In Vivo Forward Genetic Screen to Identify Novel Neuroprotective Genes in Drosophila melanogaster
Summary
We present a protocol using a forward genetic approach to screen for mutants exhibiting neurodegeneration in Drosophila melanogaster. It incorporates a climbing assay, histology analysis, gene mapping and DNA sequencing to ultimately identify novel genes related to the process of neuroprotection.
Abstract
There is much to understand about the onset and progression of neurodegenerative diseases, including the underlying genes responsible. Forward genetic screening using chemical mutagens is a useful strategy for mapping mutant phenotypes to genes among Drosophila and other model organisms that share conserved cellular pathways with humans. If the mutated gene of interest is not lethal in early developmental stages of flies, a climbing assay can be conducted to screen for phenotypic indicators of decreased brain functioning, such as low climbing rates. Subsequently, secondary histological analysis of brain tissue can be performed in order to verify the neuroprotective function of the gene by scoring neurodegeneration phenotypes. Gene mapping strategies include meiotic and deficiency mapping that rely on these same assays can be followed by DNA sequencing to identify possible nucleotide changes in the gene of interest.
Introduction
Neurons are for the most part post-mitotic and incapable of dividing1,2. In most animals, neuroprotective mechanisms exist to maintain these cells throughout the organism's lifespan, especially at old age when neurons are most vulnerable to damage. Genes underlying these mechanisms can be identified in mutants exhibiting neurodegeneration, a phenotypic indicator for the loss of neuroprotection, using a forward genetic protocol. Forward genetic screens using chemical mutagens such as ethyl methanesulfonate (EMS) or N-ethyl-N-nitrosourea (ENU) are particularly useful due to the random point mutations they induce, resulting in an inherently unbiased approach that has shed light on numerous gene functions in eukaryotic model organisms3,4,5 (in contrast, X-ray mutagenesis creates DNA breaks and can result in rearrangement rather than point mutations6).
The common fruit fly Drosophila melanogaster is an ideal subject for these screens due to its high quality, well annotated genome sequence, its long history as a model organism with highly developed genetic tools, and most significantly, its shared evolutionary history with humans7,8. A limiting factor in the applicability of this protocol is early lethality caused by the mutated genes, which would prevent testing at old age9. However, for non-lethal mutations, a climbing assay, which takes advantage of negative geotaxis, is a simple, although extensive, method of quantifying impaired motor functioning10. To exhibit sufficient locomotor reactivity, flies depend on neural functions to determine direction, sense its position, and coordinate movement. The inability of flies to sufficiently climb in response to stimuli can therefore indicate neurological defects11. Once a particular defective climbing phenotype is identified, further testing using a secondary screen such as histological analysis of brain tissue, can be used to identify neurodegeneration in climbing-defective flies. Subsequent gene mapping can then be used to reveal the genomic region on the chromosome carrying the defective neuroprotective gene of interest. To narrow down the chromosomal region of interest, meiotic mapping using mutant fly lines carrying dominant marker genes with known locations on the chromosome can be performed. The marker genes serve as a reference point for the mutation as the frequency of recombination between two loci provides a measurable distance that can be used to map the approximate location of a gene. Finally, crossing the mutant lines with lines carrying balanced deficiencies on the meiotically mapped chromosomal region of interest creates a complementation test in which the gene of interest can be verified if its known phenotype is expressed5. Polymorphic nucleotide sequences in the identified gene, possibly resulting in an altered amino acid sequences, can be evaluated by sequencing the gene and comparing it to the Drosophila genome sequence. Subsequent characterization of the gene of interest can include testing of additional mutant alleles, mutation rescue experiments and examination of additional phenotypes.
Protocol
1. Preparation and Aging of Flies
- Obtain or generate6 a collection of Drosophila mutants that will be used for the genetic screen. Here, ENU-mutagenized lines mapped to the second chromosome and balanced over CyO are used.
- Amplify experimental genotype lines in an incubator set at 25 °C, 12 h light/dark cycle on cornmeal-molasses medium.
- Collect around 20 homozygous progeny between 0-2 days of adult eclosion from each experimental genotype. Eliminate sex bias by consistently collecting either male or female flies.
- Age the flies on cornmeal-molasses medium as desired at 29 °C, flipping the vials every 2-3 days in order to maintain the flies on fresh food as they age. In the screen presented here, flies were aged for 10-12 days.
- Assemble a climbing assay testing chamber using two vials and tape as previously described10. The vials should be connected at both open ends. Use a ruler to mark a 5 cm line on one end of the chamber.
2. Protocol 1: Climbing Assay (Adapted from Ali et al.10)
- Validate the climbing assay. In this study, test the climbing assay based on previously described methodology by Ali et al.10, using ElavC155>UAS-TauR406W (exhibiting lower climbing pass rates) and ElavC155>UAS-GFP (control) and ElavC155>UAS-mCherry (control) flies.
- Flip the flies into a testing chamber, one line at a time (do not use CO2 anesthesia) and allow them to recover for 5 min. If testing is done on different days, perform the assay at the same time of the day. In this study, all climbing assays were performed in the afternoon to control for circadian effects on locomotion12.
NOTE: Avoid exposing the flies to direct sunlight when testing; this will interfere with their ability to climb. - Forcibly tap the chamber (marked side down) onto a mouse pad placed on a solid surface 3 times so all flies begin the assay at the bottom. Observe fly locomotion over a period of 10 s.
- Record the number of flies that reach or pass the 5 cm line within this time, as well as the total number of flies tested. Repeat the protocol, waiting 1 min between each trial, for a minimum of 3 trial replicates per line.
NOTE: In the present study, each vial contained between 2 and 19 flies for the initial screening (male flies) and between 6 and 21 flies for the mutant deficiency analysis of crossed female lines (Section 5.3.).
3. Protocol 2: Selection of Drosophila Strains for Further Analysis
- Enter the climbing assay data into Microsoft Excel or similar software and calculate the climbing pass rate for each line over all replicates as a percentage.
- Perform statistical analysis to detect mutant lines that deviate significantly from the mean climbing percent value of all lines using the R software package13.
- Read in data files curated for R (.csv or .txt files with a column for each variable or factor, and rows representing the specific values).
NOTE: Here, data for males (initial assay) and females (line 867 crossed to deficiency lines) were entered into separate .xlsx sheets as two columns, one where the rows of the column identify the mutant line and how many times the assay was replicated and the other where rows represent the mean climbing success (as a percentage) for each replicate vial of flies. - Read the data into R using the following code:
mali<-read.csv("../male_init.csv")
femdef<-read.csv("../fem_def_cross.csv")
NOTE: The ".." in the path directories are absolute paths from the root directory of the user's computer and would need to be specified by the individual user for their computer's directory paths. - Perform linear modeling
- Perform dummy variable regression with the lm function in R. For the initial analysis of mutant lines, evaluate the significance of factor level effects (mutant lines) on percent climbing success against the grand mean for a zero mean centered response variable (percent success).
NOTE: The "-1" at the end of the lm function call in section 3.2.3.2. removes the intercept and allows us to test all factor levels against the zero mean centered mean of percent climbing pass rate. - Print the results to the screen using the R summary function:
mali_anova<-lm(scale(mali$P_Success)~mali$Mutant_line -1)
summary(mali_anova) - Use control lines (if present) as the baseline to test factor level effects against using the following R code: x<-relevel(femdef$Mutant_line, ref="a867_plus")
femdef_anova<-lm(femdef$P_success~x)
summary(femdef_anova)
NOTE: The first line of code reorders the factor levels so that the negative control line becomes the baseline intercept to which all other factor levels (mutant lines) are tested against with the built-in t-tests in the lm function.
- Perform dummy variable regression with the lm function in R. For the initial analysis of mutant lines, evaluate the significance of factor level effects (mutant lines) on percent climbing success against the grand mean for a zero mean centered response variable (percent success).
- Read in data files curated for R (.csv or .txt files with a column for each variable or factor, and rows representing the specific values).
- Choose a threshold for candidate lines depending on age at the time of testing. To determine the threshold, perform a preliminary test at the desired age using known mutants that exhibit neurodegeneration and low climbing pass rates in comparison to wild type, control flies10.
NOTE: A passing rate below 50% may be appropriate for 10 days-old flies as previously reported for Drosophila lines overexpressing the human neurodegeneration-related gene Tau in the nervous system10. Older flies should have a higher passing rate threshold since they are expected to exhibit decreased locomotion, thus, increased neurodegeneration.
4. Protocol 3: Histology Analysis
- Wearing gloves, sever fly heads with a surgical blade following the climbing assay and use a paintbrush to gently place them in 1 mL of Carnoy's fixative (6:3:1 of 100% EtOH: chloroform: glacial acetic acid) in a 1.5 mL micro centrifuge tube O/N at 4 °C. Make sure the heads sink in the fixative solution.
- Replace the fixative solution the following day with 1 mL of 70% EtOH and maintain the samples still at 4 °C for future analysis.
NOTE: The protocol may be paused at this step. - Place the heads from step 4.2. at a maximum of five per microbiopsy cassette. Immediately place the cassettes into a container filled with 70% EtOH. Store the container at 4 °C prior to shipping to a histology facility for processing and paraffin embedding of the heads. If equipment for tissue processing and embedding is available, follow step 4.4.
- Embed the heads in paraffin for microtome sectioning:
- Follow the program settings for an automated tissue-processing machine in the order presented in Table 1 to dehydrate, clear and infiltrate with paraffin the heads.
- Transfer the samples into metal base molds that fit the cassette using paraffin heated at 60 °C using a paraffin embedding station.
- Orient the heads (antero-posterior orientation facing the top) in the base mold using fine forceps to allow proper visualization of the brain tissue following sectioning. This can be done by reheating the paraffin at 60 °C and repositioning the heads if necessary.
NOTE: Metal molds can be replaced by disposable base molds. - Position the cassette on the top of the corresponding base mold and gently move these to the refrigerated side of the paraffin embedding station (4 °C). Wait until the block hardens prior removing it from the station.
- Remove the block form the mold by gently dissociating the mold from the cassette.
- Trim each paraffin block prior sectioning to minimize the surface that will be sectioned.
- Set the microtome to cut 5 µm sections of each block.
- Place sectioned paraffin ribbon containing the tissue in a heated water bath (35 °C) for a maximum of 5 min.
- Immerse a poly-lysine coated slide (helps to retain the tissue) in the heated water bath, placing the sectioned ribbon on the slide using wood applicators. Let the slides air dry O/N at room temperature.
- Heat the slides for 15 min at 63 °C using a slide warmer.
- Place the slides on a slide staining rack and stain with hematoxylin and eosin (H&E) under a fume hood by respecting the following steps.
- Place the rack in Histochoice (non-toxic replacement of Xylene, which helps to remove the paraffin from the sample) for 5 min.
- Transfer the rack in a container filled with Histochoice for 5 min.
- Transfer the rack in a container filled with 100% EtOH 1 for 5 min.
- Transfer the rack in a container filled with 100% EtOH 2 for 5 min.
- Transfer the rack in a container filled with 95% EtOH for 3 min.
- Transfer the rack in a container filled with 70% EtOH for 3 min.
- Transfer the rack in a container filled with Distilled H2O for 5 min.
- Transfer the rack in a container filled with Harris Hematoxylin for 2-5 min.
- Take the rack out of the Hematoxylin solution and place it under running tap water for 10 min.
- Transfer the rack in a container filled with distilled water by doing a short dunk.
- Transfer the rack in a container filled with Eosin for 1-2 min.
- Transfer the rack in a container filled with 95% EtOH by doing a short dunk.
- Transfer the rack in a container filled with 95% EtOH for 5 min.
- Transfer the rack in a container filled with 100% EtOH for 5 min.
- Transfer the rack in a container filled with 100% EtOH for 5 min.
- Transfer the rack in a container filled with Histochoice (or Xylene) for 5-10 min until all slides are mounted.
- Mount the slide and coverslip (24 mm x 60 mm of size). Using forceps, take the slide out of the Histochoice or Xylene solution and make a thin strip of mounting medium on the top of it. Gently place the coverslip on the top, trying to avoid the formation of bubbles.
- Let the mounting medium on mounted slides harden O/N under the fume hood prior to analysis.
- Store stained slides in a box at room temperature.
- Quantify neurodegeneration by looking at all serial sections on the slide at 20x magnification under a light microscope. Examine the entire brain, taking qualitative note of size and number of vacuoles that appear in three consecutive sections.
- Obtain images of representative brain sections at approximately mid-brain using a light microscope equipped with imaging software.
5. Protocol 4: Gene Mapping of Recessive Mmutant Phenotype
- Outcross the candidate line to a wild type CantonS (BL_ 9517) or another wild type or isogenized strain (e.g., OregonR (BL_ 2376), w1118 (BL_5905) or yw (BL_ 6599) to determine whether the phenotype is dominant or recessive.
NOTE: Results from this step will guide the next steps of the mapping. If the phenotype is recessive, then deficiency mapping can be performed.- Using a paintbrush, make a cross by placing in a food-containing vial CO2-anesthetized flies of the mutant line of interest (5 males) and the CantonS line (10-15 virgin females). Place the vial at 25 °C.
- Collect heterozygous F1 progeny and repeat climbing and histology experiments.
- Compare the obtained results to the control line alone and to the homozygous mutants. A good indication of recessive phenotype will be if heterozygous mutants exhibit similar phenotypes as wild type flies.
- Perform meiotic mapping to roughly estimate the location of the mutation on the second chromosome using the wgSp-1 J1 L2 Pin1/CyO (BL_3227) or an equivalent strain with associated dominant markers at known cytological location. In this case, the mutation was previously known to be located on the second chromosome.
- Using a paintbrush, make a cross by placing in a food-containing vial CO2-anesthetized flies of the mutant line of interest (5 males) and the wgSp-1 J1 L2 Pin1/CyO line (10 females). Place the vial at 25 °C.
NOTE: The goal of this cross is to generate heterozygous females carrying the chromosome with the new mutation and the marker chromosome, which will recombine during meiosis6,14. Female F1 progeny are chosen rather than males since recombination does not occur in males. - Collect at least 15 virgin female progeny carrying the chromosome with the new mutation and the marker chromosome and cross them to 5 males from a balanced line that carries another visible dominant marker (e.g., CyO/snaSco (BL_2555) or equivalent).
- Collect heterozygous male progeny carrying either Sco or CyO and the potentially recombined chromosome from the cross in step 5.2.2. and individually mate them to virgin females from the stock carrying the mutated chromosome.
- Collect the progeny from final cross described in step 5.2.3. and age the flies at 29 °C (in this study flies were aged for 10-14 days).
- Collect heads, perform histology analysis as described in Protocol 3 and look at the slides with brain sections to determine the degree of neuropathology as described in step 4.11. Histology analysis is performed rather than locomotion analysis due to phenotypes that may affect the climbing assay, such as Jammed (J), which causes fluid buildup in the wings15.
- Using a paintbrush, make a cross by placing in a food-containing vial CO2-anesthetized flies of the mutant line of interest (5 males) and the wgSp-1 J1 L2 Pin1/CyO line (10 females). Place the vial at 25 °C.
- Perform deficiency mapping on narrowed region of the chromosome to refine the location of the recessive mutation using the climbing assay. Each deficiency line has a deletion of different region of the chromosomes, allowing them to be used for complementation testing.
- Start with large deficiencies that span the region identified in the meiotic mapping, which can then be further narrowed down by the use of smaller deficiencies. If the deficiency analysis results in more than one candidate gene, the use of RNA interference (RNAi) stocks are recommended to target them.
- Cross lines from the Drosophila deficiency kit16 for the second chromosome (DK2L in this study) and look for non-complementation of the phenotype of interest (in this study: climbing pass rate of 8.5% which corresponds to the pass rate of homozygous flies from line 867 used as positive control).
- Confirm the neurodegeneration phenotype using histology verification as described in Section 4 (Protocol 3).
6. Protocol 5: DNA Sequencing and Analysis
NOTE: Keep in mind that ENU mutagenesis introduces point mutations11.
- Design forward and reverse primer sequences flanking the exon-coding region of the brat gene for amplification by Polymerase Chain Reaction (PCR) of the region of interest (in the case of line 867 a DNA fragment of the size of 3,247 bp is amplified for sequencing17).
- Extract DNA from a single fly18 :
- Gently squish the fly in a 0.5 mL micro centrifuge tube containing 50 µL of Squishing buffer (10 mM Tris-Cl pH 8.2, 1 mM EDTA, 25 mM NaCl, and 200 µg/mL Proteinase K; the enzyme has to be diluted fresh from a frozen stock prior each use) using a P100 or P200 pipet tip.
- Incubate for 30 min at 37 °C
- Deactivate the Proteinase K by placing the tube at 95 °C for 2 min.
NOTE: The extracted DNA can be stored at 4 °C for several months.
- Perform the PCR reaction using a high-fidelity DNA Taq Polymerase (reagents and thermal cycling conditions used in this study are shown in Table 2 and Table 3, respectively) by respecting manufacturer's instructions and run the PCR product on a 1% agarose gel containing Invitrogen SYBR Safe DNA Gel Stain, which is a non-toxic alternative of Ethidium bromide.
- Excise the band of the correct size from the gel with a scalpel and purify the PCR product using the Wizard SV Gel and PCR Clean-Up System (Promega) or equivalent kit, by respecting manufacturer's instructions.
- Design forward and reverse primers that will be used for DNA sequencing in order to cover the entire region of interest, if possible. Sequence the gel-purified DNA from the previous step to identify point mutations. High accuracy in automated fluorescent Sanger sequencing could be achieved for up to 700 bp.
NOTE: The Ex Taq polymerase (TaKaRa) can be used to amplify PCR products from genomic DNA templates up to 20 kb of size. - Analyze obtained DNA sequences using the A plasmid Editor (ApE, by M. Wayne Davis) or similar software by aligning them and comparing them to the sequences obtained following sequencing of the same genomic region of a reference strain (e.g., originally mutagenized line).
- Open sequencing files with ApE. As reference, use an ApE file containing the genomic consensus sequence of the brat gene downloaded in a FASTA format from Flybase, or a file containing results for sequence of genetic background control (e.g., originally mutagenized Drosophila line).
- Go to Tools, then Align sequences. Using the computer's mouse, select all sequences to compare. Remember to indicate the reference sequence used for this alignment in the drop menu.
- Inspect the sequenced region for nucleotide changes in comparison with the reference sequence. If desired, save the alignment on the computer in .rtf format by clicking on Text, then Save.
NOTE: If no mutations were identified, the DNA sequencing and analysis protocol would be repeated with primers designed to include non-coding regions of the gene.
7. Protocol 6: Further Analysis of Candidate Gene Function
- Perform rescue experiments in neuroblasts to determine whether the gene brat is responsible for the neurodegeneration phenotype.
- Double balance a UAS-brat19 and a neuroblast-specific worniu-Gal4 (wor-G4) stocks carrying the constructs on the third chromosome, as well as the 867 line, which is mutant for the gene brat located on the second chromosome.
- Make three separate crosses by placing in three different food-containing vials 5 males from UAS-Brat, wor-G4 and 867 lines and add to each set of males 10-15 virgin females from a line carrying markers and balancers for the second and third chromosomes (Sp/CyO; Dr/Tm3,sb; BL_59967).
- Collect 10-15 virgin females from the F1 of each cross with the following genotypes: +/CyO; UAS-brat/Tm3,sb, +/CyO; wor-G4/Tm3,sb and 867/CyO;+/Tm3,sb and 5 males of each of the following genotypes: +/Sp;UAS-brat/Tm3,sb, +/Sp; wor-G4/Tm3,sb and 867/CyO; +/Dr.
- Cross collelcted +/CyO; UAS-brat/Tm3,sb virgin females to +/Sp; UAS-brat/Tm3,sb males; in a separate vial make a second cross with +/CyO; wor-G4 /Tm3,sb virgin females to +/Sp; wor-G4/Tm3,sb males and then in a third vial cross 867/CyO;+/Tm3,sb virgin females and 867/CyO; +/Dr males.
- From the F1 of each of these crosses, collect both males and females of the following genotypes: Sp/CyO; UAS-brat/Tm3,sb from the first cross, Sp/CyO; wor-G4/Tm3,sb from the second cross and 867/CyO; Dr/Tm3,sb from the second cross.
- Perform a final cross between brothers and sisters collected from each F1 progeny to establish permanent double balanced stocks for all three lines.
- Combine UAS-brat and wor-G4 with the 867 mutation.
- Collect sufficient number (~20-30 flies) of virgin females from the 867/CyO; Dr/Tm3,sb double balanced stock to perform two crosses.
- Place 10-15 virgin females with ~5 males from the Sp/CyO; UAS-brat/Tm3,sb (cross #1) and in a second vial 10-15 virgin females with ~5 Sp/CyO; wor-G4/Tm3,sb males.
- Collect brothers and sisters from the F1 of each cross with the following genotypes: 867/CyO; UAS-brat/Tm3,sb from cross #1 and 867/CyO;wor-G4/Tm3,sb from cross #2. Use collected F1 progeny from each cross to establish permanent stocks by crossing brothers and sisters between them.
NOTE: Following this final step, stocks for 867/CyO; UAS-brat/Tm3,sb and 867/CyO; wor-G4/Tm3,sb should be generated.
- Perform the rescue experiment.
- Collect ~15-20 virgin females from the 867/CyO; wor-G4/Tm3,sb stock and cross them to 5-10 males from the stock 867/CyO; UAS-brat/Tm3,sb. As control, cross ~15-20 virgin females from the 867/CyO; wor-G4/Tm3,sb and ~15-20 virgin females from the 867/CyO; UAS-brat/Tm3,sb line to the 867/CyO line alone.
NOTE: 867 flies carry a temperature-sensitive allele of brat and the rescue cross should be performed at 29 °C. - Collect the following F1 progeny from each cross by carefully selecting away marked balancers: 867/867; wor-G4/UAS-brat (rescue), 867/867; wor-G4/+ (control) and 867/867; UAS-brat/+ (control).
- Age the flies as desired at 29 °C and perform histology analysis on brains to look for neurodegeneration by using the protocol described in Section 4 (Protocol 3).
- Collect ~15-20 virgin females from the 867/CyO; wor-G4/Tm3,sb stock and cross them to 5-10 males from the stock 867/CyO; UAS-brat/Tm3,sb. As control, cross ~15-20 virgin females from the 867/CyO; wor-G4/Tm3,sb and ~15-20 virgin females from the 867/CyO; UAS-brat/Tm3,sb line to the 867/CyO line alone.
- Double balance a UAS-brat19 and a neuroblast-specific worniu-Gal4 (wor-G4) stocks carrying the constructs on the third chromosome, as well as the 867 line, which is mutant for the gene brat located on the second chromosome.
- Perform age-dependent analysis of neurodegeneration in 867 mutants.
- Collect 867;pcna-GFP and yw control flies, which carry a reporter gene (pcna-GFP) and are viable at 25°C and exhibit both higher penetrance and higher expressivity of the neurodegeneration phenotype than 867 alone at this temperature17.
- Age the flies up to 5, 15 and 25 as described in Section 1.4 and perform subsequent histology analysis by following the protocol in Section 4 (Protocol 3).
Representative Results
In this aerticle, we present the steps used to identify the gene brain tumor (brat) as playing a role in maintenance of neuronal integrity (e.g., neuroprotection) in adult flies17; a methodology that can be used to identify genes involved in neuroprotection. We used a forward genetic approach (the strategy is outlined in Figure 1A) to screen through a collection of chemically mutagenized flies using a climbing assay (the apparatus used for this assay is shown in Figure 1B). Among 235 homozygous lines, about 37% of tested lines exhibited a climbing pass rate below 50% when tested at the age of 10-12 days (Figure 1C). 58% of tested lines exhibited significantly different climbing behavior when their percent climbing pass rate (CPR) was compared to the mean climbing percent value of all tested lines using one-way ANOVA (Figure 1D). Subsequent histological screen on 51 of the lines exhibiting the lowest climbing pass rate revealed that 29 of these lines showed visible appearance of holes in the brain neuropil (ranging from mild to severe) indicative of neurodegeneration20 (Figure 2). Among the lines showing defective climbing behavior and severe neurodegeneration, we choose to map the mutation underlying the phenotype in line 867 (0% CPR and P value= 1.36e-14 based on one-way ANOVA). The neurodegeneration phenotype in 867 flies is recessive because the brains of heterozygous 867 flies that have been outcrossed to a wild type strain are comparable to these of controls (Figure 3A). Using histology, meiotic mapping situated the mutation in the 31-51 cytological locations, between the phenotypic markers J (Jammed) and L (Lobe) on the second Drosophila chromosome. Using the climbing assay, deficiency mapping between cytological locations 31 and 51 with lines from the Drosophila Deficiency Kit for chromosome 2L (DK2L)16 mapped the mutation in a region encompassing 118 genes (Figure 3B; where 867/+ serves as control for the statistical analysis). Examining the brain phenotype of 867 mutants crossed to additional deficiency lines (Figure 3C) confirmed that the mutation is contained in a region of the genome that includes the gene brat. A complete list of all additional genes contained within these deletions can be found in Flybase by clicking on the link associated with the deleted segment (e.g., for Df(2L)ED1272 the deleted segment is 37C5-38A2). Based on previous observations that brat mutants have supernumerary cells in their brains21, a phenotype also observed in 867 mutants, brat was selected for DNA sequencing analysis. Sanger sequencing of the brat gene containing the exon-coding region identified G/A nucleotide change at position 37,739 of the gene (Figure 4A), leading to glycine to glutamic acid (G/E) change at position 470 of the Brat protein17 (Figure 4B). Rescue experiments confirm that Brat plays a neuroprotective role, as the overexpression of the functional gene in the 867 line suppresses the neurodegeneration phenotype (Figure 5A). Moreover, the phenotype in 867 mutants worsens over time, suggesting that brat plays an age-dependent role in neuroprotection17 (Figure 5B).
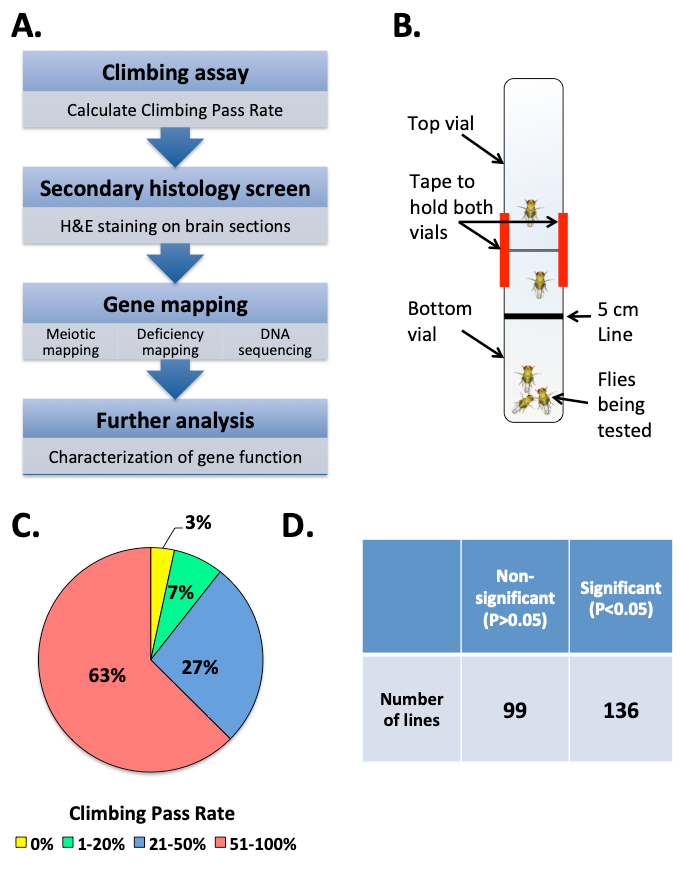
Figure 1: Forward genetic screen to identify novel neuroprotective genes. (A) Screen strategy. (B) Climbing assay testing apparatus. (C) Climbing assay results for males from 235 homozygous ENU-mutagenized lines. Pie chart represents the proportions of tested fly lines that fall into 4 groups of climbing pass rates: 0% , 1-20% , 21-50% , and 51-100% . (D) Results from 1 way ANOVA statistical analysis in which mutant lines were compared to the mean climbing percent value of all tested lines. Represented is the number for two P value categories: P < 0.05 (significant change) and P > 0.05 (not significant change).
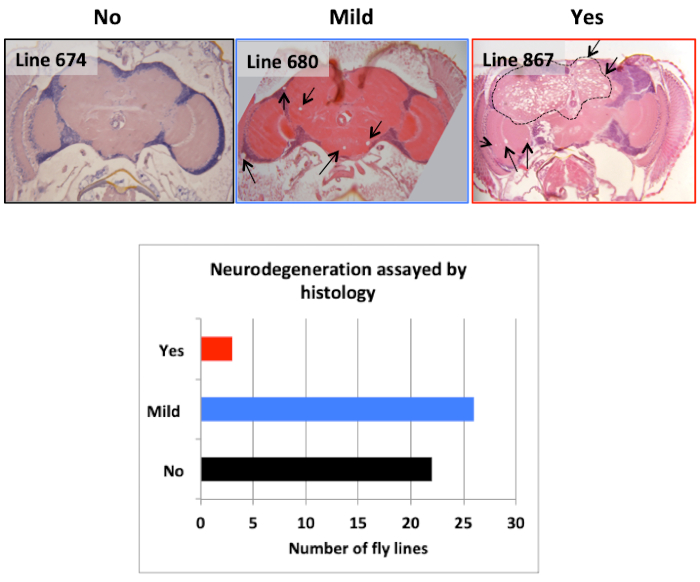
Figure 2: Secondary histology screen. H&E-stained mid-brain sections illustrating, from left to right, no neurodegeneration, mild neurodegeneration, and confirmed neurodegeneration. A total of 51 homozygous lines were retested by histology after identification of candidates using the climbing assay. Arrows indicate the holes in the brain indicative of neurodegeneration. The region circled by the dotted lines shows the severe neurodegenration observed in line 867.
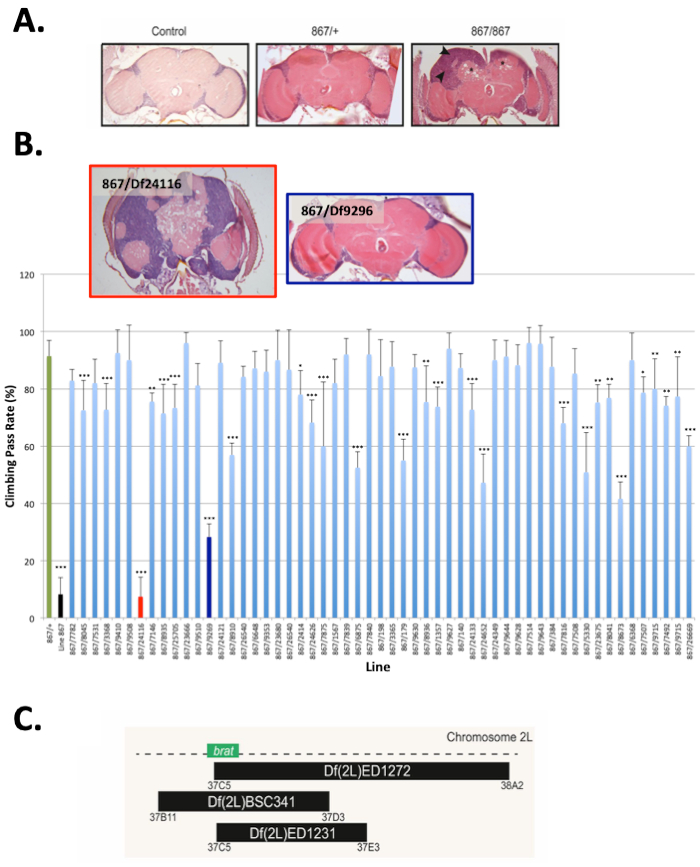
Figure 3: Identification of the neuroprotective gene brat following deficiency mapping using the climbing assay. (A) Shown are representative images of H&E-stained mid-brain sections of 18-20 days old w1118 (Control), heterozygous 867 (867/+) and homozygous 867 mutant flies. (B) The deficiency line Df(2L)ED1272 (BL_24116) does not complement the 867 mutant phenotype (black histogram) based on the climbing assay. Histological verification shows that Df(2L)ED1272 line doesn't complement the 867 neurodegeneration phenotype, whereas Df(2L)ED1315 (BL_9269) does. Graph represents mean and standard deviation of 5 climbing trials for each line. Asterisks indicate significance based on one-way ANOVA, in which the mean climbing percent value of each line was compared to the mean climbing percent value of line 867/+ (green histogram). * P < 0.05, ** P < 0.01 and ***P < 0.001. (C) Schematic representation of additional deficiency lines showing non-complementation of the 867 phenotype by histology. This figure has been modified from Loewen et al.17; an article published under the Creative Commons Attribution 4.0 International License.
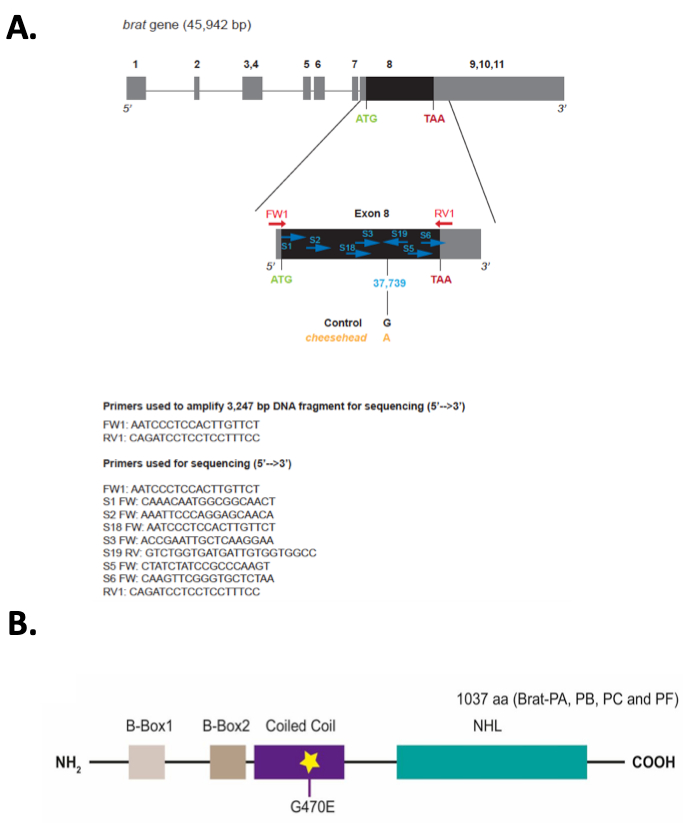
Figure 4: Point mutation in the brat gene leads to an amino acid change in the Coiled-coil domain of the Brat protein. (A) brat gene model and primers used for the amplification and sequencing of the coding region. (B) DNA sequencing of the brat locus identifies a nucleotide change that leads to an amino acid change in the Coiled Coil domain of the Brat protein. This figure has been modified from Loewen et al.17; an article published under the Creative Commons Attribution 4.0 International License.
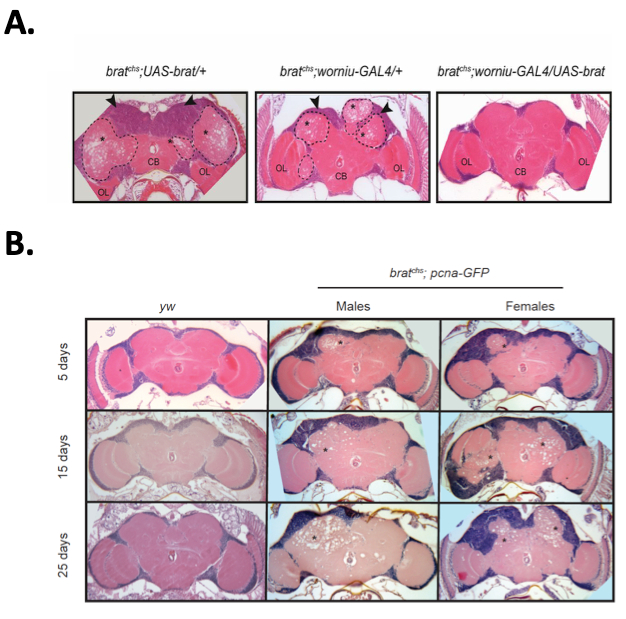
Figure 5: Further analysis of brat mutants confirms its neuroprotective role in Drosophila. (A) Using the Gal-4>UAS system22 , neuroblast-specific expression of the functional brat gene rescues the neurodegeneration phenotype in 867 mutants. (B) Age-dependent neurodegeneration is observed in 867 mutants carrying a reporter gene pcna-GFP. bratchs corresponds to the name of the brat allele in line 867, which was named cheesehead (chs). Shown are representative H&E-stained midbrain sections of yw (control) and homozygous bratchs; pcna-GFP flies of the indicated ages. This figure has been modified from Loewen et al.17; an article published under the Creative Commons Attribution 4.0 International License.
| Station | Time | Temperature | Vacuum/Pressure | Solution |
| 1 | NO DELAY-QUICK START | |||
| 2 | OFF | OFF | OFF | |
| 3 | : 15 | 37 °C | ON/ON | 80%EtOH |
| 4 | : 15 | 37 °C | ON/ON | 95% EtOH |
| 5 | : 15 | 37 °C | ON/ON | 100% EtOH |
| 6 | : 15 | 37 °C | ON/ON | 100% EtOH |
| 7 | : 15 | 37 °C | ON/ON | 100% EtOH |
| 8 | : 15 | 37 °C | ON/ON | Xylenes |
| 9 | : 15 | 37 °C | ON/ON | Xylenes |
| 10 | : 15 | 37 °C | ON/ON | Xylenes |
| 11 | : 30 | 58 °C | ON/ON | Paraffin |
| 12 | : 15 | 58 °C | ON/ON | Paraffin |
| 13 | OFF | 58 °C | OFF | Paraffin |
| 14 | : 15 | 58 °C | ON/ON | Paraffin |
Table 1: Recommended program settings for automated tissue processor. The steps in the order listed here can be used with an automated tissue processor for fixed heads.
| Reagent | Volume (μL) |
| 10x ExTaq Buffer (Mg2+ plus) (20 mM) | 2.5 |
| Forward primer (10 μM) | 2.5 |
| Reverse primer (10 μM) | 2.5 |
| 2.5 mM dNTPs | 2 |
| Template DNA | 2 |
| TaKaRa Ex Tq (5 U/μL) | 0.25 |
| Nuclease-free water | 13.25 |
| Total volume | 25 |
Table 2: Reagents used for PCR reaction. Reagents and respective volumes used in the PCR reaction to amplify the brat-coding region.
| Step | Temperature | Time |
| 35 cycles | 95 °C | 15 s |
| 55 °C | 45 s | |
| 72 °C | 3 min 10 s | |
| Hold | 4 °C | ∞ |
Table 3: Thermal cycling conditions used for PCR. The steps in the order listed here can be used to program a thermal cycler using the reaction mix shown in Table2.
Discussion
Forward genetic screens in Drosophila have been an effective approach to identify genes involved in different biological processes, including age-dependent neuroprotection5,23,24,25. Using this strategy, we were successful in identifying brat as a novel neuroprotective gene17.
One critical step in this protocol involves the proper orientation of heads (as described in Section 4.4.3.) for histology analysis. Additionally, markers of the line used for the meiotic mapping should not interfere with the phenotype of the new mutation (in this study: neurodegeneration). We recommended an initial test to confirm that the chosen markers do not cause neurodegeneration of their own. For instance, Jammed (J) causes wing blisters that could possibly interfere with climbing as is the case for amyloid precursor protein (APP)-overexpressing flies that also have blistered wings26. Lobe (L) leads to reduction in eye size; however, we do not observe central brain degeneration and this marker can be used to map central brain neurodegeneration. Pin and Sternoplural (Sp) are also appropriate to use, as brains of flies carrying these markers are comparable to brains of wild type controls.
One disadvantage of the forward genetic methodology in flies is the length of time required to narrow regions of DNA to the gene of interest3,5. The use of improved mapping strategies27 along with the availability of next generation sequencing technologies 28 should allow even easier identification of mutations associated with phenotypes of interest. Forward genetic screens can be labor-intensive. For instance, histology confirmation, which assays directly for neurodegeneration in the brain, alone requires a considerable amount of time. However, this approach will be much more difficult and expensive to perform in mammalian models, not necessarily allowing extensive genetic studies. Chemical mutagenesis screens are advantageous because the identification of point mutations could provide critical information about the function of affected genes' products. The identification of a point mutation causing an amino acid change in a conserved domain of the Brat protein (Figure 4B) illustrates the importance of the coiled-coil domain of this TRIM-NHL protein in neuroprotection18. The climbing assay, which measures locomotor behavior, is certainly advantageous by allowing rapid testing of large numbers of flies for defects in mobility, also often seen in human neurodegeneration25. This assay could also be performed in another screen setting such as genome-wide in vivo RNA interference (RNAi) screen that could be used to identify neuroprotective genes. Although we decreased the distance between the bottom of the test chamber and the passing line from 8 cm10 to 5 cm to set a stricter passing rate in the present study, low climbing rates did not mirror neurodegeneration for large number of lines. Neurodegeneration may become apparent after the onset of behavioral defects, meaning that flies may need to be aged for longer before vacuoles appear. Another possibility is that the locomotor defects we observe in certain lines are not due to defective neurological functioning but rather to muscle weakness or more general unfitness of the flies. Additionally, it is possible that we are missing potential candidates, in which climbing defects do not manifest at younger age (e.g., 10 days old) but rather become prominent later in life. This screening strategy could certainly be applied to identify age-dependent genes, thus eliminating genes associated with potential defects of neural development. The protocol presented here can be adjusted depending on the desired probability of candidate lines to contain the mutated gene involved in neuroprotection. Lowering the passing rate threshold directly is another means of achieving the same result. These candidates would theoretically exhibit greater neurodegeneration, which could save time and resources during confirmation and mapping.
In general, the protocol describes a straightforward way to screen for neurodegeneration and subsequently identify neuroprotective gene candidates. This genetic approach is not limited to the behavior and histological assays described here, and may also be used to screen for other phenotypes like temperature-sensitive (ts) paralysis, egg laying or fertility phenotypes, just to cite a few. For instance, Drosophila ts-paralytic mutants are known to be enriched for neurodegeneration29 and could represent an additional source for the identification of neuroprotective genes.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We are particularly grateful to Dr. Barry Ganetzky, in who's lab the genetic screen was performed, allowing the identification and characterization of brat as a neuroprotective gene. We thank Dr. Steven Robinow for kindly providing the collection of ENU-mutagenized flies used in the genetic screen presented in this article. We thank the members of Ganetzky lab, Drs. Grace Boekhoff-Falk and David Wassarman for helpful discussions throughout the duration of this project, Ling Ling Ho and Bob Kreber for technical assistance, Dr. Aki Ikeda for the use of his microtome facility at the University of Wisconsin and Dr. Kim Lackey and the Optical Analysis Facility at the University of Alabama.
Materials
| Major equipment | |||
| Fume hood for histology | |||
| Light Microscope | Nikon | Eclipe E100 | Preferred objective for imaging is X20 |
| Imaging software | Nikon | ||
| Microscope Camera | Nikon | ||
| Thermal cycler | Eppendorf | ||
| Fly pushing and climbing assay | |||
| VWR® Drosophila Vial, Narrow | VWR | 75813-160 | |
| VWR® General-Purpose Laboratory Labeling Tape | VWR | 89097-912 | |
| Standard mouse pad | |||
| Stereoscope | Motic | Model SMZ-168 | |
| CO2 anesthesia station (Blowgun, foot valve, Ultimate Flypad) | Genesee Scientific | 54-104, 59-121, 59-172 | Doesn’t iinclude CO2 tank |
| Fine-Tip Brushes | SOLO HORTON BRUSHES, INC. | ||
| Drosophila Incubator | VWR | 89510-750 | |
| Gene mapping | |||
| CantonS | Bloomington Drosophila Stock Center | 9517 | |
| w1118 | Bloomington Drosophila Stock Center | 5905 | |
| yw | Bloomington Drosophila Stock Center | 6599 | |
| Drosophila line used for recombination mapping | Bloomington Drosophila Stock Center | 3227 | Genotype: wg[Sp-1] J[1] L[2] Pin[1]/CyO, P{ry[+t7.2]=ftz/lacB}E3 |
| CyO/sno[Sco] | Bloomington Drosophila Stock Center | 2555 | Drosophila balancer line used for recombination mapping |
| Deficiency Kit for chromosome 2L | Bloomington Drosophila Stock Center | DK2L | Cook et al., 2012 |
| Histology analysis | |||
| Ethanol, (100%) | Thermo Fischer Scientific | A4094 | |
| Chloroform | Thermo Fischer Scientific | C298-500 | |
| Glacial Acetic Acid | Thermo Fischer Scientific | A38-500 | |
| Fisherbrand™ Premium Microcentrifuge Tubes: 1.5mL | Thermo Fischer Scientific | 05-408-129 | |
| Histochoice clearing agent 1X | VWR Life Sciences | 97060-934 | |
| Harris Hematoxylin | VWR | 95057-858 | |
| Eosin | VWR | 95057-848 | |
| Thermo Scientific™ Richard-Allan Scientific™ Mounting Medium | Thermo Scientific™ 4112 | 22-110-610 | CyO/sna[Sco] |
| Unifrost Poly-L-Lysine microscope slides, 75x25x1mm, EverMark Select Plus | Azer Scientific | ||
| Fisherbrand™ Cover Glasses: Rectangles | Fisherbrand | 12-545M | Dimensions: 24×60 mm |
| Traceable timer | VWR | ||
| Slide Warmer | Barnstead International | model no. 26025 | |
| Slide tray and racks | DWK Life Sciences | Rack to hold 20 slides | |
| Fisherbrand™ General-Purpose Extra-Long Forceps | Fisherbrand | 10-316A | |
| Kimwipes™ | Kimberly-Clark™ Professional | ||
| 6 inch Puritan applicators | Hardwood Products Company, Guilford, Maine | 807-12 | |
| VWR® Razor Blades | VWR | 55411-050 | |
| Tupperware or glass containers for histology liquids | 16 + 1 for running water | ||
| High Profile Coated Microtome Blades | VWR | 95057-834 | |
| Corning™ Round Ice Bucket with Lid, 4L | Corning™ | ||
| Beaker | Or other container for ice water and cassettes | ||
| Tissue Bath | Precision Scientific Company | 66630 | |
| Microtome | Leica Biosystems | ||
| Molecular analysis | |||
| Wizard® SV Gel and PCR Clean-Up System | Promega | A9282 | |
| Ex Taq DNA polymerase | TaKaRa | 5 U/μl | |
| Invitrogen™ SYBR™ Safe™ DNA Gel Stain | Invitrogen™ | ||
| UltraPure™ Agarose | Invitrogen™ | ||
| 1 Kb Plus DNA Ladder | Invitrogen™ | ||
| ApE-A plasmid Editor software | Available for free download | ||
| Statistical analysis | |||
| R software package | |||
| Further analysis | |||
| y[1] w[*]; wg[Sp-1]/CyO; Dr[1]/TM3, Sb[1] | Bloomington Drosophila Stock Center | 59967 | |
References
- Frade, J. M., Ovejero-Benito, M. C. Neuronal cell cycle: the neuron itself and its circumstances. Cell Cycle. 14 (5), 712-720 (2015).
- Aranda-Anzaldo, A. The post-mitotic state in neurons correlates with a stable nuclear higher-order structure. Communicative & Integrative Biology. 5 (2), 134-139 (2012).
- Haelterman, N. A., et al. Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Res. 24 (10), 1707-1718 (2014).
- Moresco, E. M., Li, X., Beutler, B. Going forward with genetics: recent technological advances and forward genetics in mice. The American Journal of Pathology. 182 (5), 1462-1473 (2013).
- St Johnston, D. The art and design of genetic screens: Drosophila melanogaster. Nature Reviews Genetics. 3 (3), 176-188 (2002).
- Greenspan, R. J. . Fly pushing: The theory and practice of Drosophila genetics. , (2004).
- Reiter, L. T., Potocki, L., Chien, S., Gribskov, M., Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Research. 11 (6), 1114-1125 (2001).
- Rubin, G. M., et al. Comparative genomics of the eukaryotes. Science. 287 (5461), 2204-2215 (2000).
- Nichols, C. D., Becnel, J., Pandey, U. B. Methods to assay Drosophila behavior. Journal of Visualized Experiments. (61), (2012).
- Ali, Y. O., Escala, W., Ruan, K., Zhai, R. G. Assaying locomotor, learning, and memory deficits in Drosophila models of neurodegeneration. Journal of Visualized Experiments. (49), (2011).
- Karres, J. S., Hilgers, V., Carrera, I., Treisman, J., Cohen, S. M. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 131 (1), 136-145 (2007).
- Helfrich, C., Engelmann, W. Circadian-Rhythm of the Locomotor-Activity in Drosophila-Melanogaster and Its Mutants Sine Oculis and Small Optic Lobes. Physiological Entomology. 8 (3), 257-272 (1983).
- R Development Core Team. . R: A language and environment for statistical computing. , (2008).
- Bokel, C. EMS screens : from mutagenesis to screening and mapping. Methods in Molecular Bioogyl. 420, 119-138 (2008).
- Lindsley, D. L., Zimm, G. G. . The Genome of Drosophila Melanogaster. , (1992).
- Cook, R. K., et al. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biology. 13 (3), R21 (2012).
- Loewen, C., Boekhoff-Falk, G., Ganetzky, B., Chtarbanova, S. A Novel Mutation in Brain Tumor Causes Both Neural Over-Proliferation and Neurodegeneration in Adult Drosophila. Genes Genomes Genetics G3 (Bethesda). 8 (10), 3331-3346 (2018).
- Gloor, G. B., et al. Type I repressors of P element mobility. Genetics. 135 (1), 81-95 (1993).
- Komori, H., Xiao, Q., McCartney, B. M., Lee, C. Y. Brain tumor specifies intermediate progenitor cell identity by attenuating beta-catenin/Armadillo activity. Development. 141 (1), 51-62 (2014).
- Kretzschmar, D., Hasan, G., Sharma, S., Heisenberg, M., Benzer, S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. The Journal of Neuroscience. 17 (19), 7425-7432 (1997).
- Kang, K. H., Reichert, H. Control of neural stem cell self-renewal and differentiation in Drosophila. Cell and Tissue Research. 359 (1), 33-45 (2015).
- Brand, A. H., Perrimon, N. Targeted Gene-Expression as a Means of Altering Cell Fates and Generating Dominant Phenotypes. Development. 118 (2), 401-415 (1993).
- Loewen, C. A., Ganetzky, B. Mito-Nuclear Interactions Affecting Lifespan and Neurodegeneration in a Drosophila Model of Leigh Syndrome. Genetics. 208 (4), 1535-1552 (2018).
- Cao, Y., Chtarbanova, S., Petersen, A. J., Ganetzky, B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proceedings of the National Academy of Sciences of the United States of America. 110 (19), E1752-E1760 (2013).
- Lessing, D., Bonini, N. M. Maintaining the brain: insight into human neurodegeneration from Drosophila melanogaster mutants. Nature Reviews Genetics. 10 (6), 359-370 (2009).
- Peng, F., et al. Loss of Polo ameliorates APP-induced Alzheimer’s disease-like symptoms in Drosophila. Scientific Reports. 5, 16816 (2015).
- Venken, K. J., Bellen, H. J. Chemical mutagens, transposons, and transgenes to interrogate gene function in Drosophila melanogaster. Methods. 68 (1), 15-28 (2014).
- Gonzalez, M. A., et al. Whole Genome Sequencing and a New Bioinformatics Platform Allow for Rapid Gene Identification in D. melanogaster EMS Screens. Biology (Basel). 1 (3), 766-777 (2012).
- Palladino, M. J., Hadley, T. J., Ganetzky, B. Temperature-sensitive paralytic mutants are enriched for those causing neurodegeneration in drosophila. Genetics. 161 (3), 1197-1208 (2002).

