Analyzing Satellite Cell Function During Skeletal Muscle Regeneration by Cardiotoxin Injury and Injection of Self-delivering siRNA In Vivo
Summary
We describe an in vivo method to investigate the functional loss of a specific gene in satellite cells by using a combination of cardiotoxin mediated injury of skeletal muscle and the injection of a self-delivering siRNA.
Abstract
Skeletal muscle possesses an enormous capacity to regenerate after injury. This process is mainly driven by muscle stem cells, also termed satellite cells. Satellite cells are characterized by the expression of the transcription factor Pax7 and their location underneath the basal lamina in the resting skeletal muscle. Upon injury, satellite cells get activated, undergo self-renewal or differentiation to either form new myofibers or to fuse with damaged ones. The functionality of satellite cells in vivo can be investigated using a cardiotoxin based injury model of skeletal muscle. To study the function of one gene during the regeneration of skeletal muscle, transgenic mouse models are mostly used. Here, we present an alternative method to transgenic mice, to investigate the gene function in satellite cells during regeneration, e.g., in cases where transgenic mice are not available. We combine the cardiotoxin mediated injury of a specific skeletal muscle with the injection of a self-delivering siRNA into the regenerating muscle which is then taken up by satellite cells among other cells. Thereby, we provide a method to analyze gene function in satellite cells during regeneration under physiological conditions without the need for transgenic mice.
Introduction
Skeletal muscle is the body’s largest tissue representing approximately 40% of total body weight enabling voluntary locomotion. The tissue architecture of skeletal muscle is mainly composed of postmitotic, terminally differentiated, multinucleated myofibers as well as various other cells from the peripheral nervous system, vascular system, and interstitial cells1. Importantly, skeletal muscle has a tremendous capacity to regenerate and restore function upon injury or damage2. This process depends on the tissue resident muscle stem cells also called satellite cells3,4. Satellite cells are located between the myofiber and the basal lamina and characterized by the expression of the transcription factor Pax75,6,7. Under homeostatic conditions, satellite cells are quiescent but get activated due to traumatic injury, e.g., through eccentric exercise or experimentally through injection of the snake venom cardiotoxin6,8. Once activated, Pax7 positive satellite cells co-express MyoD and Myf5, which commits the stem cells to myogenic differentiation. Satellite cells that resist the upregulation of commitment factors will retain their stemness potential and return to quiescence to replenish the stem cell pool for future demands. After the expansion of the myogenic progenitor pool, transcriptional networks are activated by differentiation factors like Myogenin to initiate the cell cycle exit and terminal differentiation. These myoprogenitors then fuse to each other or to existing myofibers contributing myonuclei to maintain the size of the myonuclear domain. The myofibers express terminal muscle differentiation genes like Myosin Heavy Chain. Finally, the newly formed myofibers grow and mature to build the functional units of skeletal muscle9,10.
The regeneration of skeletal muscle can be affected by various conditions including muscle diseases or aging11,12, ranging from mild impairments to life threatening conditions, e.g., in Duchenne muscular dystrophy13,14. Therefore, regenerative medicine aims to restore damaged or malfunctioning skeletal muscle tissue using its inherent regenerative power by targeting satellite cell function15,16. To use its full potential a comprehensive understanding of satellite cells in their endogenous niche during regeneration of skeletal muscle is required. Although experimental approaches exist to isolate satellite cells adjacent to their myofibers17, the full complexity of cellular and systemic interactions of satellite cells with their environment can only be recapitulated in vivo. In that regard, great knowledge about skeletal muscle regeneration has been acquired using mouse injury models2,18.
Here we introduce a specific experimental mouse injury model to study the stem cell mediated regeneration of cardiotoxin-induced damage of the tibialis anterior muscle in vivo. Cardiotoxin, a snake derived cytolytic toxin that causes myofiber depolarization and necrosis, is injected into the tibialis anterior muscle, which in turn will trigger tissue degeneration followed by regeneration. To analyze the function of genes during acute regeneration, self-delivering siRNAs are injected at day 3 after injury, at the peak of the satellite cell expansion. Experimental animals are sacrificed at various timepoints and the tibialis anterior muscles are collected. The dissected muscles are frozen and processed for further cryo-sectioning. Immunofluorescence microscopy is then used to analyze markers of regeneration. This method allows for the investigation of the function of a single gene during satellite cell driven regeneration of skeletal muscle using wild type mice.
Protocol
All animal procedures were approved by the Animal Welfare Department of the Thüringer Landesamt für Lebensmittelsicherheit und Verbraucherschutz (03-048/16; TLV; Bad Langensalza, Germany).
1. Cardiotoxin-induced Muscle Injury
NOTE: Perform all experiments involving living mice in accordance with the national Animal Welfare Act and under appropriate aseptic conditions. Also, sacrificing of animals must be performed in accordance with the national Animal Welfare Act.
- Disinfect the inhalation box used for inhalation anesthesia and the surgery area with 70% ethanol. Place a sterile surgical cloth on the heating pad where the surgery will take place.
- Turn on the heating pad at 37 °C.
- Administer analgesics (e.g., 1 mg/kg body weight meloxicam subcutaneously) 15 min prior to start of the surgery.
NOTE: For a typical experiment, use mice which are at least 6 weeks old, sex-matched and in a good general physical condition. - Prepare the cardiotoxin solution (20 µM in 0.9 % NaCl), store the solution at -20 °C.
- Allow the cardiotoxin solution to reach room temperature (RT).
- Transfer the mouse into the inhalation box and induce anesthesia with isoflurane (initiation 3.5 – 5 % in pure oxygen). Check the depth of anesthesia with a lack of response to a toe pinch.
- Place the mouse on a clean paper towel and maintain anesthesia with a nose mask (1.5 – 3 % isoflurane in pure oxygen). Shave the injection area of the cranial lower leg (from knee to paw, Figure 1A). Remove all excessive loose hair.
NOTE: Physically separating the shaving and injection area in the room will provide more sterile conditions for the injections. - Place the mouse in the lateral recumbent position on the heating pad covered with a sterile surgical cloth and maintain anesthesia with a nose mask (1.5 – 3% isoflurane in pure oxygen).
- Disinfect the injection area of the cranial lower leg (from knee to paw) with 70% ethanol.
- Perform a toe pinch before starting the intramuscular injection to ensure an adequate depth of anesthesia.
- Inject 50 µL cardiotoxin (20 µM in 0.9% NaCl) into the tibialis anterior muscle using an insulin syringe with a 29-gauge needle. First, pierce the skin just distal of the knee.
- Insert the needle fully into the muscle (in myofiber orientation/parallel to the tibia bone) and inject the cardiotoxin slowly (10-20 s) along the full length of the muscle while moving the needle back and forth to allow an even distribution of the cardiotoxin thereby injuring the whole tibialis anterior muscle (Figure 1B,C).
NOTE: Injure the tibialis anterior muscle from only one leg, the contralateral tibialis anterior muscle can serve as an internal control to determine that skeletal muscles were not pathologically affected before cardiotoxin injury. - Transfer the mouse back to its cage placed on a heating pad and monitor the recovery process until the animal is conscious and becomes ambulatory.
NOTE: The mice will only show a slight limp. If mice are limping heavily and do not put weight on the leg at all, sacrifice the mouse. - Administer analgesics during the following 2 days (e.g., 1 mg/kg body weight meloxicam subcutaneously, every 24 h) and monitor them on a weekly basis.
2. Injection of Self-delivering siRNA into the Regenerating Tibialis Anterior Muscle (At Day 3 Post Cardiotoxin Injury)
- Prepare the siRNA solution by resuspending the siRNA (e.g., siRNA smart pool) in 0.9% NaCl (final concentration of 2 µg/µL) according to the manufacturer’s instruction.
NOTE: The siRNA is chemically modified to facilitate the passive uptake by cells and protect it from nuclease degradation. No transfection reagents are needed, thereby reducing toxicity. Using a smart pool of four independent siRNA sequences targeting the same gene increases knockdown efficiency. - Store the siRNA at -20 °C and transfer it on ice to the surgery room.
- Disinfect the inhalation box used for inhalation anesthesia and the surgery area with 70% ethanol. Place a sterile surgical cloth on the heating pad where the surgery will take place.
- Turn on the heating pad at 37 °C.
- Transfer the mouse into the inhalation box and induce anesthesia with isoflurane (initiation 3.5 – 5% in pure oxygen). Check the depth of anesthesia with a lack of response to a toe pinch.
- Place the mouse in the lateral recumbent position on the heating pad covered with a sterile surgical cloth and maintain anesthesia with a nose mask (1.5 – 3% isoflurane in pure oxygen).
- Disinfect the injection area of the cranial lower leg (from knee to paw).
- Perform a toe pinch before starting the intramuscular injection to ensure an adequate depth of anesthesia.
- Inject up to 50 µL siRNA solution (up to 100 µg total in 0.9% NaCl; directed against the target gene; use scrambled siRNA as control) into the injured tibialis anterior muscle using an insulin syringe with a 29 G needle. First, pierce the skin just distal of the knee, then insert the needle into the tibialis anterior muscle.
- Insert the needle fully into the muscle (in myofiber orientation/parallel to the tibia bone) and inject the siRNA solution slowly (10 – 20 s) along the full length of the muscle while moving the needle back and forth to allow an even distribution of the siRNA along the whole tibialis anterior muscle.
- Transfer the mouse back to its cage placed on a heating pad and monitor the recovery process until the animal is conscious and becomes ambulatory.
NOTE: Analgesia is not necessarily following the injection of siRNA.
3. Dissection of Tibialis Anterior Muscle
- Prepare the freezing solution (2 parts OCT compound and 1 part 30% sucrose in deionized water) at least 12 h before usage to avoid air bubbles.
- Prepare freezing molds by wrapping an aluminum foil around a pencil and seal it with tape. It is important that the bottom of the mold provides an even, closed surface.
NOTE: Smaller freezing molds allow the faster freezing of the muscle avoiding freezing artifacts. - Sacrifice the mouse, e.g., by CO2 inhalation, at the respective time point of regeneration and spray the whole mouse and dissection tools with 70% ethanol.
NOTE: Cervical dislocation can be applied, in addition, to avoid excessive bleeding at the leg when isolating the tibialis anterior muscle. - Remove the fur and skin from the injured hindlimb by cutting the skin at the ankle with extra fine sharp scissors (cutting edge: 13 mm) and pulling up the skin towards the knee using forceps.
- To expose the tibialis anterior tendon at the ankle, pull the remaining skin towards the foot or cut with sharp scissors.
NOTE: The mouse can be pinned to a support board to allow better fixation. - Before harvesting the tibialis anterior muscle, remove the fascia using fine forceps (Dumont 5 or 7, straight or curved). Pinch the closed forceps through the fascia next to the tibia bone at the ankle of the injured leg (Figure 2A). Move the forceps towards the knee thereby tearing the fascia and exposing the tibialis anterior muscle (Figure 2B).
- To isolate the tibialis anterior muscle, expose the distal tendon and grab it with fine forceps (Dumont 7, curved). Cut the tendon using spring scissors (cutting edge: 5 mm, tip diameter: 0.35 mm) and pull the muscle (holding it at the tendon) towards the knee.
- To harvest the tibialis anterior muscle, cut it as close to the knee as possible using sharp scissors.
- Prior to freezing, cut the tibialis anterior muscle at the mid belly region of the muscle with straight scissors into two halves of equal size to allow the analysis of the mid belly region (Figure 2C,D).
- Fill the freezing mold halfway with the freezing solution.
- Insert the two halves of the tibialis anterior muscle into the freezing mold with the mid belly region facing the bottom of the mold (Figure 2E). Make sure the tibialis anterior halves are inserted in an upright position leaning against the wall of the freezing mold to avoid tipping of the muscle.
NOTE: The more freezing solution is in the mold, the longer the freezing will take, thus increasing the risk of cryo-artifacts. - Hold the freezing mold using forceps and transfer it halfway into liquid nitrogen (Figure 2F). Make sure that no liquid nitrogen is entering the freezing mold during the freezing process.
- Observe the freezing process, the freezing medium changes color from transparent to white and becomes solid. Submerse the freezing mold in liquid nitrogen for a few seconds and transfer the freezing mold to a -80 °C freezer or to dry ice for future processing.
- Store the frozen muscles in the freezing molds at -80 °C until further usage.
NOTE: The freezing molds fit well in 24-well plates or 1.5 mL reaction tubes allowing labeling and organized storage. - Handle the muscle in the freezing molds for further usage always on dry ice.
4. Cryosectioning of Regenerating Tibialis Anterior Muscle
- Before starting sectioning, set the chamber temperature of the cryostat to -21 °C, the object temperature to -20 °C and the cutting thickness to 10, 12 or 14 µm (depending on the future applications) (Figure 3A).
- Transfer the muscle sample in the freezing mold into the cryostat and let it adjust to the temperature for several minutes. During this time, label the microscope slides.
- Remove the foil around the embedded muscle using precooled forceps inside the cryostat. Avoid touching of the sample as the cryomedium starts to thaw easily.
- Mount the sample with the mid belly region of the tibialis anterior muscle directed upwards onto the metal sample holder of the cryostat using cryomedium. This ensures that the cutting site of the muscle (bottom of the mold) is facing the experimenter.
- Insert the sample holder with the mounted sample into the sectioning mechanism.
NOTE: To ensure proper sectioning of the sample, use a new blade before starting. - First, trim the sample block (30 µm) to get an even sectioning plane and to reach the respective region of the muscle (Figure 3B).
- After trimming, cut consecutive sections of the respective thickness (e.g., 14 µm).
- Collect the sections on microscope glass slides by holding the slide face down over the section. The section is attaching to the slide (Figure 3C).
- Store sections at -80 °C or -20 °C until further use or directly use for immunofluorescence or immunohistochemistry.
5. Immunostaining for Markers of Regeneration
- Perform all further steps in a humidified chamber.
- Briefly, equilibrate the sections at room temperature.
- Fix sections with 1 mL of 2% PFA (in PBS, pH 7.4) per slide for 5 min at RT.
- Remove the 2% PFA solution by pouring it into the respective waste container.
- Wash 3 times with 1 mL of PBS (pH 7.4) for 5 min at RT.
- Remove the PBS by pouring it into a waste container.
- Per slide, add 1 mL of the permeabilization solution (0.1 % Triton X-100, 0.1 M Glycine in PBS pH 7.4) for 10 min.
- Remove the permeabilization solution by pouring it into a waste container.
- Add 150 µL of the blocking solution (M.O.M. 1:40 in PBS pH 7.4) per slide and cover with the coverslip. Incubate for 1 h at RT.
- Remove the coverslip and the blocking solution, apply 100 µL of primary antibody solution [PAX7, DSHB, mouse IgG1, undiluted or devMHC, DSHB, undiluted, mouse IgG1and laminin (rabbit, 1:1,000)] per slide. Cover with coverslip. Incubate O/N (overnight) at 4 °C.
NOTE: Perform a control staining by omitting the primary antibodies, incubate the section with blocking solution instead. - Wash 3 times with 1 mL of PBS (pH 7.4) for 5 min at RT.
- Remove PBS by pouring it into a waste container.
- Add 100 µL of the secondary antibody solution (Alexa Fluor 546 goat anti-mouse IgG1 and Alexa Fluor 488 donkey anti-rabbit IgG in blocking solution, 1: 1,000) per slide and cover with the coverslip. Incubate for 1 h at RT.
- Incubate the slides in the dark, e.g., use an aluminum foil to cover or use a black humidified chamber.
NOTE: From now on all steps should be performed in reduced light conditions since some secondary antibodies are light sensitive. - Remove the coverslip and the secondary antibody solution.
- Wash 3 times with 1 mL of PBS (pH 7.4) for 5 min at RT.
- Remove the PBS by pouring it into a waste container.
- Perform DAPI staining by adding 1 mL of the solution per slide to a final concentration of 10 µg/mL for 5 min at RT.
- Remove the DAPI staining solution by pouring it into a waste container.
- Wash 3 times with 1 mL PBS (pH 7.4) for 5 min at RT.
- Completely remove the PBS used for washing.
- Apply 2 – 3 drops of aqueous mounting medium and directly cover the slide with a new coverslip.
- Let the slides dry for 1 h at RT in the dark.
- Store the stained sections at 4 °C in the dark until further analysis using a fluorescence microscope.
6. Hematoxylin and Eosin Staining
- Perform all further steps in a humidified chamber.
- Fix sections with 1 mL of 2% PFA (in PBS, pH 7.4) per slide for 5 min at RT.
- Remove the 2% PFA solution by pouring it into the respective waste container.
- Transfer the slides into a Coplin jar.
- Gently rinse the slides with tap water.
NOTE: Do not turn on the faucet too high since that might damage the sections. - Transfer the slides to the humidified chamber. Remove the liquid.
- Apply 1 mL of Hematoxylin staining solution (Gill No 3) for 2 min.
- Transfer the slides into a Coplin jar.
- Gently rinse the slides with the tap water until the nuclei turn blue.
NOTE: Do not turn on the faucet too high since that might damage your sections. - Transfer the slides to the humidified chamber. Remove the liquid.
- Apply 1 mL of Eosin Y solution and incubate for 2 min.
- Transfer the slides into a Coplin jar.
- Gently rinse the slides with tap water until the tap water coming off the slides is clear.
- Remove all liquid and let the slides dry for 2 min.
- Mount in mounting medium suitable for immunohistochemistry.
Representative Results
A typical result of a hematoxylin and eosin (H&E) staining of a tibialis anterior muscle before and after cardiotoxin mediated injury is presented in Figure 4A,B. In the control muscles, the architecture of the muscle is intact as seen by the localization of the nuclei in the periphery of the myofibers and the lack of accumulation of mononucleated cells in the interstitial space (Figure 4A). 7 days after cardiotoxin mediated injury new myofibers are formed marked by centrally located nuclei (Figure 4B). Furthermore, an accumulation of mononucleated cells can be observed, consisting mostly of satellite cells but also non-myogenic cells like immune cells. The whole muscle should be injured characterized by the central location of all myonuclei and the accumulation of mononucleated cells on the whole muscle section.
To characterize the progression and success of the regeneration process, several immunofluorescent staining using markers of regeneration can be performed. The number of satellite cells can be evaluated by staining for Pax7, the canonical marker for satellite cells (Figure 4C,D). Three days after injury the number of satellite cells increases (Figure 4D), satellite cells are not located under the basal lamina anymore. To further analyze the regeneration process, newly formed myofibers can be stained with antibodies directed to developmental myosin (Figure 4E). Newly formed myofibers display centrally located nuclei and a strong expression of developmental myosin. As the myofibers mature, expression of developmental myosin decreases, the diameter of the myofiber increases while nuclei are migrating to the periphery of the myofibers.
The influence of a single gene on the process of regeneration can be investigated using transgenic mice or as shown here by injection of a self-delivering siRNA. Fluorescently labeled self-delivering scrambled control siRNA was injected into the regenerating tibialis anterior muscle at day 3 after injury, the time point when the proliferation of satellite cells peaks. Two days after injection of the siRNA satellite cells were analyzed for the presence of the fluorescently labeled siRNA (Figure 4F). We analyzed how many satellite cells had taken up the fluorescently labeled siRNA two days after injection into the regenerating muscle (Figure 5). About 75% of all satellite cells in the regenerating muscle were positive for the fluorescently labeled siRNA. Furthermore, we determined that about 74% of all regenerating myofibers were positive for the fluorescently labeled siRNA suggesting that either 74% of the regenerating myofibers had taken up the siRNA or that siRNA-positive satellite cells either had fused with each other to form new myofibers or that fusion with already existing regenerating myofibers occurred (Figure 5). This suggests that satellite cells take up the self-delivering siRNA and that the siRNA persists in the satellite cells for at least two days.
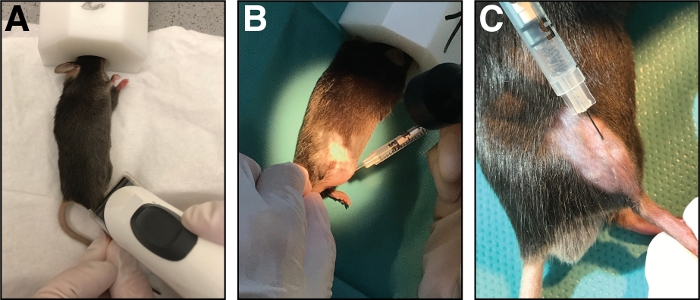
Figure 1: Injection of cardiotoxin into the tibialis anterior muscle. (A) Mice are anesthetized by inhalation of isoflurane and the lower limb is shaved. (B) A 29 G needle is injected into the tibialis anterior muscle (C) and moved along the tibia bone during injection of the cardiotoxin solution. Please click here to view a larger version of this figure.
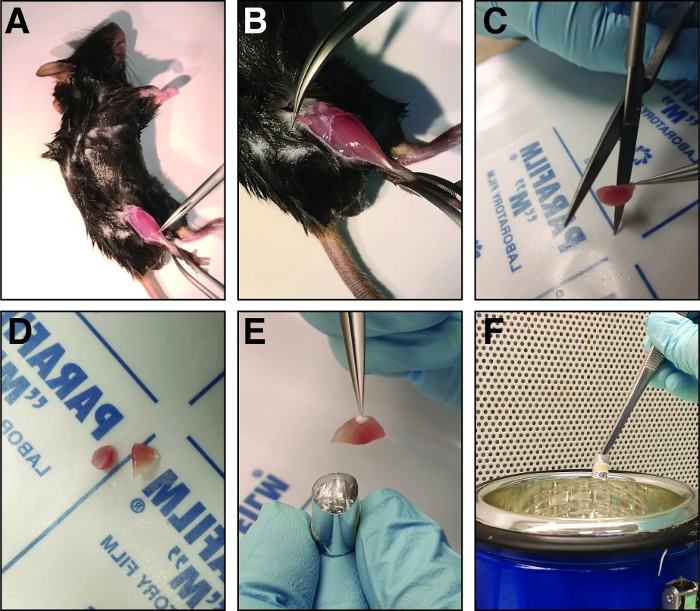
Figure 2: Steps involved in dissection and freezing of the injured tibialis anterior muscle. (A) The hind limb muscles are exposed (B) and the fascia surrounding the tibialis anterior muscle is ripped to expose the muscle. (C) After removing the muscle, it is cut at the mid belly region into two halves of similar size using sharp straight scissors (D). (E) The halves of the dissected muscle are embedded in an aluminum foil mold filled with freezing solution and frozen in liquid nitrogen (F). Please click here to view a larger version of this figure.
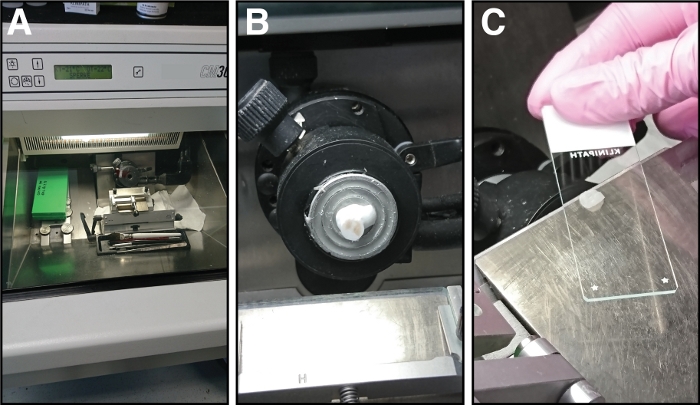
Figure 3: Equipment and steps involved in cryosectioning of tibialis anterior muscles. (A) The chamber temperature of the cryostat is set to -21 °C. (B) The tibialis anterior muscle in the freezing solution is mounted on the sample holder. (C) Sections of 14 µm thickness are attached to glass microscope slides. Please click here to view a larger version of this figure.
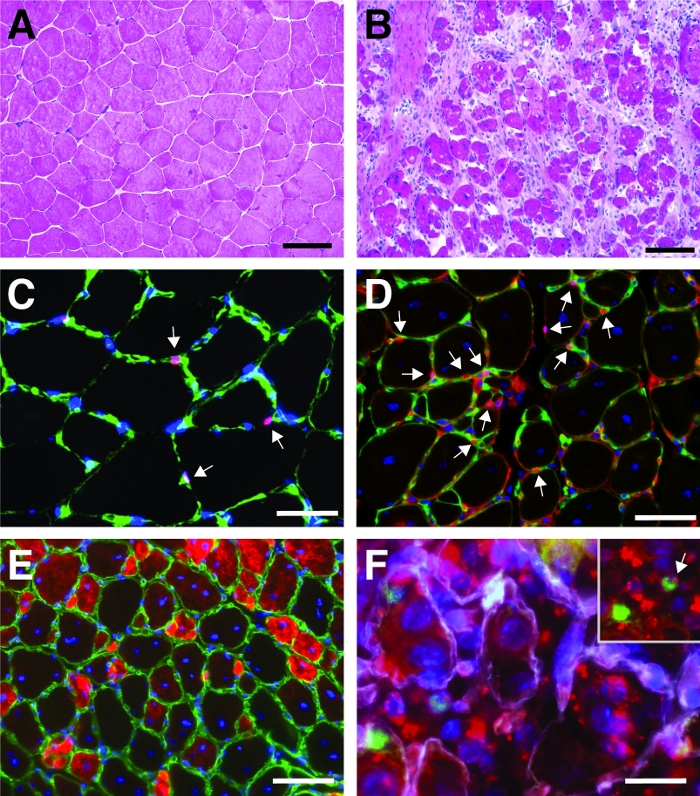
Figure 4: Representative images of regenerating tibialis anterior muscles. H&E staining of tibialis anterior muscles before (A) and 7 days after cardiotoxin injury (B). (C) In uninjured muscle Pax7 positive (in red) satellite cells are located underneath the basal lamina marked by laminin (in green). Nuclei are counterstained with DAPI (in blue). Pax7 positive cells are marked by an arrow. (D) After 10 days of regeneration, myofibers are characterized by centrally located nuclei (in blue), Pax7 is shown in red, laminin in green. Pax7 positive cells are marked by an arrow. (E) Newly formed myofibers express developmental myosin (in red), laminin in green, nuclei are counterstained with DAPI (in blue). (F) Fluorescently labeled self-delivering siRNA (siRED, depicted in red) is still found in satellite cells (Pax7 positive, in green, marked by an arrow in the inset) 2 days after injection of the self-delivering siRNA into regenerating tibialis anterior muscle (injection at day 3 after cardiotoxin mediated injury). Laminin is shown in white, nuclei are counterstained with DAPI (in blue). Scale bar = 100 µm (A and B), 50 µm (C-E), 25 µm (F). Please click here to view a larger version of this figure.
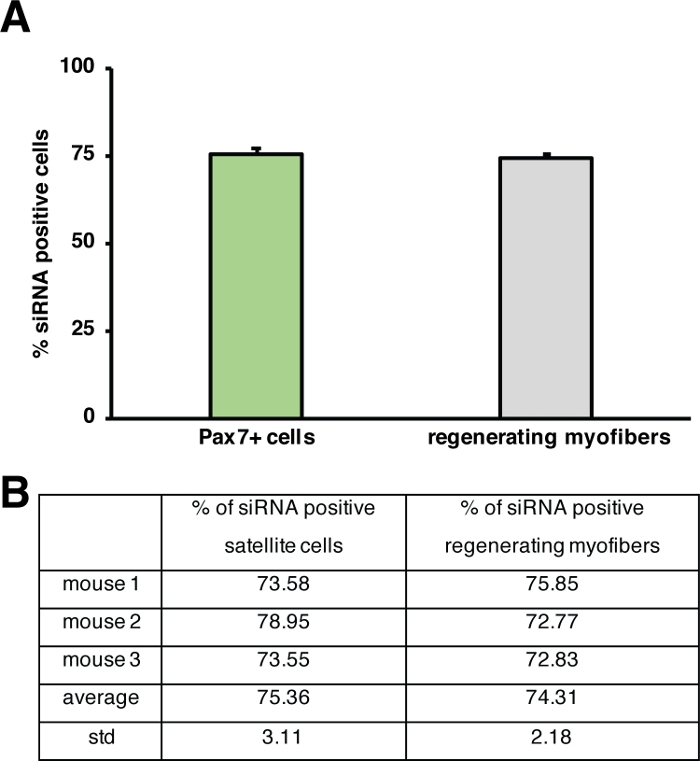
Figure 5: Quantification of uptake of fluorescently labeled siRNA. (A) Quantification of siRNA uptake by satellite cells (Pax7+ cells) and regenerating myofibers 2 days after injection into regenerating skeletal muscle. Injection was performed at day 3 after cardiotoxin mediated injury. n = 3, error bars show SEM. (B) Quantification underlying the graph depicted in (A). Please click here to view a larger version of this figure.
Discussion
Here we present a method to investigate the function of a specific gene during regeneration of skeletal muscle without the need of transgenic animals. This is accomplished by the combination of cardiotoxin induced muscle injury with the injection of a self-delivering siRNA into the regenerating skeletal muscle at day 3 after injury. We have described in detail the procedures of muscle injury by cardiotoxin, the injection of the self-delivering siRNA and processing of the harvested muscles to analyze the progress of regeneration. We demonstrate that injection of the snake venom cardiotoxin into skeletal muscle effectively injures the whole muscle and that self-delivering siRNAs are found in around 75% of all satellite cells among other cell types two days after their injection into the regenerating skeletal muscle (Figure 4, Figure 5).
Particular attention should be focused on a homogeneous injury of the tibialis anterior muscle since varying degrees of injury influence the regeneration outcome and thereby also the effect of the siRNA might be affected. Furthermore, it is critically important that the whole regenerating area is injected with the self-delivering siRNA. For the analysis of the regeneration process, it is recommended to always compare similar areas of the regenerating skeletal muscle, therefore, the tibialis anterior muscle should be cut in half to always compare the mid-belly region of the muscle. When analyzing the muscle, the whole cryosection needs to be analyzed since myofiber composition differs in the tibialis anterior muscle and might therefore regenerate differently.
The function of satellite cells can be investigated by various experimental procedures including their culture on the adjacent isolated single myofibers17, by transplantation and by analyzing regeneration of skeletal muscle after induced injury18,19. Investigating the function of satellite cells by using an in vivo induced injury model, e.g., injection of cardiotoxin, provides the ability to analyze satellite cell function also in terms of their interaction with other cell types such as macrophages and investigation of the influence of systemic factors2. Injury of skeletal muscle can be accomplished by various means, e.g., eccentric exercise, freeze injury, injection of BaCl2 or injection of snake venoms such as cardiotoxin or notexin18. While eccentric exercise is probably the most physiological injury method, the injury to one particular muscle is only limited20. Freeze injuries can be applied when the migration of satellite cells towards the site of injury is the aim of the study or only a specific part of the muscle should be injured. Experimentally the disadvantage of freeze injuries is the open surgery which needs to be performed to apply the precooled metal probe. Injection of BaCl2 or snake venoms is the most dramatic method of injury, thereby challenging satellite cell function the most. Furthermore, the injection is minimally invasive, surgery time, in general, is less than five minutes and does not involve suturing, etc. thereby minimizing the risk of infections.
Muscle injury is mostly used to investigate the functional consequences of loss of gene functions, e.g., loss of Pax77,21. Especially if aged mice are the focus of the scientific question, the generation or use of transgenic mice is often not feasible. Injection of self-delivering siRNAs targeting a specific gene is a viable alternative in those cases and has been used successfully22. In brief, the tibialis anterior muscle of mice was injured by injection of cardiotoxin and self-delivering siRNAs directed against fibronectin (FN) were injected at day 3 after injury. Muscles were analyzed 10 days after injury and a significant decrease in satellite cell numbers was observed in siFN versus scrambled siRNA control conditions. The knockdown efficiency was determined in whole muscle lysates by quantitative Real Time PCR 2 days after siRNA injection, a reduction in expression levels of 58% was achieved suggesting that delivery and knockdown efficiency are sufficient for functional analyses22. Alternatives for testing the knockdown efficiency are either immunoblot or immunofluorescence analyses with antibodies directed against the target gene. The efficiency and specificity of the siRNA used for in vivo injections should be determined before injection into mice, e.g., by testing the efficiency in isolated satellite cells or primary myoblasts. The use of a smart pool consisting of 4 different siRNAs versus a single siRNA increases the efficiency of knockdown but also increases the risk of unspecific targeting. The specificity of all siRNA sequences used should be tested in cell culture to avoid off-target effects. As a control, a non-targeting scrambled siRNA should be used since the injection of a siRNA per se might influence the regeneration process due to the injection and, thereby, additional damage of the muscle. The time point of injecting the siRNA is depending on the scientific question and on the expression profile of the target gene. Generally, an injection of self-delivering siRNA at day 3 after cardiotoxin injury targets most genes important for satellite cell proliferation since satellite cell proliferation peaks around day 3 after injury. The time point for the first siRNA injection should not be less than 48 h after cardiotoxin injury since the injection volume of cardiotoxin is quite high and reabsorption of the liquid should have taken place before injecting additional solutions into the muscle. In general, multiple injections of siRNAs or a combination of different siRNAs is possible although one must consider that each injection into the regenerating muscle is causing additional damage.
One limitation of the described method is the fact, that the effect observed is not necessarily only depending on knockdown of the target gene in satellite cells but might be attributed to other cell types such as immune cells or fibro-adipogenic progenitor cells. Therefore, it is necessary to combine those experiments with experiments investigating a pure population of satellite cells. One can either perform experiments using floating myofiber cultures, where satellite cells are cultured on their adjacent myofibers or perform transplantation experiments using isolated satellite cells17.
An alternative to the injection of self-delivering siRNAs is the injection of small molecule inhibitors or recombinant proteins which can be performed, depending on the scientific question. For instance, the injection of the extracellular matrix protein fibronectin or small molecule inhibitors of JAK/STAT signaling have been successfully performed in aged mice15,16. The analysis of a particular gene function in one specific cell type during regeneration of skeletal muscle, e.g., in satellite cells, is only possible through the use of an inducible genetic mouse model. Injections of self-delivering siRNAs, recombinant proteins or small molecule inhibitors might affect multiple cell types in regenerating skeletal muscle.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors declare no competing financial interests.
Materials
| isoflurane | Henry Schein | Isothesia | inhalation narcotics |
| Hot Plate 062 | Labotect | 13854 | |
| Anesthesia System (Tec 7) with inhalation box + nose masks | Tem Sega | Minihub | |
| shaver for rodents | isis | GT420 | |
| cardiotoxin | Latoxan | L8102 | snake venom needed for muscle injury |
| Accell siRNA smart pool | Dharmacon | depending on your target gene | self delivering siRNA |
| Alexa Fluor 488 donkey anti-rabbit IgG | Thermo Fisher | A-21206 | secondary antibody |
| Alexa Fluor 546 goat anti-mouse IgG1 | Thermo Fisher | A-21123 | secondary antibody |
| Coverslips | VWR | 631-1574 | |
| CV Mount | Leica | 14046430011 | mounting medium for immunohistochemistry |
| DAPI | Sigma Aldrich | D9542 | nuclear staining |
| devMHC antibody | DHSB | F.1652 | |
| Eosin Y | Thermo Fisher | 73104 | |
| Haematoxylin Gill No3 | Sigma-Aldrich | GHS316-500ML | |
| insulin syringe (29g) | Terumo | 3SS05M2813 | syringue used for muscle injections |
| laminin antibody | Sigma Aldrich | L9393 | |
| M.O.M blocking reagent | Vector labs | MKB-2213 | blocking for immunofluorescent staining |
| Meloxicam | Boehringer Ingelheim | Metacam | analgesics |
| OCT | Thermo Fisher | 6502 | tissue embedding |
| Pax7 antibody | DSHB | PAX7 | satellite cell specific antibody |
| ProLong Gold Antifade Mountant | Thermo Fisher | P36934 | aequos mounting medium |
| Sucrose | Carl Roth | 4621.1 | tissue embedding |
| Superfrost plus | Thermo Scientific | J1830AMNZ | microscope slides |
| TritonX-100 | Amresco | 0694-1L | permeabilization reagent |
| Dissection tools | |||
| Dumont 5, straight | Fine Science Tools | 11295-10 | |
| Dumont 7, curved | Fine Science Tools | 11272-40 | |
| Extra fine Bonn scissors (cutting edge: 13 mm) | Fine Science Tools | 14084-08 | |
| Narrow pattern forceps | Fine Science Tools | 11002-16 | |
| Spring scissors (cutting edge: 5 mm, tip diameter: 0.35 mm) | Fine Science Tools | 91500-09 | |
References
- Frontera, W. R., Ochala, J. Skeletal muscle: a brief review of structure and function. Calcified Tissue International. 96 (3), 183-195 (2015).
- Schmidt, M., Schuler, S. C., Huttner, S. S., von Eyss, B., von Maltzahn, J. Adult stem cells at work: regenerating skeletal muscle. Cellular and Molecular Life Sciences. , (2019).
- Lepper, C., Partridge, T. A., Fan, C. M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 138 (17), 3639-3646 (2011).
- Murphy, M. M., Lawson, J. A., Mathew, S. J., Hutcheson, D. A., Kardon, G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 138 (17), 3625-3637 (2011).
- Mauro, A. Satellite cell of skeletal muscle fibers. The Journal of Biophysical and Biochemical Cytolology. 9, 493-495 (1961).
- Shea, K. L., et al. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 6 (2), 117-129 (2010).
- von Maltzahn, J., Jones, A. E., Parks, R. J., Rudnicki, M. A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proceedings of the National Academy of Sciences of the United States of America U S A. 110 (41), 16474-16479 (2013).
- Sambasivan, R., et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 138 (17), 3647-3656 (2011).
- Yin, H., Price, F., Rudnicki, M. A. Satellite cells and the muscle stem cell niche. Physiological Reviews. 93 (1), 23-67 (2013).
- Bentzinger, C. F., Wang, Y. X., Rudnicki, M. A. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspectives in Biology. 4 (2), (2012).
- Schworer, S., et al. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature. 540 (7633), 428-432 (2016).
- Sousa-Victor, P., et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 506 (7488), 316-321 (2014).
- Dumont, N. A., et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nature Medicine. 21 (12), 1455-1463 (2015).
- von Maltzahn, J., Renaud, J. M., Parise, G., Rudnicki, M. A. Wnt7a treatment ameliorates muscular dystrophy. Proceedings of the National Academy of Sciences of the United States of America U S A. 109 (50), 20614-20619 (2012).
- Price, F. D., et al. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nature Medicine. 20 (10), 1174-1181 (2014).
- Lukjanenko, L., et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nature Medicine. 22 (8), 897-905 (2016).
- Huttner, S. S., et al. Isolation and Culture of Individual Myofibers and Their Adjacent Muscle Stem Cells from Aged and Adult Skeletal Muscle. Methods in Molecular Biology. , (2019).
- Hardy, D., et al. Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLoS One. 11 (1), e0147198 (2016).
- Hall, M. N., et al. Transplantation of Skeletal Muscle Stem Cells. Methods in Molecular Biology. 1556, 237-244 (2017).
- Dueweke, J. J., Awan, T. M., Mendias, C. L. Regeneration of Skeletal Muscle After Eccentric Injury. Journal of Sport Rehabilitation. 26 (2), 171-179 (2017).
- Lepper, C., Conway, S. J., Fan, C. M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 460 (7255), 627-631 (2009).
- Bentzinger, C. F., et al. Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell. 12 (1), 75-87 (2013).

