Utilization of Grafix for the Detection of Transient Interactors of Saccharomyces cerevisiae Spliceosome Subcomplexes
Summary
Here, we describe the utilization of Grafix (Gradient Fixation), a glycerol gradient centrifugation in the presence of a crosslinker, to identify interactions between splicing factors that bind transiently to the spliceosome complex.
Abstract
Pre-mRNA splicing is a very dynamic process that involves many molecular rearrangements of the spliceosome subcomplexes during assembly, RNA processing, and release of the complex components. Glycerol gradient centrifugation has been used for the separation of protein or RNP (RiboNucleoProtein) complexes for functional and structural studies. Here, we describe the utilization of Grafix (Gradient Fixation), which was first developed to purify and stabilize macromolecular complexes for single particle cryo-electron microscopy, to identify interactions between splicing factors that bind transiently to the spliceosome complex. This method is based on the centrifugation of samples into an increasing concentration of a fixation reagent to stabilize complexes. After centrifugation of yeast total extracts loaded on glycerol gradients, recovered fractions are analyzed by dot blot for the identification of the spliceosome sub-complexes and determination of the presence of individual splicing factors.
Introduction
Splicing is a highly dynamic process that requires binding and release of a multitude of factors in a coordinated manner. These splicing factors include RNA binding proteins, ATPases, helicases, protein kinases and phosphatases, ubiquitin ligases, among others1,2,3; and to allow for the molecular rearrangements to take place, some of these factors bind very transiently to the spliceosome subcomplexes, making the isolation and identification of these RNP intermediate complexes very challenging.
Here, we used the Grafix method4,5 to stabilize the interaction of the yeast splicing factor Cwc24 with the Bact complex6 to allow for the identification of other factors bound concomitantly to that subcomplex and determine whether the ubiquitin ligase Prp19 plays any role in the binding or release of Cwc24 to the Nineteen (NTC) complex and to the 5’ end of the intron before activation and the first transesterification reaction takes place. The advantage of exposing the macromolecules to an increasing concentration of the crosslinker along the glycerol gradient is that it avoids inter-complexes crosslinks4,5, and therefore, the formation of aggregates.
This method was used as a complementation to protein coimmunoprecipitation and pull-down assays, which, despite allowing the isolation of large complexes, may not be reliable for maintaining transient interactions within large dynamic complexes7,8. The use of fixation reagents in the glycerol gradient stabilizes the binding of such factors, allowing the confirmation of interactions of specific proteins with splicing subcomplexes. Because the chosen crosslinker was chemically irreversible, proteins present in the recovered fractions were analyzed by dot blot after the gradient centrifugation.
Protocol
1. Yeast total extract preparation
- Grow the yeast cells expressing one of the splicing factors fused to the TAP tag9 in 1 L YNB-glu media (Yeast Nitrogen Basis supplemented with 2% m/v glucose) with the appropriate amino acids or nucleic bases, in this case, adenine (20 µg/mL), leucine (30 µg/mL), tryptophan (30 µg/mL), up to OD600 = 1.0.
- Collect the cells from the 1 L culture by centrifuging in three 500 mL centrifuge bottles at 17,000 x g for 10 min at 4 °C and wash twice with 10 mL cold sterile water.
- Resuspend the collected yeast cells in 1/10 of the cell volume of cold buffer A (10 mmol L-1 HEPES pH = 7.9, 1.5 mmol L-1 MgCl2, 50 mmol L-1 KCl, 5% v/v glycerol, 0.5 mmol L-1 DTT, EDTA-free Protease Inhibitor Cocktail).
- Freeze small drops of cell suspension in liquid nitrogen. The small drops can be obtained by pipetting the resuspended cell solution and dropping 50 µL directly into liquid nitrogen.
CAUTION: Liquid nitrogen can cause injuries in contact with skin or eyes. Handle using appropriated safety equipment.
NOTE: The protocol can be paused here. The frozen drops of cell suspension can be stored at -80 °C. - Prepare yeast cells lysates by grinding in a Ball Mill device through six cycles at 20 Hz/s for 3 min.
NOTE: After each cycle, immerse the container harboring the frozen cells in liquid nitrogen to avoid melting. The protocol can be paused here. The frozen extracts can be stored at -80 °C. Yeast splicing extracts can also be prepared using homogenizers to lysate spheroplasts10, or using mortar and pestle11,12. - Melt the extract by placing the tubes containing them in water at room temperature, shaking occasionally.
- Centrifuge extracts at 45,000 x g for 1 h at 4 °C.
- Quantify the protein content of the cleared supernatant by the BCA method13.
- Prepare aliquots of the extracts, fast freeze them in liquid nitrogen and store at -80 °C.
NOTE: The protocol can be paused here.
2. Glycerol gradient preparation
- Prepare two glycerol solutions in Buffer A, one containing 10% v/v and the other containing 30% v/v glycerol.
- Add the crosslinking agent glutaraldehyde to 0.1% v/v in the 30% v/v glycerol solution and mix to homogenize.
CAUTION: Handle glutaraldehyde inside the fume hood using appropriate safety equipment. - Add 6 mL of the cold 10% v/v glycerol solution at the bottom of the 12 mL centrifuge tube (14 x 89 mm).
- Add 6 mL of the cold 30% v/v glycerol solution supplemented with glutaraldehyde with a syringe attached to a long needle provided with the Gradient Master device at the bottom of the tube, just beneath the 10% v/v glycerol solution.
NOTE: Alternatively, the 10% v/v glycerol solution can be carefully pipetted on the top of the 30% v/v glycerol/glutaraldehyde solution. - Use the Gradient Master device to generate a continuous density gradient.
NOTE: In the Gradient Master device, the tubes are placed into an appropriate rack and rotated briefly, following the manufacturer’s recommendations for determining the parameters (time/angle/speed). In this work we used: 2:25 min/81.5°/11 rpm. - Carefully add 200 μL of a 7% v/v glycerol/buffer A cushion on the top just before the addition of the cell extracts.
NOTE: The glycerol concentration of the cushion must be lower than the less concentrated glycerol solution used to create the linear gradient.
3. Extracts centrifugation
- Load approximately 2 mg of total protein on the top of each 12 mL linear glycerol gradient 10%–30% glutaraldehyde with cushion.
- Place the tubes in a pre-cooled swing-bucket rotor.
NOTE: Handle the tubes gently to avoid mixing before ultracentrifugation. - Centrifuge at 194,000 x g for 16 h at 4 °C.
- Aliquot each 12 mL tube in twenty-four 500 μL fractions using an adapted EconoSystem or by carefully pipetting.
NOTE: An adapted EconoSystem consists of a peristaltic pump, the UV detector, and the fraction collector connected to the tube-perforating device. The sedimentation profile can be monitored by the measurement of the absorbance at 280 nm. - Use a 40% v/v glycerol solution through the peristaltic pump to push the glycerol 10%–30% gradient from the tube to the fraction collector.
NOTE: The protocol can be paused here. Store the fractions at -80 °C until use. - Load 50 μL of each fraction directly on nitrocellulose membranes on a dot blot or slot blot device. Detect protein by immunoblot using antibody against CBP (1:6,000, to detect the CBP portion of the TAP tag) and anti-rabbit IgG conjugated with Horseradish Peroxidase (1:15,000) as the secondary antibody.
Representative Results
To analyze the sedimentation profile of Cwc24-TAP and determine whether the Grafix method was effective to stabilize its binding to splicing subcomplexes, we separated total yeast extracts of cells expressing Cwc24-TAP through centrifugation on glycerol gradients, in the presence or absence of glutaraldehyde as a crosslinking agent. Samples of twenty-four 500 μL fractions were then analyzed by slot blot with antibody against the CBP portion of the TAP tag. The results show that in the absence of the crosslinker, Cwc24 is concentrated between fractions 5 and 13 (Figure 1B), corresponding to complexes of about 80 to 200 MDa14. In the presence of the crosslinker, however, the position of Cwc24 on the gradient is shifted to fractions at the bottom of the gradient, corresponding to larger complexes (Figure 1). These results indicate that glutaraldehyde stabilizes the association of Cwc24 with splicing complexes.
In order to establish whether the complexes retaining Cwc24 correspond to the Bact complex, which is formed by snRNPs U2, U5, and U6 and the NTC complex, we compared Cwc24 sedimentation to those of the U5 snRNP subunit Prp8 and the NTC subunit Prp19. Yeast strains expressing either of these proteins fused to the TAP tag were subjected to Grafix, followed by dot blot for the detection of the proteins. These three spliceosome subunits showed similar profiles, sedimenting with larger complexes at the bottom of the gradient. Cwc24 is concentrated in the same fractions, but because it associates transiently with the spliceosome, it is also present in the lighter fractions (Figure 2). Quantification of the dots shows these profiles (Figure 2B).
Interestingly, fractions 11 and 12 are those where Prp8 and Prp19 start to concentrate, suggesting that this is the portion of the gradient where Bact complex starts to sediment. To ascertain that these proteins are bound to complexes in these fractions, aliquots of fractions 11 and 12 were subjected to electrophoresis on native gels and subsequently to western blot. Signals of Prp8 and Prp19 appear as smears that barely enter an 8% acrylamide gel, showing that indeed they are part of large complexes (Figure 3). Cwc24 is also present in fraction 12, but in much lower concentration, consistent with its transient binding to the Bact complex. These results show that glutaraldehyde can be used as a crosslinker to stabilize the binding of transient factors to splicing subcomplexes.
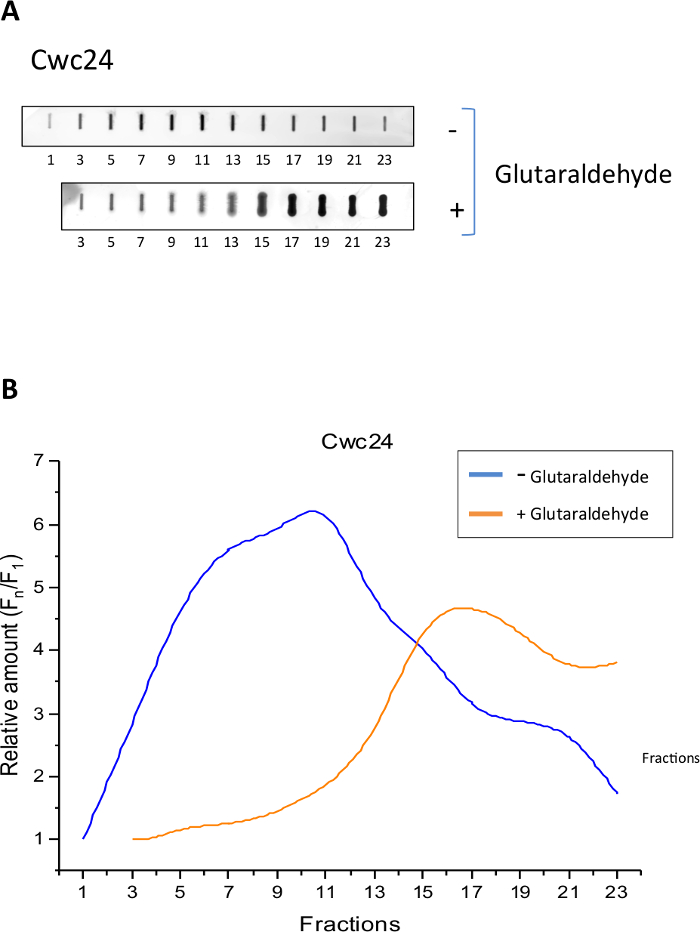
Figure 1: Grafix holds Cwc24 associated to larger complexes. Total yeast extract of cells expressing Cwc24-TAP was separated by centrifugation on glycerol gradients, either in the absence or presence of the crosslinker glutaraldehyde. (A) Odd fractions of the gradient were subjected to slot blot for the immunodetection of Cwc24-TAP with antibody against the CBP portion of the TAP tag. (B) Quantification of Cwc24-TAP using Image J shows the concentration of the protein through the fractions of the glycerol gradient. Presence or absence of glutaraldehyde is indicated. Y axis shows the relative amounts of Cwc24 through the gradient, calculated as the intensity of the signal in the fractions, relative to the first fraction (Fn/F1). Please click here to view a larger version of this figure.

Figure 2: Sedimentation of splicing factors through glycerol/glutaraldehyde gradients. Extracts of yeast cells expressing either Cwc24, Prp8, or Prp19 fused to the TAP tag were loaded on glycerol gradients containing glutaraldehyde and centrifuged for the separation of splicing complexes. (A) Samples of the twenty-four fractions of the gradient were analyzed by dot blot and immunodetection of the TAP-fused proteins with antibody against the CBP portion of the TAP tag. (B) Quantification of the proteins using Image J shows their concentration at the bottom fractions of the glycerol/glutaraldehyde gradient. Y-axis shows the relative amounts of the proteins through the gradient, calculated as the intensity of the signal in the fractions, relative to the first fraction showing a signal above background (Fn/F1). Please click here to view a larger version of this figure.

Figure 3: Detection of splicing complexes by native PAGE and immunoblot. Aliquots of fractions 11 and 12 of the gradient shown in Figure 2 were separated by electrophoresis on 8% acrylamide native gel and subjected to immunoblot with antibody against CBP. Proteins are not detected as bands because complexes are too large to separate on native gels. Please click here to view a larger version of this figure.
Discussion
Protein-protein and ribonucleic acids-protein interactions can be stabilized using crosslinking agents. It is important that the resulting complex is stable to withstand ultracentrifugation on glycerol gradient. Additionally, the buffer conditions should allow the interaction, but be stringent enough to avoid non-specific binding. In the experiments shown here, we used a buffer solution already established for in vitro splicing reactions15.
The speed and time of centrifugation should be optimized for the complexes being analyzed. We tested different sedimenting conditions, centrifuging samples at 94,000 x g and 194,000 x g for 16 h at 4 °C, with similar results. These controls are important to determine the conditions in which the complexes being analyzed do not precipitate.
We also tested different blot methods, pipetting directly on the membrane, using slot blot or dot blot systems, which showed that dot blot gave more reproducible results.
A major limitation of using glutaraldehyde as a crosslinker is that the crosslink cannot be reverted. Therefore, proteins cannot be analyzed by immunoblot after SDS-PAGE, but only by native gels or dot blot. However, reversible crosslinking agents, such as formaldehyde, could potentially also be used.
We have previously used co-immunoprecipitation of proteins to identify splicing complexes, but in the case of transient or weak interactions7, crosslink may help the isolation of intermediate complexes for later determination of their components by mass spectrometry. Although our focus here was the spliceosome, this method can certainly be used for the isolation of other dynamic processes that include intermediate complexes with varying compositions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a FAPESP grant (15/06477-9).
Materials
| Anti-Calmodulin Binding Protein Epitope | Millipore | 07-482 | |
| ECL anti-Rabbit IgG | GE Healthcare | NA934 | |
| EconoSystem | Bio-Rad | 1-800-424-6723 | Parts of the EconoSystem used: peristaltic pump, the UV detector and the fraction collector |
| EDTA-free Protease Inhibitor Cocktail | Roche | 11873580001 | |
| Fraction Recovery System | Beckman Coulter | 270-331580 | Tube-perforating device that was connected to the parts of the EconoSystem |
| Gradient Master Model 107ip | Biocomp | 107-201M | |
| Mixer Mill MM 200 | Retsch | 207460001 | Ball Mill device |
| Rotor F12-6x500Lex | Thermo Scientific | 096-062375 | |
| Sorvall RC 6 Plus Centrifuge | Thermo Scientific | 36-101-0816 | |
| Swinging Bucket Rotor P40ST | Hitachi | ||
| Ultracentrifuge CP 80 NX | Hitachi | 901069 | |
| Ultra-Clear Centrifuge Tubes (14 x 89 mm) | Beckman Coulter | 344059 |
References
- Kastner, B., Will, C. L., Stark, H., Lührmann, R. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harbor Perspectives in Biology. 11 (11), 032417 (2019).
- Yan, C., Wan, R., Shi, Y. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harbor Perspectives in Biology. 11 (1), 032409 (2019).
- Cordin, O., Hahn, D., Beggs, J. D. Structure, function and regulation of spliceosomal RNA helicases. Current Opinion in Cell Biology. 24, 431-438 (2012).
- Kastner, B., et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nature Methods. 5 (1), 53-55 (2008).
- Stark, H. Grafix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods in Enzymology. 481, 109-126 (2010).
- Yan, C., Wan, R., Bai, R., Huang, G., Shi, Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 353 (6302), 895-904 (2016).
- Ohi, M. D., et al. Proteomics analysis revelas stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Molecular and Cellular Biology. 22 (7), 2011-2024 (2002).
- Lourenco, R. F., Leme, A. F. P., Oliveira, C. C. Proteomic analysis of yeast mutant RNA exosome complexes. Journal of Proteome Research. 12 (12), 5912-5922 (2013).
- Rigaut, G., et al. A generic protein purification method for protein complex characterizatioin and proteome exploration. Nature Biotechnology. 17, 1030-1032 (1999).
- Cheng, S. C., Newman, A., Lin, R. J., McFarland, G. D., Abelson, J. N. Preparation and fractionation of yeast splicing extract. Methods in Enzymology. 181, 89-96 (1990).
- Umen, J. G., Guthrie, C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Development. 9, 855-868 (1995).
- Dunn, E. A., Rader, S. D., Hertel, K. J. Preparation of yeast whole cell splicing extract. Spliceosomal Pre-mRNA Splicing: Methods and Protocols. Methods in Molecular Biology. 1126, 123-135 (2014).
- Hill, H. D., Straka, J. G. Protein determination using bicinchoninic acid in the presence of sulfhydryl reagents. Analalytical Biochemistry. 170 (1), 203-208 (1988).
- Cepeda, L. P., et al. The ribosome assembly factor Nop53 controls association of the RNA exosome with pre-60S particles in yeast. The Journal of Biological Chemistry. 294 (50), 19365-19380 (2019).
- Lin, R. J., Newman, A. J., Cheng, S. C., Abelson, J. Yeast mRNA splicing in vitro. The Journal of Biological Chemistry. 260 (27), 14780-14792 (1985).

