increased interest in the characterization of materials with optical microscopy has taken place.
Many optical techniques are certainly available for nanomaterial characterization (e.g., dark field, brightfield, polarized light, fluorescence, etc.). Dark field is widely popular in nanoparticle research, but it relies solely on the collection of scatter and provides limited information about complex samples2. Fluorescence can be useful, but only with samples that luminesce or that can be properly stained. DIC microscopy has several traits that make it a valuable tool for the analysis of nanoparticles. The most frequently stated advantages of DIC in comparison to other methods and in regards to plasmonic nanoparticles are: no sample staining required, no halo effects, shallow depth of field, and high lateral resolution3. DIC has additional strengths that are valuable to plasmonic nanoparticle research. First of all, two inherent and orthogonal polarization fields are present, and they can be measured simultaneously for spectroscopy purposes2. Secondly, the depolarized signal of nanoparticles is not captured in the final image2, which can be a cause for serious concern in dark field spectroscopy measurements.
The purpose of this article is to provide a clear methodology for utilizing transmitted-light Nomarski DIC microscopy to perform spectroscopy on plasmonic nanoparticles. Although DIC is a powerful technique that can be applied to highly diverse materials, it is also a technique that requires great skill and understanding to operate it properly when imaging nanoparticles. Transmission-based Nomarski DIC microscopy has a complex light path1 that will only be briefly reviewed here. The optical train of DIC is displayed in Figure 1. Light is transmitted through the microscope by first being passed through a polarizer and a beam-splitting Nomarski prism before being focused by the condenser onto the sample plane. After passing through the objective, the light encounters a beam-combining Nomarski prism and an analyzer before exiting to the detector. The two polarizers and Nomarski prisms are critical to formation of the DIC image and are responsible for producing DIC’s two orthogonal polarization fields1. For the reader interested in knowing more about the working principles and optical path of Nomarski DIC microscopes, or the differences between Nomarski DIC and other styles of DIC, please refer to other well-written accounts on these topics1,4,5,6,7.
It is equally important to understand the basic nature of plasmonic nanoparticles before attempting to perform spectroscopy on them, whether it be with Nomarski DIC, dark field, or any other microscopy technique. In the field of plasmonics, nanoparticles are defined as particulates with dimensions on the scale of 10-100 nm8,9. Nanoparticles can take on many shapes (e.g., spheres, rods, stars, dumbbells, etc.), and most of their important properties arise from interactions with light in the ultraviolet-visible-near infrared range of the electromagnetic spectrum. The term “plasmonic” is not restricted to nanoparticles10; however, when discussing nanoparticles, it is used in reference to localized surface plasmon resonance (LSPR). LSPR is a phenomenon in which the conduction electrons in a nanoparticle oscillate due to a Coulombic interaction with electromagnetic radiation of a highly specific and relatively narrow frequency band8. At these same frequencies, plasmonic nanoparticles exhibit increased absorption and scattering of light, making them observable with optical microscopy. In many cases, it is preferred to observe the nanoparticles while placing bandpass filters before the condenser2, to improve imaging contrast and to eliminate light that fails to induce the LSPR effect. Using filters also makes it possible to perform single particle spectroscopy experiments.
LSPR-related optical behavior is highly dependent on the size and shape of the nanoparticles, and it can be investigated with many optical microscopy techniques. However, in order to decipher orientation information of plasmonic nanoparticles with an anisotropic (i.e., non-spherical) shape, it is necessary to utilize polarization of the light field. By carefully rotating the polarization field or the sample substrate at small increments, it is possible to monitor the orientation-dependent spectroscopic properties of individual nanoparticles. Rotation and polarization can also aid in determining whether a spectral feature is due to a dipolar or higher order oscillation of the nanoparticle’s surface electrons. However, in the case of isotropic (i.e., spherical) nanoparticles, the spectral profile remains essentially unchanged upon rotating the sample under polarized light.
When viewed through a DIC microscope (Figure 2), nanoparticles have an airy disk with a shadow-cast white-and-black appearance against a gray background. Spherical nanoparticles will retain this appearance under rotation and with the changing of bandpass filters; however, the particles will gradually fade from view as the filter’s central wavelength becomes further separated from the sphere’s only dipolar LSPR wavelength11. The appearance of nanorods can change quite dramatically as they are rotated2. Nanorods have two LSPR bands with dipolar behavior, the location of which are based on the physical dimensions of the nanorods. When the longitudinal axis of a nanorod is oriented parallel to one of the DIC polarization fields, the airy disc will appear all white or all black if viewed with a bandpass filter associated with that LSPR wavelength. After rotating the sample 90°, it will take on the opposite color. Alternatively, since the transverse axis of a nanorod is perpendicular to the longitudinal axis, the rod will take on the opposite color when switching between filters that match the LSPR wavelengths for the two axes. At other orientations and filter settings, nanorods will appear more like spheres, presenting a variety of shadow-cast airy disc patterns. For nanorods with a transverse axis < 25 nm, it can be difficult to detect signal at that LSPR’s wavelength using DIC microscopy.
To perform single particle spectroscopy, it is important to use the correct optical components and to align them properly. An objective capable of DIC microscopy must be used. For single particle experiments, 80x or 100x oil objectives are ideal. Nomarski DIC prisms ordinarily come in three varieties: standard, high contrast, and high resolution. The ideal type highly depends on the purpose of the experiment and the size of the nanoparticles. Standard prisms are fine for many experiments; but when working with smaller nanoparticles (< 50 nm), high contrast prisms can be beneficial, since particle contrast decreases as the particles decrease in size11. Adjusting the DIC contrast is achieved either by rotating a polarizer or by translating one of the DIC prisms, depending on the microscope brand or model6.
After setting Kohler illumination and the polarizer settings, it is critical to not readjust these settings while collecting spectroscopy data. Furthermore, a constant average background signal must be maintained at all times during data collection, even when switching between filters and angle settings. The actual ideal background value depends on the dynamic range of the scientific camera, but in general, the background should be in the range of 15%–40% of the maximum detection level of the camera. This reduces the likelihood of saturating the camera sensor while enabling optimal particle contrast. For collecting spectroscopy data, it is necessary to work with a scientific camera that captures images in black and white, as opposed to a color camera.
Sample preparation is another critical aspect of imaging plasmonic nanoparticles. It is imperative that operators of DIC microscopy have an understanding of the sample’s optical properties and of the sample’s substrate. “Pre-cleaned” microscope glass is not sufficiently prepared for imaging nanoparticles, and it must be properly re-cleaned before sample deposition to ensure unobstructed observation of the sample. Many cleaning protocols for microscope slides have been previously documented12, but it is not a step that is normally reported in experimental studies.
Finally, data analysis methods are the final component to single particle spectroscopy. The maximum and minimum intensities for each nanoparticle must be measured, as well as the local background average. Particles of interest should be located in areas with no background debris, substrate defects, or uneven illumination. One method for determining the spectral profile of a nanoparticle is by calculating particle contrast at each wavelength, using the equation below11,13,14,15:
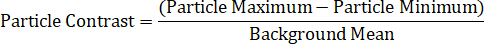
Alternatively, a single particle’s spectrum can be split into its individual maximum and minimum signal components, which represent DIC’s two polarization fields, thereby displaying the two simultaneously-collected directionally-dependent spectra, through the two equations:
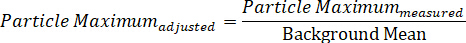
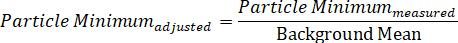
Protocol
1. Sample preparation with standard glass microscopy slides
- Prepare glass microscope slides for sample deposition.
NOTE: In some circumstances, it may be more appropriate to store the glass in ultrapure water instead of ethanol. However, storing in water or air makes the glass hydrophobic over time.- For best results, purchase glass or quartz microscope slides and cover glass.
- Using a scribing pen, place a shallow and short scratch mark onto the center of each glass cover slip.
- Clean all microscope glass, even if it is purchased “pre-cleaned”, to remove glass shards, dust, powder, organic residue, and any other contaminants that affect imaging quality or sample deposition.
NOTE: This cleaning method below works well for the types of samples described here and avoids the use of harsh chemicals. Harsher chemicals can etch the glass and require more care in handling and disposal.- Place microscope glass onto storage racks and then into a beaker, or into a staining jar. Do not place microscope glass at the bottom of beakers and other lab glassware without racking, because each piece and surface of microscope glass should be fully exposed to the cleaning agents.
- Pour ~1 mL of liquid detergent (Table of Materials) into the container and top off the container with water. Sonicate for 30 min.
NOTE: Once the cleaning process begins, only handle the glass while wearing gloves, to avoid leaving fingerprint residue on the glass. - Pour out the liquid contents of the cleaning container into a sink. Rinse container several times with ultrapure water to remove all appearance of detergent. Refill the container with ultrapure water. Sonicate the container with microscope glass for another 30 min.
- Repeat the previous step at least once more. Perform additional rounds of sonication in water until it is obvious that all traces of the detergent have been removed.
- Pour out the contents of the cleaning container. Rinse the container with ultrapure water. Refill the container with ethanol. Sonicate microscope glass for 30 min.
- Pour out the contents of the cleaning container into a waste container. Refill with ethanol. Cover the container to prevent loss of ethanol through evaporation. Store the microscope glass in this container until time of experiment. Slides remain clean and usable as long as they remain submerged in ethanol inside a covered container.
- Preparation of nanoparticle solution
- Using a micropipette, remove a 100 µL aliquot of 0.05 mg/mL gold nanoparticle solution from its original storage container and eject the solution into a 1.5 mL centrifuge tube.
- Centrifuge the sample for 10 min at 6,000 x g.
- Remove the supernatant with a micropipette, in order to remove excess surfactant.
- Using a micropipette, place 100 µL of ultrapure water into the centrifuge tube.
NOTE: If not all of the supernatant can be removed on the first attempt, repeat the centrifugation and resuspension steps. - Briefly vortex the sample to resuspend the pellet. Sonicate immediately afterwards for 20 min to fully resuspend and break up nanoparticle aggregates.
NOTE: If the sample is not used immediately, it should be sonicated again for 20 min before depositing solution onto microscope glass.
- Sample deposition
- Remove cleaned cover slips and microscope slides from their storage containers. Blow dry the glass with pressurized nitrogen or argon.
- Using a micropipette, drop-cast 6 µL of nanoparticle solution from step 1.2.5 onto the cover slip. To spread out the droplet evenly, carefully place a second, larger piece of microscope glass on top of the cover slip, such as a second cover slip or a microscope slide. Avoid getting air bubbles trapped between the two pieces of glass.
- Turn the sample substrate over, and seal off the edges of the cover slip with a narrow line of nail polish in order to prevent evaporation of the medium solution.
- Alternatively, to image the sample “dry”, allow the solution to stand for 5–15 min on the cover slip, before removing the unwanted piece of glass. Gently blow the cover slip dry with pressurized nitrogen or argon.
- If possible, image samples immediately after preparation. If that is not possible, store samples in a covered container, such as a Petri dish until imaging.
2. DIC imaging
- Align objective and condenser.
- After placing the sample onto the microscope, find the focal plane with the sample on it. First locate and focus on the scratch mark created earlier. Then fine tune the focus until nanoparticles come into view.
- To determine the accurate placement of the condenser, utilize the Kohler Illumination method.5 Kohler Illumination at high magnification (80x, 100x) is more easily achieved by first setting the Kohler Illumination at a lower magnification, such as 20x.
NOTE: Normally, Kohler Illumination does not need to be re-adjusted during imaging of a single sample. However, it is good practice to verify that Kohler Illumination is properly set when switching to a new microscope slide.
- Optimize contrast settings.
- Select a region of interest within the sample for imaging. Center the region in the camera’s field of view and adjust focus as necessary.
- If the microscope has the de Senarmont design, start with the polarizer set near to maximum background extinction and gradually rotate the polarizer towards decreasing background extinction. The background intensity will gradually increase.
- If the microscope does not have a de Senarmont design, start with the optical train set at maximum background extinction. In this case, gradually adjust the objective prism position towards decreasing background extinction.
NOTE: The ideal setting is achieved when the nanoparticles reach their greatest intensity difference (i.e., contrast) from the averaged local background value. For plasmonic nanoparticles, optimal contrast is normally achieved with a relatively dark background, thus at settings near maximum background extinction.
- Select a region of interest within the sample for imaging. Center the region in the camera’s field of view and adjust focus as necessary.
- Image the sample.
- Turn off room lighting to prevent stray illumination from interacting with process.
- While viewing the nanoparticles with a scientific imaging camera, determine the optimal background level. Using a 10 nm full width at half maximum (FWHM) bandpass filter with its central wavelength co-located with the main LSPR wavelength, view the region of interest. Adjust the lamp intensity or exposure time until the background level is in the range of 15%–40% of the camera’s maximum capacity level and no objects within the region of interest exhibit signal intensities that exceed 90% of the camera’s maximum intensity level.
NOTE: The goal of step 2.3.2 is to prevent saturating the sensor when switching between filters. The ideal background level will vary between samples and cameras. Once this step is completed, exposure time can be adjusted but not the lamp intensity. - Image the sample with a series of bandpass filters that each has a FWHM of 10 nm and that as a whole enable imaging across the entire wavelength range of interest. Ensure that the background intensity remains consistent from image to image (within ~5% of one another) by adjusting the exposure time. After switching filters, re-focus the sample before image capture.
- Save the Images as uncompressed TIFF files and/or in the software’s native file format, in order to preserve all information.
- Rotate the sample.
- After collecting images of the sample at its original position, the sample can now be rotated and imaged at additional orientations in the light path. Perform rotation at regular intervals (e.g., 10° or 15°) across either a 180° or 360° range.
NOTE: Rotation requires a rotatable sample stage. - As in sections 2.1-2.3, adjust camera settings to provide a consistent background level from image to image.
NOTE: No adjustment to Kohler Illumination should be made.
- After collecting images of the sample at its original position, the sample can now be rotated and imaged at additional orientations in the light path. Perform rotation at regular intervals (e.g., 10° or 15°) across either a 180° or 360° range.
3. Data analysis using ImageJ
NOTE: The following calculations can be performed in a variety of software packages, and sometimes in the native program used to collect the images. ImageJ is a freely available software from the National Institutes of Health (NIH).
- Calculate particle contrast or intensity.
- Open the image with ImageJ.
- Select the Rectangle tool and draw a rectangle around the main region of interest.
- On the Tool bar, select Image, then Zoom, then To Selection. The imaging window will zoom in on the selected area.
- On the Tool bar, select Image, then Adjust, then Brightness/Contrast. A new window appears. To enable better viewing of the sample region, adjust the four settings: Minimum, Maximum, Brightness, and Contrast. These adjustments do not alter the scientific data, they merely enable better visibility of the sample region.
NOTE: Steps 3.1.3 and 3.1.4 may be performed multiple times and in reverse order. - Using the rectangle tool again, draw a box around the first nanoparticle to be measured. The box should be only slightly larger than the nanoparticle’s airy disc.
- On the Tool bar, select Analyze, then Measure. A new window appears that reports the Minimum, Maximum, and Mean Intensities for the pixels located inside of the selected box.
- Drag the box used to measure the nanoparticle to an area immediately adjacent to the particle, where the background contrast is relatively even and no particles or contaminants are present. Retain the original size of the box.
- Use the Measure tool to determine the Mean Intensity for the background area.
- Measure the remaining particles and an adjacent background area for each.
- Repeat the process for each particle in all of the images in the series.
- Export the data to a spreadsheet to calculate the contrast or intensity of each particle, across all wavelengths and angles.
- Calculate each particle’s contrast, using the following equation13,14,15:
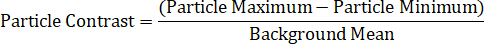
NOTE: Using this equation, particle contrast should always be > 0. - Calculate the particle’s background-adjusted maximum value by dividing the measured maximum particle intensity by the background mean:
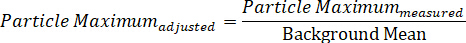
- Likewise, calculate the background-adjusted minimum value by dividing the measured minimum particle intensity by the background mean:
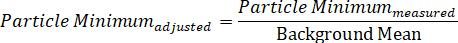
NOTE: As calculated, the maximum should have a value greater than one, while the minimum will be less than one. It is acceptable to subtract each value by “1”, so that the average background is essentially zero, the maximum is represented as a positive value, and the minimum value is assigned a negative value16. This latter approach allows the analyst to separately consider what is occurring along each of the polarization fields, which is useful when studying anisotropic particles. - To graph the spectral profile at a given nanoparticle position, plot data with the wavelength along the x-axis and the contrast or intensity along the y-axis.
- To graph the rotational profile at a given wavelength, plot the rotation angle along the x-axis and the contrast or intensity along the y-axis.
Representative Results
When working with samples that are large enough to be seen with the naked eye, placing landmarks on the glass substrate is not normally required. However, when working with nanomaterials or when rotation of the sample is required, landmarks can provide an easy method for locating, distinguishing, and tracking the orientation of the sample. Although more sophisticated techniques can be utilized for leaving landmarks on glass substrates17, scratching the glass with a scribing pen is an economical and simple method that works in many situations. It is important to avoid examining sample regions that are immediately adjacent to these landmarks, since scratch marks create a complex background with the potential of impacting data (Figure 3). However, at the tips of the scratch marks, “spider webs” often extend outward from the scratch. These lines are quite valuable as landmarks, but again, nanoparticles should be avoided if they overlap with these defects.
In order to achieve optimal imaging with DIC microscopy, it is of vital importance to determine the proper focal plane. Objects that are slightly out of focus will appear fuzzy, have blurred edges, and have decreased contrast. Figure 4 displays gold nanoparticles that are out of focus to varying degrees. Nanoparticles in the bottom right corner are in focus, while nanoparticles become farther out of focus as they approach the upper left corner of this image. Because DIC has a shallow depth of field, it is not uncommon for some nanoparticles to be in focus while others are out of focus when imaging them on a glass substrate. As a result, it is critical to consistently focus on the same exact particles when making adjustments to the microscope during an experiment.
Figure 5 provides an example of the effect of adjusting the polarizer settings while imaging gold nanospheres. Five nanoparticles are in focus, while one is slightly out of focus. A 540 nm bandpass filter with 10 nm FWHM was also in the optical path. In this series of images, the background brightness was adjusted with ImageJ after image acquisition in order to make the five particles more apparent against the background. When the polarizer is set at 0° in a de Senarmont designed Nomarski DIC microscope, it is orthogonal to the analyzer (Figure 5A). At 0°, the particles appear mostly white, with a dark stripe running across their mid-section. This is indicative of cross-polarization for nanosphere samples. When the polarizer is rotated to different angles (Figure 5B-E), the particles appear to be casting dark shadows towards the southwest. The black and white components to the signal arise as a result of DIC’s two polarization fields and provide information about the orientation of plasmonic nanoparticles when working with bandpass filters. As the polarizer is rotated towards higher angles, the shadow pattern remains similar. However, the particle contrast values change dramatically. This is best demonstrated by measuring the contrast values for the individual particles, using the equation provided above. The particle highlighted with the black box has contrast values of 0.65 (crossed polarizers), 0.84 (polarizer shift of 5°), 1.10 (10°), 0.44 (20°), and 0.23 (45°). Therefore, for this sample, the optimal imaging setting is with a polarizer shift of 10°. Plasmonic nanoparticles often require a polarizer setting in the range of 5°–15°, and smaller increments than these should normally be used to identify the ideal setting. For further information on the imaging and analysis of spherical gold nanoparticles, readers are referred to the prior work by Sun et al.11.
Anisotropic-shaped nanoparticles produce patterns of higher complexity than nanospheres. Gold nanorods were imaged (Figure 6) at their longitudinal LSPR wavelength, 650 nm. In the initial image (Figure 6A), five bright nanorods and several dimmer particles are apparent. Instead of having a shadow-cast appearance, three of the rods have predominately black airy discs while two are mostly white. The polarizer was set at 10° to the left of the crossed polarization setting. In Figure 6B, crossed polarization was used; only three of the particles appear, as fully white airy discs. The others have disappeared or appear to be slightly out of focus. With the polarizer set at 10° to the right of crossed polarization (Figure 6C), the patterns are now reversed of what was observed in Figure 6A. The polarizer was next turned to 45° right of crossed polarization (Figure 6D), the maximum setting, to demonstrate that particles retain their colors at this setting, but contrast has declined significantly. In the remaining figure panels, the collection of nanorods was incrementally rotated a full 90° clockwise while the polarizer was set at 10° to the right of crossed polarization. The pattern gradually changes for each nanorod, and after a full 90° rotation, the particles have reversed their colors from the initial setting. In brief, if one of the axes of a plasmonic nanorod is lined up with one of the two polarization fields, and if the nanorod is imaged at that axis’ LSPR wavelength, the nanorod will appear to be mostly white or mostly black, depending on which polarization field it is aligned with (Figure 6A,C)2. If the nanoparticle is rotated a full 90° (Figure 6H), it will now be lined up with the opposite polarization field and take on the opposite color. If instead the nanoparticle was rotated only 45° (Figure 6F), then it will be in a position where the particle will exhibit its greatest shadow-cast appearance, showing striking similarity to what is observed with the plasmonic nanospheres. As a result of this optical behavior, plasmonic nanoparticles with an anisotropic shape often look flat instead of having the three-dimensional shadow-cast appearance of nanospheres. The result of this difference in optical behavior is that it can be exploited in order to distinguish between plasmonic nanoparticles that are spherical and anisotropic in shape, as has been previously discussed in multiple research studies2,3,6,7,11,13,16.
Finally, Figure 7 displays representative single particle spectroscopy data, as contrast of a gold nanosphere (Figure 7A)11, intensity of a single gold nanorod with its longitudinal axis oriented parallel to one of the polarization fields (Figure 7B)6, and intensity profile of a single gold nanorod at its LSPR wavelength and during rotation of the stage (Figure 7C)6. Either method of presentation reveals the width and location of the LSPR effect. For plasmonic nanoparticles with an anisotropic shape, the intensity and rotation data reveal the directionality of the effect, and hence, the orientation of the particle on the sample substrate, which has been previously proven through correlative studies on such particles using DIC and transmission electron microscopy2,16,18.
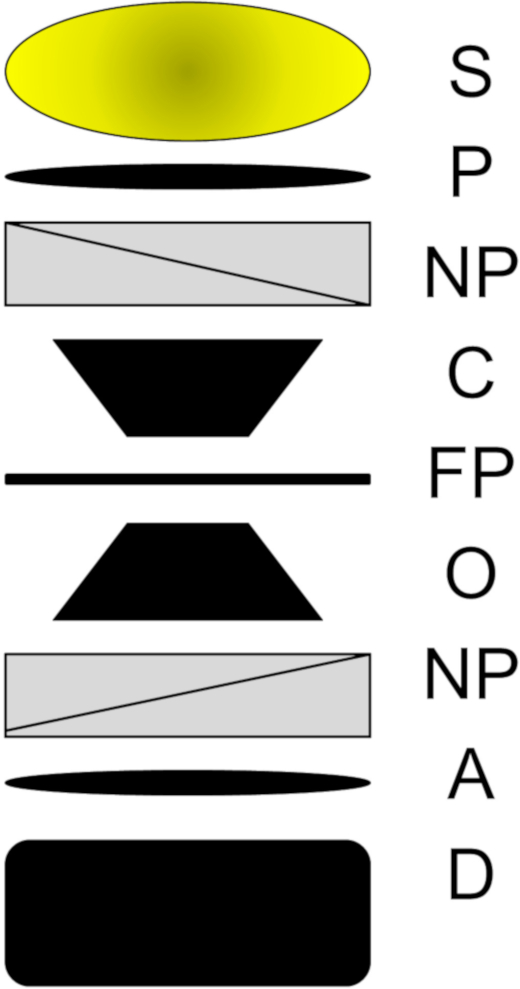
Figure 1: The light path in transmitted-light Nomarski-based DIC microscopy. After leaving the light source (S), light passes through a polarizer (P), a beam-shearing Nomarski prism (NP), the condenser (C), the focal plane (FP), the objective (O), a beam-combining Nomarski prism (NP), the analyzer (A), and finally the detector (D). Please click here to view a larger version of this figure.
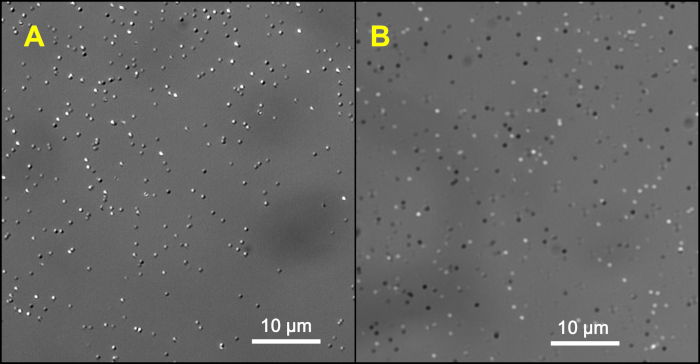
Figure 2: Examples of plasmonic nanoparticles imaged at their LSPR wavelengths with 10 nm bandpass filters, using a DIC microscope. Both images are collected at 100x. (A) Silver nanospheres with 40 nm diameter imaged at 480 nm with a bandpass filter having 10 nm FWHM. (B) Rod-like gold nanoparticles imaged at 700 nm using a bandpass filter with 10 nm FWHM. Please click here to view a larger version of this figure.
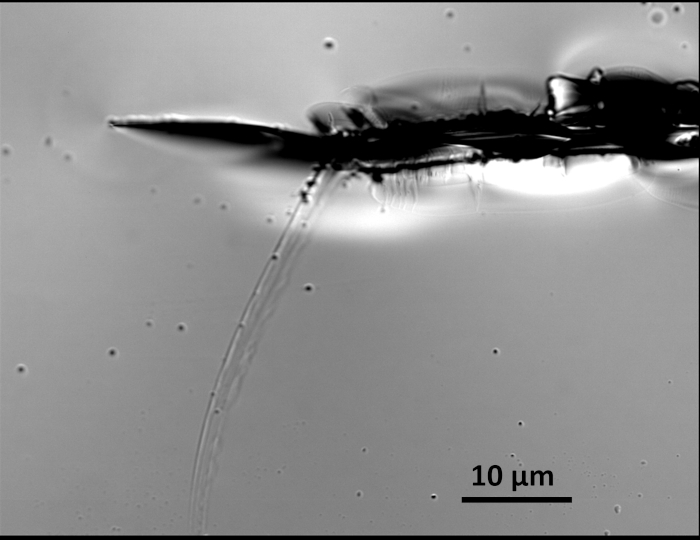
Figure 3: Image of a scratch made into a glass cover slip with a scribing pen. Near the end of the actual indentation, a series of narrow and shallow “spider web” lines branch out from the scratch itself, resulting in a pattern that can be utilized as an imaging landmark. This image was collected using 100x magnification and broadband white light. Please click here to view a larger version of this figure.

Figure 4: Gold nanospheres imaged with broadband white light at 100x. Particles in the lower right are in focus but particles drift farther from the focal plane towards the upper left corner. Object in middle of image is debris. Please click here to view a larger version of this figure.
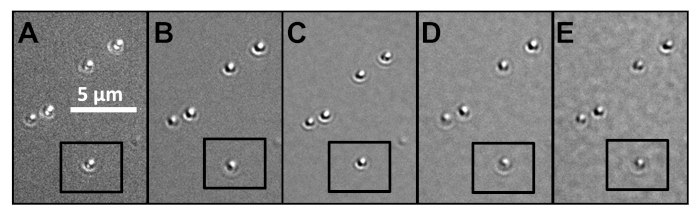
Figure 5: Gold nanospheres imaged under a series of different polarizer settings at a wavelength of 540 nm and a magnification of 100x. Background brightness was adjusted post-imaging with ImageJ to make particles more apparent. Polarizer setting of (A) 0° (polarizer orthogonal to analyzer), (B) 5°, (C) 10° (the best contrast of this series of images), (D) 20°, and (E) 45°. Measured contrast of particle in black box is (A) 0.65, (B) 0.84, (C) 1.10, (D) 0.44, and (E) 0.23. Please click here to view a larger version of this figure.
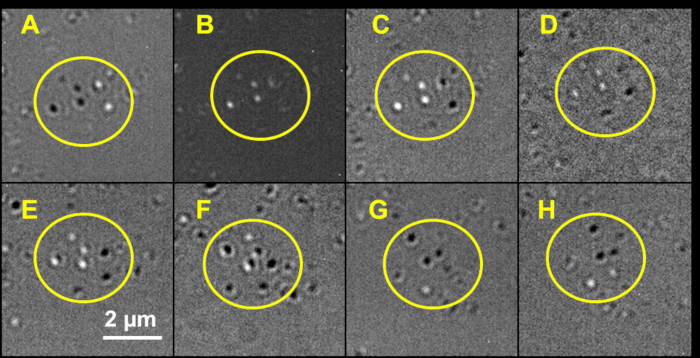
Figure 6: Example of imaging anisotropic plasmonic nanoparticles: gold nanorods at their longitudinal LSPR wavelength of 650 nm and a magnification of 100x. Particles of main interest are enclosed in yellow box. Polarizer settings are: (A) left 10°, (B) 0°, (C) right 10°, (D) right 45°. With the polarizer set to right 10°, the stage was rotated clockwise by (E) 20°, (F) 45°, (G) 70°, and (H) 90°. Please click here to view a larger version of this figure.
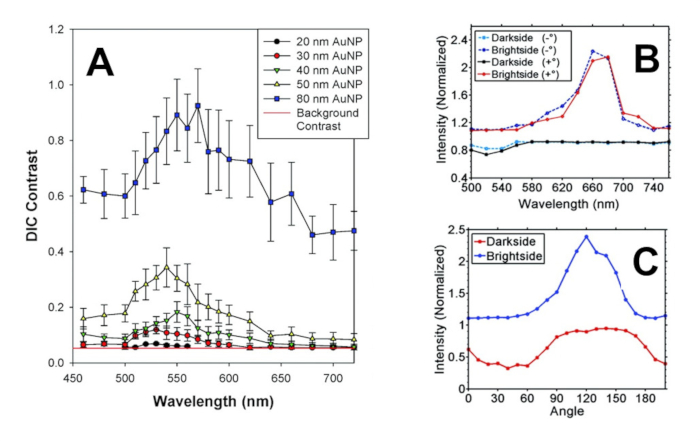
Figure 7: Representative results of single particle spectroscopy data. (A) Gold nanosphere spectroscopy displayed in terms of DIC Contrast. Each data point represents an average of 20 nanospheres for each particle diameter, and data capture relied on 10nm FWHM bandpass filters. (B) A single gold nanorod displayed as DIC Intensity data, using two different polarizer settings (2° on either side of crossed polarization). (C) DIC Intensity data for a single gold nanorod at the LSPR wavelength of 680 nm, while it was rotated 180° and the polarizer was held at 2° off the crossed polarization position. Figure 7A is adapted with permission from Sun et al., Analytical Chemistry. 81 (22), 9203-9208 (2009), and Figure 7B,C from Stender et al., Analytical Chemistry. 84 (12), 5210-5215 (2012). Copyright American Chemical Society. Please click here to view a larger version of this figure.
Discussion
When imaging with DIC microscopy, it is critical to optimize the optical components before collecting data. Even minor adjustments to the polarizer in the middle of an experiment can result in significant impacts to the final data6. Moreover, different materials require different polarizer settings. Although large step sizes were utilized here to demonstrate the effect of polarization angle, in an actual experiment, it is imperative to optimize the polarizer setting within 1°–2° of the optimal contrast setting. The polarizer setting should also be recorded for future reference. It is also recommended to always work on the same side of the crossed polarizer (0°) point. Switching back and forth does not provide any advantages, but it can lead to confusion, due to a reversal in the shadow pattern.
Next in importance, it is critical to monitor the background intensity when planning to perform spectroscopy. This is best accomplished by adjusting the camera exposure time, or by adding neutral density filters to the light path. Adjusting apertures or lamp intensities can impact the Kohler Illumination and alter contrast values. The background needs to be relatively even across the sample, so that the selection of a background region does not alter the contrast calculation. Sample specimens that are not adjacent to a clean background space should be avoided. Moreover, the background intensity cannot be initially set too high or too low. If the background intensity is set too high, there is an increased risk that some signals will exceed the maximum range of the camera, making it impossible to calculate the contrast in those regions. If the background intensity is set too low, it will be extremely difficult to achieve good contrast between the dark component of the DIC signal and the background signal. Understanding the typical or expected behavior of a sample can aid in selecting the proper background intensity.
Finding the proper focal plane is also essential. One of Nomarski DIC’s advantages is that it has a shallow depth of field. However, this makes it more challenging to focus on thin samples, such as nanoparticles. With thicker samples, the challenge is in finding the actual focal plane of greatest interest. Many focal planes may be interesting and have nanoparticles on them, so it is important to determine early on the nanoparticles of greatest interest.
In the case of nanoparticles, it is important for the microscopist to recognize that they are viewing an airy disc or “point spread function” of the object2. In general, the airy disc is useful in determining whether a plasmonic nanoparticle has a shape that is isotropic or anisotropic, but nanoparticle imaging is in fact much more complex than what is discussed here. Complex nanoparticle aggregates can sometimes resemble isotropic particles, and as a result, electron microscopy methods are then necessary to characterize the nanoparticle patterns2,16,18,19. To image plasmonic nanoparticles with a DIC microscope, it is crucial to use filtered imaging and to image the particles at one of their highly-absorbing plasmonic wavelengths6. Imaging at an improper wavelength or without filters can result in the capture of shadow-cast patterns that are difficult to decipher.
When imaging nanoparticles alongside objects that are larger than the diffraction limit of light, it is important to remember that the microscope’s objective “sees” a relatively flat focal plane. A common misconception of DIC is that it enables viewing of an object in actual 3D relief. This is caused by the shadow-cast patterning, which indeed makes many objects appear to be three-dimensional. However, to collect vertical information on multiple focal planes, it would be necessary to raise or lower the stage and collect a sequence of images. This can be very difficult to perform and to interpret, especially for thicker samples, such as cells. Thus, the microscopist needs a deep understanding of all materials involved when performing such experiments and must record the positions of the individual focal planes that were utilized.
Finally, the data analysis step is as critical as data collection. When measuring the contrast or intensity values of the sample, several factors should be kept in mind. Typically, the analyst is primarily interested in the minimum and maximum values for the particle of interest. When the contrast to noise ratio for the sample is sufficiently high, and if the background area is clean and evenly illuminated, then a simple geometric shape can be drawn around the sample region without concern of signal being introduced by contaminants. Furthermore, if the background is clean and evenly illuminated, a background measurement can be made in any area immediately adjacent to the sample. However, if there are contaminants or if the background is uneven, then the analyst must make a critical review of the sample’s environs, and the analyst needs to assess whether it is even possible to make a reasonable background measurement. It is also critical to measure the sample and background areas with the same-sized and shaped tool, in order to avoid the introduction of bias into the calculation. In general, smaller-sized measurement areas have a lower likelihood of detecting outliers (e.g., contaminants, bad pixels, etc.) but larger sampling areas often provide a more reliable measurement of the background’s mean value.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Dr. Anthony S. Stender wishes to acknowledge technical support through the Nanoscale and Quantum Phenomena Institute (NQPI) at Ohio University. This article was made possible through start-up funding provided to Dr. Stender by Ohio University.
Materials
| Contrad 70 | Decon Labs, Inc. | 1002 | For cleaning microscope glass, Available through many chemical suppliers |
| Ethanol | Fisher Scientific | A962-4 | For cleaning and storing microscope glass |
| Glass microscope cover slips | Ted Pella | 260148 | |
| Glass microscope slides | Ted Pella | 26007 | |
| Gold nanorods | Nanopartz | DIAM-SPR-25-650 | |
| Gold nanospheres (80 nm) | Sigma Aldrich | 742023-25ML | |
| ImageJ | NIH | N/A | Free Software availabe for data analysis from NIJ |
| Nail polish | Electron Microscopy Sciences | 72180 | |
| Nikon Ti-E microscope | Nikon | N/A | |
| Nitrogen gas | Airgas | N/A | |
| ORCA Flash 4.0 V2+ digital sCMOS camera | Hamamatsu | 77054098 | |
| Scribing pen | Amazon | N/A | Many options available online for under $10. Not necessary to buy an expensive version. |
| Ultrapure water | 18 megaohm |
References
- Pluta, M. Ch 7: Differential Interference Contrast in. Advanced Light Microscopy. 2, 146-197 (1989).
- Stender, A. S., Wang, G., Sun, W., Fang, N. Influence of Gold Nanorod Geometry on Optical Response. ACS Nano. 4 (12), 7667-7675 (2010).
- Stender, A. S., et al. Single Cell Optical Imaging and Spectroscopy. Chemical Reviews. 113 (4), 2469-2527 (2013).
- Mehta, S. B., Sheppard, C. J. R. Partially coherent image formation in differential interference contrast (DIC) microscope. Optics Express. 16 (24), 19462-19479 (2008).
- Murphy, D. B., Davidson, M. W. Ch 1: Fundamentals of Light Microscopy. Fundamentals of Light Microscopy and Electronic Imaging, Second edition. , 1-20 (2012).
- Stender, A. S., Augspurger, A. E., Wang, G., Fang, N. Influence of Polarization Setting on Gold Nanorod Signal at Nonplasmonic Wavelengths Under Differential Interference Contrast Microscopy. Analytical Chemistry. 84 (12), 5210-5215 (2012).
- Wang, G., Sun, W., Luo, Y., Fang, N. Resolving Rotational Motions of Nano-objects in Engineered Environments and Live Cells with Gold Nanorods and Differential Interference Contrast Microscopy. Journal of the American Chemical Society. 132 (46), 16417-16422 (2010).
- Kelly, K. L., Coronado, E., Zhao, L. L., Schatz, G. C. The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. The Journal of Physical Chemistry B. 107 (3), 668-677 (2003).
- Mulvaney, P. Not All That’s Gold Does Glitter. MRS Bulletin. 26 (12), 1009-1014 (2012).
- Maier, S. A. . Plasmonics: Fundamentals and Applications. , (2007).
- Sun, W., Wang, G., Fang, N., Yeung, E. S. Wavelength-Dependent Differential Interference Contrast Microscopy: Selectively Imaging Nanoparticle Probes in Live Cells. Analytical Chemistry. 81 (22), 9203-9208 (2009).
- Cras, J. J., Rowe-Taitt, C. A., Nivens, D. A., Ligler, F. S. Comparison of chemical cleaning methods of glass in preparation for silanization. Biosensors and Bioelectronics. 14 (8), 683-688 (1999).
- Augspurger, A. E., Sun, X., Trewyn, B. G., Fang, N., Stender, A. S. Monitoring the Stimulated Uncapping Process of Gold-Capped Mesoporous Silica Nanoparticles. Analytical Chemistry. 90 (5), 3183-3188 (2018).
- Murphy, D. B., Davidson, M. W. Ch 2: Light and Color. Fundamentals of Light Microscopy and Electronic Imaging, Second Edition. , 21-33 (2012).
- Wayne, R. Ch 3: The Dependence of Image Formation on the Nature of Light. Light and Video Microscopy (Second Edition). , 43-78 (2014).
- Stender, A. S., Wei, X., Augspurger, A. E., Fang, N. Plasmonic Behavior of Single Gold Dumbbells and Simple Dumbbell Geometries. The Journal of Physical Chemistry C. 117 (31), 16195-16202 (2013).
- Hu, M., et al. Dark-field microscopy studies of single metal nanoparticles: understanding the factors that influence the linewidth of the localized surface plasmon resonance. Journal of Materials Chemistry. 18 (17), 1949-1960 (2008).
- Choo, P., et al. Wavelength-Dependent Differential Interference Contrast Inversion of Anisotropic Gold Nanoparticles. The Journal of Physical Chemistry C. 122 (47), 27024-27031 (2018).
- Funston, A. M., Novo, C., Davis, T. J., Mulvaney, P. Plasmon Coupling of Gold Nanorods at Short Distances and in Different Geometries. Nano Letters. 9 (4), 1651-1658 (2009).

