A Spin-Tip Enrichment Strategy for Simultaneous Analysis of N-Glycopeptides and Phosphopeptides from Human Pancreatic Tissues
Summary
Post-translational modifications (PTMs) change protein structures and functions. Methods for the simultaneous enrichment of multiple PTM types can maximize coverage in analyses. We present a protocol using dual-functional Ti(IV)-immobilized metal affinity chromatography followed by mass spectrometry for the simultaneous enrichment and analysis of protein N-glycosylation and phosphorylation in pancreatic tissues.
Abstract
Mass spectrometry can provide deep coverage of post-translational modifications (PTMs), although enrichment of these modifications from complex biological matrices is often necessary due to their low stoichiometry in comparison to non-modified analytes. Most enrichment workflows of PTMs on peptides in bottom-up proteomics workflows, where proteins are enzymatically digested before the resulting peptides are analyzed, only enrich one type of modification. It is the entire complement of PTMs, however, that leads to biological functions, and enrichment of a single type of PTM may miss such crosstalk of PTMs. PTM crosstalk has been observed between protein glycosylation and phosphorylation, the two most common PTMs in human proteins and also the two most studied PTMs using mass spectrometry workflows. Using the simultaneous enrichment strategy described herein, both PTMs are enriched from post-mortem human pancreatic tissue, a complex biological matrix. Dual-functional Ti(IV)-immobilized metal affinity chromatography is used to separate various forms of glycosylation and phosphorylation simultaneously in multiple fractions in a convenient spin tip-based method, allowing downstream analyses of potential PTM crosstalk interactions. This enrichment workflow for glyco- and phosphopeptides can be applied to various sample types to achieve deep profiling of multiple PTMs and identify potential target molecules for future studies.
Introduction
Protein post-translational modifications (PTMs) play a major role in modulating protein structures and consequently their functions and downstream biological processes. The diversity of the human proteome increases exponentially due to the combinatorial variability afforded by various PTMs. Different variants of proteins from their canonical sequences as predicted by the genome are known as proteoforms, and many proteoforms arise from PTMs1. Studying proteoform diversity in health and disease has become an area of research of great interest in recent years2,3.
The study of proteoforms and more specifically PTMs with great depth has become more facile through the development of mass spectrometry (MS)-based proteomics methods. Using MS, analytes are ionized, fragmented, and identified based on the m/z of fragments. Enrichment methods are often necessary due to the low relative abundance of PTMs compared to non-modified forms of proteins. Though analysis of intact proteins and their PTMs, called top-down analyses, have become more routine, the enzymatic digestion of proteins and the analysis of their component peptides in bottom-up analyses is still the most widely used route for PTM analysis. The two most widely studied PTMs, and the two most common PTMs in vivo, are glycosylation and phosphorylation4. These two PTMs play major roles in cell signaling and recognition and thus are important modifications to characterize in disease research.
The chemical properties of various PTMs often provides routes toward enrichment of these PTMs at the protein and peptide levels prior to analysis. Glycosylation is a hydrophilic PTM due to the abundance of hydroxyl groups on each monosaccharide. This property can be used to enrich glycopeptides in hydrophilic interaction chromatography (HILIC), which can separate more hydrophilic glycopeptides from the hydrophobic non-modified peptides5. Phosphorylation adds the phosphate moiety, which is negatively charged except at acidic pH. Due to this charge, various metal cations, including titanium, can be used to attract and bind phosphopeptides while non-phosphorylated species are washed away. This is the principle of immobilized metal affinity chromatography (IMAC). Further discussions of these and other enrichment strategies for glycosylation and phosphorylation can be found in recent reviews6,7.
Comparatively large amounts of starting peptide material (0.5 mg or more) are often needed for enrichment protocols due to the low stoichiometry of PTMs on peptides. In scenarios where this amount of sample may not be easily obtained, such as tumor core biopsy or cerebrospinal fluid analyses, it is beneficial to use facile workflows that result in maximum biomolecular information. Recent strategies developed by our lab and others have highlighted the simultaneous and parallel analysis of glycosylation and phosphorylation using the same PTM enrichment workflow8,9,10,11,12. Though the chemical properties of these two PTMs may differ, these PTMs may be analyzed in multiple steps due to the innovative separation techniques and materials used. For example, electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) overlays separations based on hydrophilic interactions between analytes and the mobile phase with charge-charge interactions between analytes and the stationary phase material13,14,15,16. At acidic pH, the attraction of phosphorylated peptides to the stationary phase can improve their retention and separation from non-modified peptides. Material consisting of Ti(IV) immobilized on hydrophilic microspheres can be used for HILIC and IMAC-based elution to separate phosphopeptides and neutral, acidic, and mannose-6-phosphorylated glycopeptides17,18. This strategy is known as dual-functional Ti(IV)-IMAC. Using these strategies for enriching multiple PTMs in a single workflow can make analyses of potential PTM crosstalk interactions more accessible. Additionally, the total sample amount and time requirements are less than the conventional enrichment methods when performed in parallel (i.e., HILIC and IMAC on separate sample aliquots).
To demonstrate the dual-functional Ti(IV)-IMAC strategy for simultaneous analysis of protein glycosylation and phosphorylation, we have applied it to analyze post-mortem human pancreatic tissues. The pancreas produces both digestive enzymes and regulatory hormones, including insulin and glucagon. The pancreatic function is impaired in pancreatic disease. In diabetes, the regulation of blood sugar is affected, leading to higher levels of glucose in the blood. In pancreatitis, inflammation results from auto-digestion of the organ3. Changes in PTM profiles, including glycosylation and phosphorylation, may result, as is often the case, in other diseases.
Here, we describe a protocol for a spin-tip based simultaneous enrichment method, based on a dual-functional Ti(IV)-IMAC strategy, for N-glycopeptides and phosphopeptides derived from proteins extracted from pancreatic tissue. The protocol includes protein extraction and digestion, enrichment, MS data collection, and data processing, as can be seen in Figure 1. Representative data from this study are available via ProteomeXchange Consortium with identifier PXD033065.
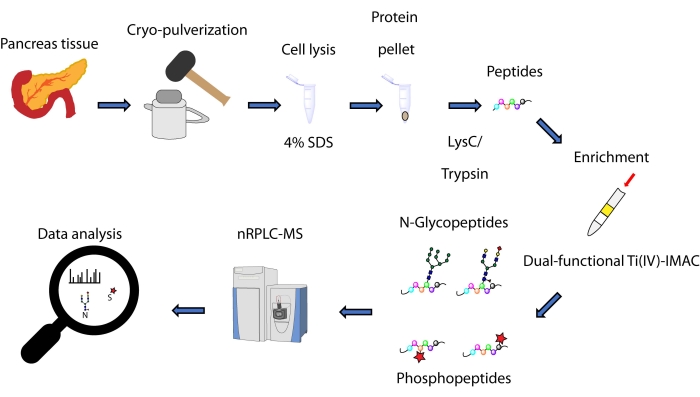
Figure 1: Workflow for simultaneous analysis of N-glycopeptides and phosphopeptides from human pancreatic tissues. Tissues are first cryo-pulverized into a fine powder before protein extraction using the detergent sodium dodecyl sulfate (SDS). Proteins are then subjected to enzymatic digestion. The resulting peptides are aliquoted prior to enrichment using dual-functional Ti(IV)-IMAC. Raw data is collected using nanoscale reversed phase liquid chromatography-mass spectrometry (nRPLC-MS) and is analyzed using database searching software. Please click here to view a larger version of this figure.
This protocol is intended to make PTM analyses more accessible and to enable more widespread analysis of multiple PTMs in the same workflow. This protocol can be applied to other complex biological matrices, including cells and biofluids.
Protocol
Consent was obtained for the use of pancreatic tissues for research from the deceased's next of kin and an authorization by the University of Wisconsin-Madison Health Sciences Institutional Review Board was obtained. IRB oversight is not required because it does not involve human subjects as recognized by 45 CFR 46.102(f).
CAUTION: Care should be taken when handling the reagents used in this protocol, which include acids (formic, acetic, trifluoroacetic), bases (ammonium hydroxide), and cryogens (liquid nitrogen). Read the safety data sheets for the reagents used to become familiar with the associated hazards and needed precautions. Concentrations denoted using percentages are volume/total volume (v/v) and are diluted with water.
1. Tissue cryo-pulverization, lysis, and protein extraction
- Fill a dewar with liquid nitrogen. Pre-chill the parts of the tissue pulverizer that will come in contact with the pancreatic tissues, namely, the chamber, pulverizer, and recovery spoon in a polystyrene container.
- Transfer the frozen tissue pieces into the pre-chilled sample holder and add a spoonful of liquid nitrogen to the tissue.
- Place the pulverizer into the chamber and strike it using a mallet five to ten times to crush the sample. Remove the pulverizer from the chamber and scrape off adherent tissue powder and pieces.
- Add a spoonful of liquid nitrogen to the chamber if tissues begin to melt. Repeat the pulverization process until the sample is a fine powder without large tissue chunks. Portion the samples into approximately 100 mg aliquots into pre-chilled tubes.
- Prepare lysis buffer containing 4% sodium dodecyl sulfate (SDS), 150 mM NaCl, and 25 mM Tris (pH = 7.4). Dissolve one tablet each of protease and phosphatase inhibitor in 500 µL of water for a 20x stock of each. Add the required volume of 20x stock of each inhibitor to the lysis buffer for a final 1x concentration.
- Add 600 µL of lysis buffer per 100 mg tissue to the tube and incubate at 95 °C in a heating block for 10 min with shaking at 800 rpm. Remove the samples from the heating block and let them cool to room temperature.
- Sonicate the samples at 60 W energy (20 kHz) for 45 s using 15 s pulses with a 30 s rest in between. Pellet the samples at 3,000 x g for 15 min at 4 °C.
- Add the supernatant to a 5x precipitation solvent, 300 µL of lysis buffer to 1.5 mL of precipitation solvent, containing 50% acetone, 49.9% ethanol, and 0.1% acetic acid. Chill overnight at -20 °C.
- Pellet the samples again at 3,000 x g for 15 min at 4 °C and remove the supernatant. Wash the pellet by breaking up with a spatula and mixing with the same amount of precipitation solvent. Pellet again, repeating the washing step 2x.
- Pellet the sample at 16,000 x g for 15 min at 4 °C. Air dry the sample pellet in a fume hood for 15 min and store at -80 °C until ready to proceed.
2. Protein digestion and desalting
- Resuspend the protein pellet in 300 µL of freshly made digestion buffer containing 50 mM triethylammonium bicarbonate (TEAB) and 8 M urea.
- Estimate the protein concentration of the solution using a protein assay according to the manufacturer's protocol.
- To the protein in the solution, add dithiothreitol (DTT) to a final concentration of 5 mM, mix, and reduce at room temperature for 1 h. Then, add iodoacetamide (IAA) to a final concentration of 15 mM, mix, and alkylate at room temperature for 30 min in dark. Quench alkylation by repeating the addition of DTT in the same volume as before and mix.
- Add LysC/trypsin at 1:100 enzyme:protein ratio and incubate at 37 °C for 4 h. Then, add 50 mM TEAB to dilute the 8 M urea used to < 1 M. Add trypsin at 1:100 enzyme:protein ratio and incubate at 37 °C overnight.
- Quench the digestion with the addition of trifluoroacetic acid (TFA) to 0.3% v/v. For every 1 mg of starting protein, condition a desalting cartridge (1 cc, 10 mg) with 1 mL of acetonitrile (ACN) and 3x with 1 mL of 0.1% TFA.
- Load the digested mixture onto the desalting cartridge. Wash the mixture 3x using 1 mL of 0.1% TFA. If the elution is slow, apply positive pressure but avoid a flow rate > one drop per second.
- Elute peptides using 1 mL of 60% ACN and 0.1% formic acid (FA) solution. Dry eluted peptides at approximately 35 °C using a centrifugal vacuum concentrator until the solvent has completely evaporated.
- Resuspend peptides in 300 µL of water and estimate peptide concentrations using a peptide assay according to the manufacturer's protocol. Portion the peptides into 500 µg aliquots and dry completely.
3. ERLIC N-glycopeptide enrichment
NOTE: Exact centrifuge speeds and times may differ based on samples and must be optimized. In general, 300 x g for 2 min is appropriate for conditioning and washing of the material and 100 x g for 5 min for eluting.
- Weigh approximately 3 mg of cotton wool and pack it into an empty 200 µL pipette tip (spin-tip; see Table of Materials).
- Transfer strong anion-exchange enrichment material into a tube and add 200 µL of 0.1% TFA per 10 mg material. For enrichment of 500 µg of peptides, use 15 mg of the material. Activate the material by shaking for 15 min.
- Using a tube adapter, place the spin-tip over a 2 mL tube and add enough slurry to the tip for 15 mg of material. Remove the liquid by spinning in a benchtop centrifuge.
- Condition the material by centrifuging 3x with 200 µL of ACN. Repeat the triplicate conditioning using 100 mM ammonium acetate (NH4Ac), 1% TFA, and 80% ACN/0.1% TFA (loading buffer).
- Resuspend 500 µg peptide samples in approximately 200 µL of loading buffer and flow through the spin-tip. Reload the flow-through 2x to ensure complete binding.
- Wash the material by centrifuging 5x using 200 µL of 80% ACN/0.1% TFA, and then 2x using 200 µL of 80% ACN/0.1% FA.
- Elute the peptides by centrifuging with 200 µL of each of the following: 50% ACN/0.1% FA (E1); 0.1% FA (E2); 0.1% TFA (E3); 300 mM KH2PO4, 10% ACN (E4).
- Wash the material by centrifuging 2x with 200 µL of 80% ACN/5% NH4OH. Elute the remaining peptides with 200 µL of 10% NH4OH (E5).
- Dry all the elutions completely using a centrifugal vacuum concentrator at approximately 35 °C.
- Perform desalting of the basic elution (E5) using a desalting tip according to manufacturer's protocol.
- Dry the elution from the desalting tip using a centrifugal vacuum concentrator at approximately 35 °C to completeness.
4. Ti(IV)-IMAC phosphopeptide enrichment
NOTE: Exact centrifuge speeds and times may differ based on samples and must be optimized. In general, 300 x g for 2 min is appropriate for conditioning and washing of the material and 100 x g for 5 min for eluting.
- Weigh approximately 3 mg of cotton wool and pack it into an empty 200 µL pipette tip (spin-tip).
- Transfer Ti-IMAC phosphopeptide enrichment material into a tube and add 200 µL of 0.1% TFA per 10 mg material. For enrichment of 500 µg peptides, use 10 mg of material.
- Using a tube adapter, place the spin-tip over a 2 mL tube and add enough slurry to the tip for 10 mg material. Remove the liquid by spinning in a benchtop centrifuge.
- Condition the material with 200 µL of 40% ACN/3% TFA using the same centrifuge settings as described above. Resuspend peptide samples in 200 µL of 40% ACN/3% TFA and flow through the spin-tip. Reload the flow-through twice to ensure more complete binding.
- Wash the material by centrifuging with 200 µL of 50% ACN, 6% TFA, and 200 mM NaCl solution, followed by washing 2x in 200 µL of 30% ACN and 0.1% TFA solution.
- Elute peptides with 200 µL of 10% NH4OH. Dry the elution completely under vacuum. Perform desalting using a desalting tip according to the manufacturer's protocol. Dry the elution from the desalting tip under vacuum to completeness.
5. Dual-functional Ti(IV) simultaneous enrichment
NOTE: Exact centrifuge speeds and times may differ based on samples and must be optimized. In general, 300 x g for 2 min is appropriate for conditioning and washing of the material and 100 x g for 5 min for eluting.
- Weigh approximately 3 mg of cotton wool and pack it into an empty spin-tip.
- Transfer approximately 1 g of Ti(IV)-IMAC material into a tube. Add 0.1% TFA to a known concentration of material, e.g., 20 mg/200 µL (material can be stored in this suspension at 4 °C).
- For enrichment from 500 µg peptides, add enough slurry to a spin-tip to transfer 20 mg of material. Wash the spin-tip by centrifuging with 200 µL of 0.1% TFA.
- Resuspend the samples in 200 µL of loading/washing solvent (80% ACN and 3% TFA) and flow through the spin-tip. Reload the flow-through 2x.
- Wash the spin-tip 6x by centrifuging with 200 µL of loading/washing solvent. Wash the spin-tip with 200 µL of 80% ACN/0.1% FA solution.
- Elute the peptides with 200 µL of each of the following: 60% ACN/0.1% FA (E1) and 40% ACN/0.1% FA (E2). Analyze each elution separately.
- Elute the peptides with 200 µL of each of the following: 20% ACN/0.1% FA and 0.1% FA. Combine the two elutions and analyze as one sample (E3).
- Elute the peptides with 200 µL of each of the following: 40% ACN/3% TFA; 50% ACN/6% TFA, 200 mM NaCl; and 30% ACN/0.1% TFA. Combine these elutions and analyze as one (E4) after desalting using a packed tip.
- Condition the material to basic pH using 200 µL of 90% ACN/2.5% NH4OH for 3x. Discard these washes.
- Elute the peptides with 200 µL of each of the following: 60% ACN/10% NH4OH (E5) and 40% ACN/10% NH4OH (E6). Analyze these elutions separately after desalting using a packed tip.
- Elute the peptides with 200 µL of each of the following: 20% ACN/10% NH4OH; 10% ACN/10% NH4OH; and 10% NH4OH. Combine these elutions and analyze as one (E7) after desalting using a packed tip.
- Dry all the elutions completely under vacuum.
6. Nano-flow reversed phase liquid chromatography-mass spectrometry (nRPLC-MS)
NOTE: MS data acquisition and analysis methods are diverse, and thus only one suggested LC-MS pipeline (and its associated parameters) is described here in the following steps. Samples generated using the previously outlined sample preparation and enrichment steps can be analyzed using other instrumental set-ups, including using commercially available chromatographic columns, given sufficient data quality.
- Pull and pack a 15 cm long capillary (75 µm inner diameter) using C18 material as described in19. Prepare mobile phase A (0.1% FA in water) and mobile phase B (0.1% FA in ACN).
- Reconstitute enriched peptide samples in 15 µL of 3% mobile phase B. Load 2 µL of the sample (~13% sample volume) directly onto the column. Analyze each sample in technical duplicate.
- Elute peptides using a gradient from 3% to 30% mobile phase B over 90 min at a flow rate of 0.3 µL/min. Wash the column using 75% B for 8 min followed by 95% B for 8 min, finishing the method with equilibration at 3% B for 12 min.
- Operate the mass spectrometer in the positive ion mode using data-dependent acquisition of the top 20 peaks with the following parameters: spray voltage 2 kV; MS1 detection in LC/MS from 400-2,000 m/z at 120,000 resolution, 2E5 automatic gain control (AGC) target, 100 ms maximum injection time, 30% RF lens, quadrupole isolation with 1.6 m/z window, for charge states 2-8 and undetermined, and 30 s dynamic exclusion; MS2 detection in the LC/MS using stepped HCD fragmentation at 22%, 30%, and 38%, fixed first mass 120 m/z at 30,000 resolution, 5E4 AGC target, and 2.5E4 minimum intensity requirement.
7. MS data analysis
NOTE: One data analysis pipeline using two different software to analyze the same dataset is presented here. Phosphorylation and glycosylation can be searched at the same time using a single software instead of two separate software as described here, though in general, software search time is proportional to the search space, i.e., number of PTMs considered. For this reason, two different software are used in parallel to the search for glycopeptides and phosphopeptides.
- Process raw data files using the appropriate software (see Table of Materials).
- Search spectra against the UniProt human (or appropriate species-specific) database. Set carbamidomethylation of Cys as a fixed modification. Set the maximum number of missed enzymatic cleavages as two.
- In the raw data files, using a commercial software (see Table of Materials), search for glycopeptides to be analyzed20. Use a 10 ppm precursor mass tolerance and a 0.01 Da fragment mass tolerance with a minimum peptide length of four residues.
- Search using the software-embedded N-glycan database expanded with typical mannose-6-phosphate(M6P)glycans, including HexNAc(2)Hex(4-9)Phospho(1-2), HexNAc(3-4)Hex(4-9)Phospho(1-2), HexNAc(2)Hex(3-4)Phospho(1), and HexNAc(3)Hex(3-4)Phospho(1).
- Set N-glycosylation as a common modification. Set the maximum total modifications per peptide spectral match (PSM) as one common and two rare.
- Set the following variable modifications: oxidation of Met (rare), deamidation at Asn and Gln (rare), and phosphorylation at Ser, Thr, and Tyr (common). Set the following identification filters: score cut-off > 150, |log prob| > 1, and delta mod score > 10.
- Export and analyze search results in the Proteins and Peptide Group tabs.
- Manually screen the identifications containing M6P glycans for the presence of the PhosphoHex oxonium ion (243 m/z) in the MS/MS spectra and remove false positive identifications, which do not contain this diagnostic ion.
- In the raw data files, using an open-source software (see Table of Materials), search for phosphopeptides to be analyzed21. Use a 20 ppm first search peptide tolerance and a 4.5 ppm main search peptide tolerance.
- Set the maximum number of modifications per peptide to 5. Set PSM and protein false discovery rate (FDR) to 1% and minimum peptide length to 7 residues.
- Set variable modifications as oxidation at Met, N-terminal protein acetylation, and phosphorylation at Ser, Thr, and Tyr. Analyze search results using the modificationSpecificPeptides files.
- Determine the number of identifications of PTMs at the protein, peptide, and modification site levels.
Representative Results
Representative mass spectrometry data, including raw files and search results, have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD03306522.
In this work, duplicate injection replicates were analyzed for each enrichment elution. Identifications made from both technical replicates were collated in the final analysis. Due to the semi-stochastic nature of data-dependent acquisition in picking peptide precursors for MS/MS fragmentation, identification overlap of about 70%-80% is expected between technical duplicates. In applications using these methods, at least two technical replicates are encouraged. For downstream statistical analyses, biological replicates for different experimental conditions should be taken into consideration. Depending on the desired stringency for data reporting, it may be useful to set filters for identifications, e.g., identification in at least x number of technical or biological replicates to be reported.
An example total ion chromatogram (TIC) for each elution from each enrichment can be seen in Supplementary Figure S1, Supplementary Figure S2, and Supplementary Figure S3. Using stringent filters during database searching of raw MS files can ensure more accurate and more confident identifications of MS/MS spectra to peptide sequences with PTMs. As is evident in the TICs for each elution, the peptide samples are still complex even after fractionation from the enrichment. Nanoflow chromatography coupled to a high-resolution, high-mass accuracy, and fast-scanning mass spectrometer can help analyze complex mixtures, such as peptides from a tryptic digest, with great sensitivity and depth. Example MS/MS spectra for glycosylated and phosphorylated spectra can be seen in Figure 2. Well-annotated spectra such as these result from rich fragmentation of precursors which increase confidence in the identification as assigned by the database searching software.
Figure 3 illustrates the peptide identifications made using the dual-functional Ti(IV)-IMAC spin-tip enrichment method over each elution fraction. The majority of glycopeptides elute in the first four fractions, while the majority of phosphopeptides elute in the last three fractions. This separation of different PTMs among the fractions helps prevent any interference in ionization that may result if they were not as adequately separated across elutions.
Figure 4 compares the glycoproteomics results from the dual Ti method compared to an ERLIC glycopeptide-only enrichment. As can be seen in Figure 4A, many identifications at the protein, glycoform, PTM, and modification site levels are common to both methods. ERLIC has more unique identifications at all levels compared to the dual Ti method, however. This includes M6P (high mannose glycans with at least one phosphate group) glycoforms, which require additional manual curation of the data to remove false positive identifications. Further, the types of glycans identified using either method are similar. The proportions of each type of glycan after binning into six categories based on their compositions are similar between both methods16.
Phosphoproteomics results from the dual Ti method are compared to a conventional IMAC phosphopeptide-only enrichment in Figure 5. The dual-functional Ti(IV) enrichment performs similarly to conventional IMAC as shown by substantial overlap at the protein, peptide, and PTM levels. Using either method, the majority of phosphorylated amino acids are serine and threonine, with around 1% phosphorylated tyrosine identified as well. Both the methods can also be used to identify multi-phosphorylated peptides.
The lists of modified proteins are mapped to genes and analyzed using gene ontology (GO) enrichment as shown in Figure 6 and Figure 7. GO analyses for the modified proteins identified using ERLIC and IMAC are shown in Supplementary Figure S4 and Supplementary Figure S5. The free, online Metascape tool performs the enrichment and creates networks based on enriched pathways and processes, linking them by similarity23. The node terms for each GO analysis can be found in Supplementary Table S1, Supplementary Table S2, Supplementary Table S3, and Supplementary Table S4. Since glycosylation is a major protein PTM in the extracellular matrix, it is not surprising that several terms enriched using the list of N-glycoproteins identified are related to the extracellular matrix. Phosphorylation is involved in cell signaling and this can be seen in several enriched terms found using the list of phosphoproteins identified.
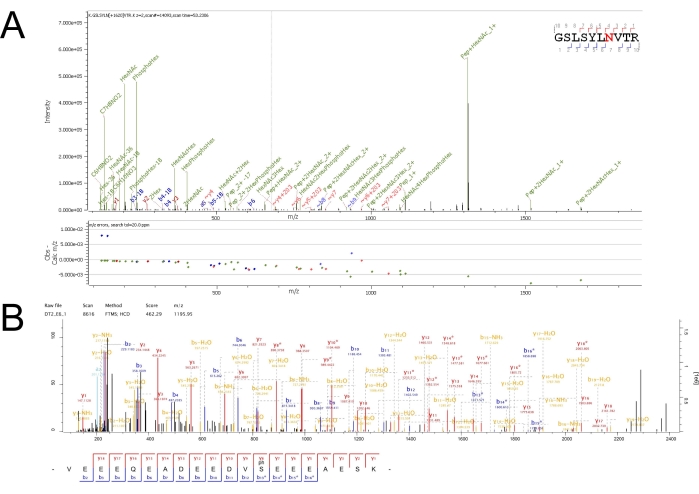
Figure 2: Annotated high confidence MS/MS spectra from N-glycosylated and phosphorylated peptides. (A) Mannose-6-phosphorylated peptide GSLSYLN(HexNAc(2)Hex(7)Phospho(1))VTR from cathepsin D (CATD) identified from retention times 53.08-53.33 min from the seventh enrichment elution. The presence of the PhosphoHex oxonium ion at 243 m/z improves confidence in this PTM assignment. (B) Phosphorylated peptide VEEEQEADEEDVS(Phospho)EEEAESK from thioredoxin-related transmembrane protein 1 (TMX1) from retention times 32.31-32.72 min from the sixth enrichment elution. The presence of -98 (-H3PO4) neutral losses between peptide fragment ions and their de-phosphorylated variants (denoted by fragment*) improves confidence in this PTM assignment. Please click here to view a larger version of this figure.
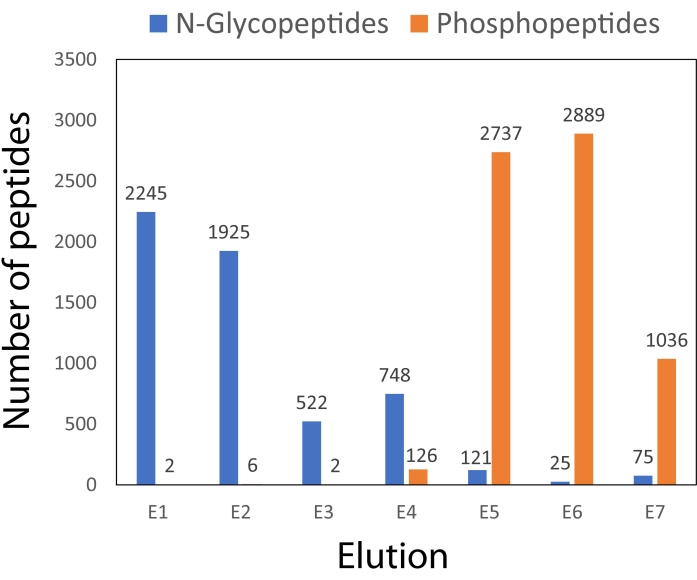
Figure 3: Comparison of peptide identifications from dual-functional Ti(IV)-IMAC enrichment across seven elutions. The number of identifications includes peptides identified in at least one of the two technical (injection) replicates. Please click here to view a larger version of this figure.
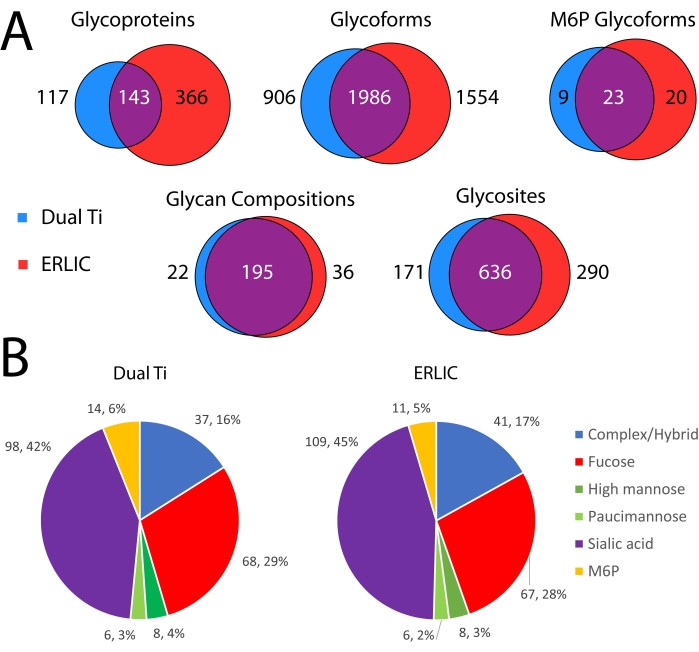
Figure 4: Comparison of glycoproteomics results from dual-functional Ti(IV)-IMAC enrichment compared to a conventional ERLIC glycopeptide enrichment. (A) Venn diagrams of glycoprotein, glycoform (protein, site, glycan), glycan composition, and glycosite identifications between enrichment methods. (B) Pie charts of glycans identified on peptide backbones between enrichment methods. Glycans are binned into six groups based on their composition16. Please click here to view a larger version of this figure.
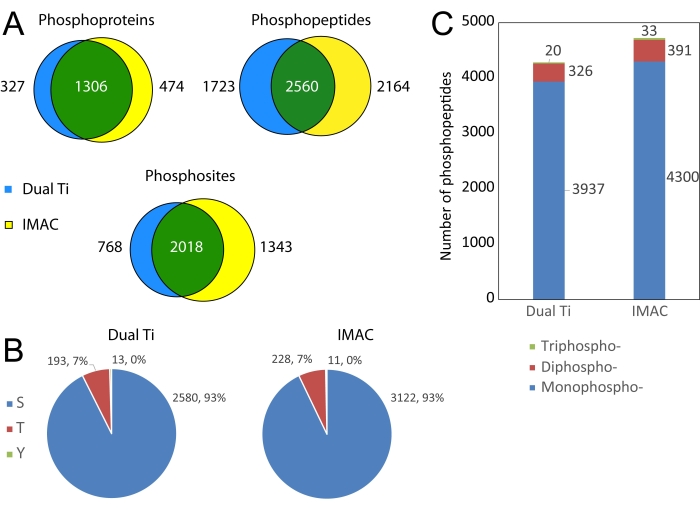
Figure 5: Comparison of phosphoproteomics results from dual-functional Ti(IV)-IMAC enrichment compared to a conventional Ti(IV)-IMAC phosphopeptide enrichment. (A) Venn diagrams of phosphoprotein, phosphopeptide, and phosphosite identifications between enrichment methods. (B) Pie charts of confidently localized (75% or higher probability) phosphosites broken down by amino acid residue. (C) Stacked bar plot of identified phosphopeptides binned by a number of phosphorylated residues. Please click here to view a larger version of this figure.
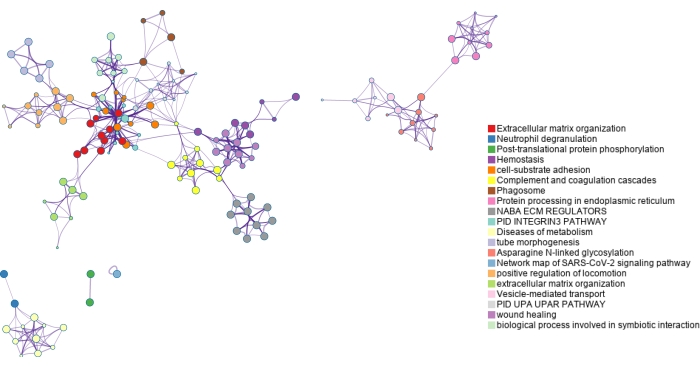
Figure 6: Gene ontology enrichment of N-glycoproteins identified using dual-functional Ti(IV)-IMAC enrichment. N-glycoproteins are mapped back to genes and significantly enriched pathways and process terms are identified using the entirety of the genome as the background. Various colors are used to label clusters of terms which are connected by term similarity. Please click here to view a larger version of this figure.
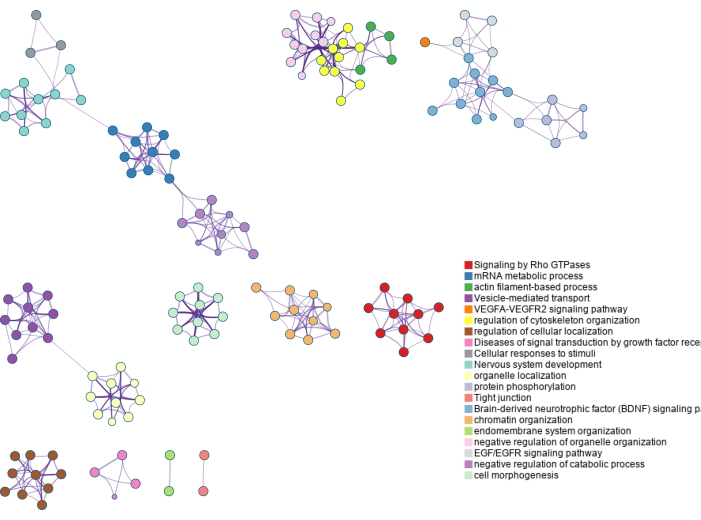
Figure 7: Gene ontology enrichment of phosphoproteins identified using dual-functional Ti(IV)-IMAC enrichment. Phosphoproteins are mapped back to genes and significantly enriched pathways and process terms are identified using the entirety of the genome as the background. Various colors are used to label clusters of terms, which are connected by term similarity. Please click here to view a larger version of this figure.
Supplementary Figure S1: Example total ion chromatograms (TICs) from each elution from dual-functional Ti(IV)-IMAC (E1 through E7, panels A-G) enrichment. Total signal (ion current) is plotted over the 117 min data collection period. Intensity of the base peak is given by the value NL. Please click here to download this File.
Supplementary Figure S2: Example total ion chromatograms (TICs) from each elution from ERLIC (E1 through E5, panels A-E) enrichment. Total signal (ion current) is plotted over the 117 min data collection period. Intensity of the base peak is given by the value NL. Please click here to download this File.
Supplementary Figure S3: Example total ion chromatogram (TIC) from the elution from Ti-IMAC enrichment. Total signal (ion current) is plotted over the 117 min data collection period. Intensity of the base peak is given by the value NL. Please click here to download this File.
Supplementary Figure S4: Gene ontology enrichment of N-glycoproteins identified using conventional glycopeptide enrichment with ERLIC. N-glycoproteins are mapped back to genes and significantly enriched pathways and process terms are identified using the entirety of the genome as the background. Various colors are used to label clusters of terms, which are connected by term similarity. Please click here to download this File.
Supplementary Figure S5: Gene ontology enrichment of phosphoproteins identified using conventional phosphopeptide enrichment with Ti(IV)-IMAC. Phosphoproteins are mapped back to genes and significantly enriched pathways and process terms are identified using the entirety of the genome as the background. Various colors are used to label clusters of terms, which are connected by term similarity. Please click here to download this File.
Supplementary Tables S1–S4: Metascape node information for gene ontology enrichment analyses. Tables contain node information for lists of N-glycoproteins and phosphoproteins identified using dual Ti (Table S1 and Table S2), N-glycoproteins using ERLIC (Table S3), and phosphoproteins using IMAC (Table S4). Please click here to download this Table.
Discussion
The dual-functional Ti(IV)-IMAC strategy is useful for the simultaneous analysis of N-glycopeptides and phosphopeptides from the same sample in a single sample preparation workflow. ERLIC-based methods have also been shown to perform simultaneous enrichment of PTMs. Both strategies have been used previously for deep coverage in PTM analyses14,18. In adapting the dual Ti method to decreasing sample incubation time by using spin-tips, we hope that this protocol has become less resource-intensive and, thus, more widely accessible.
The simultaneous analysis of multiple PTMs is achieved through synergistic interactions between analytes and the functional groups on the surface of the enrichment material. In first packing spin-tips with cotton, retention of glycopeptides in a HILIC mode is increased due to the prevalence of hydroxyl groups on cellulose24. By using water and acetonitrile (a water-miscible solvent), glycopeptides are retained and separated due to the formation of water and organic layers. The material for dual-functional Ti(IV) enrichment is dependent on the immobilized Ti(IV) cations for the binding of phosphopeptides. The hydrophilic properties of the material's side chains are used to retain glycopeptides in a HILIC mode, which are eluted first using acidic elution with increasing aqueous content. The material is further washed with conventional IMAC phosphopeptide enrichment buffers to elute sialylated glycopeptides, which have higher affinity to the stationary phase than other neutral glycopeptides. After conditioning the material to basic pH, ammonium hydroxide and ACN solution is used to break the electrostatic interactions that keep phosphopeptides bound to the material. In the meantime, the decreasing organic content allows separation of mono- and multi-phosphorylated peptides and M6P glycopeptides.
When separating PTMs using multiple elution steps in the same workflow, biomolecular information can be maximized from limited samples. A caveat with such dual PTM analysis workflows is the requirement of a relatively large amount of sample (0.5 mg of peptides) as starting material for each enrichment when at least five fractions are collected. Nonetheless, the simultaneous enrichment strategy presented here still offers significant saving of materials compared to conventional strategies. Even before enrichment, several important steps are necessary to ensure PTM integrity and to avoid artifactual loss of PTM information. These include the proper storage of samples at low temperatures when not in use (ideally -80 °C), the use of protease and phosphatase inhibitors during protein extraction, and the use of a hydrophilic peptide cleanup method, such as the HLB cartridges used here.
It has been observed that ERLIC (conventional glycopeptide enrichment) resulted in better glycoproteome coverage, while dual-functional Ti(IV)-IMAC was outperformed in unique identifications by conventional Ti-IMAC in phosphoproteomics results. The physical structure and increased hydrophilicity of the ERLIC material compared to that of the dual Ti enrichment material is likely a major factor in the improved glycoproteome coverage in ERLIC. The increased coverage of the phosphoproteome in conventional IMAC may be a result of decreased ion suppression from the removal of glycopeptides during the first elution steps and washing steps. Despite these slight decreases in coverage using the simultaneous dual Ti workflow, significant overlap with conventional single PTM enrichment is still achieved with the added benefit of only performing one enrichment workflow instead of two.
Concerning time constraints, ERLIC and conventional Ti-IMAC have fewer fractions than the dual Ti workflow. Concerning resources needed, the anion-exchange material needed for ERLIC is cheaper than the functionalized material needed for the dual Ti and conventional IMAC workflows. In these protocols, only N-linked and not O-linked glycosylation was analyzed. The enzyme PNGase F can (and should) be used to cleave N-glycans to limit the number of false positives obtained if searching for O-glycopeptides. It has been shown, however, that this strategy reduces the specificity of HILIC and ERLIC for glycopeptide enrichment25. In the protocol, a strategy using two separate software is used to analyze raw data files. Byonic, a commercial software, is considered the gold standard in analyzing glycoproteomics data. Due to the diversity of N-glycan structures, peptide search space is increased, thus increasing search times. Phosphorylation can also be added as a common PTM in addition to glycosylation during searching, though search times may skyrocket depending on whether peptide multi-phosphorylation is considered. MaxQuant, an open-source software, can be optimized to minimize FDR in phosphoproteomics results, though complex glycosylation is not as facile to search. There are also other open-source options for searching glycoproteomic data26,27,28. Depending on the experimental goals and resources available, the LC-MS and data analysis pipeline described here can be used as presented or be modified. Further optimization of the methods is expected to improve the coverage of the PTMs.
PTMs play a major role in biochemical processes due to the changes to protein structure they impart. It is ideal that all proteomic analyses take all protein PTMs into account simultaneously. The importance of the sum of all protein glycosylation, for example, has been termed meta-heterogeneity29. However, the goal of total PTM analysis is not yet possible with current MS-based technologies. Bottom-up proteomics followed by PTM-specific enrichment, i.e., HILIC and IMAC, is still the gold standard for PTM analyses, though by using simultaneous enrichment strategies such as the ones described here, biomolecular information can be better elucidated. Cross-talk interactions have been found to occur between various PTMs30,31, including glycosylation and phosphorylation32. Quantitative analysis of such interactions in disease, for example, in pancreatic diseases such as diabetes and cancer, may prove fruitful in increasing our understanding of disease mechanisms. With optimization of extraction methods among different sample types, the enrichment protocols described here can be used for in-depth profiling of the glycoproteome and the phosphoproteome in complex biological matrices.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported in part by grant funding from the NIH (R01DK071801, RF1AG052324, P01CA250972, and R21AG065728), and Juvenile Diabetes Research Foundation (1-PNF-2016-250-S-B and SRA-2016-168-S-B). Data presented here were also in part obtained through support from an NIH/NCATS UL1TR002373 award through the University of Wisconsin Institute for Clinical and Translational Research. The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. We would also like to acknowledge the generous support of the University of Wisconsin Organ and Tissue Donation Organization who provided human pancreas for research and the help of Dan Tremmel, Dr. Sara D. Sackett, and Prof. Jon Odorico for providing the samples to our lab. Our research team would like to give special thanks to the families who donated tissues for this study. L.L. acknowledges NIH grant S10OD025084, a Pancreas Cancer Pilot grant from the University of Wisconsin Carbone Cancer Center (233-AAI9632), as well as a Vilas Distinguished Achievement Professorship and the Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Materials
| Acetic Acid, Glacial (Certified ACS) | Fisher Scientific | A38S-500 | |
| Acetone (Certified ACS) | Fisher Scientific | A18-1 | |
| Acetonitrile, Optima LC/MS Grade | Fisher Scientific | A955-4 | |
| Ammonium Acetate (Crystalline/Certified ACS) | Fisher Scientific | A637-500 | |
| Ammonium Hydroxide (Certified ACS Plus) | Fisher Scientific | A669-212 | |
| Byonic software | Protein Metrics | n/a | Commercial software used for glycoproteomic analysis (https://proteinmetrics.com/byos/) |
| C18 BEH material | Waters | 186002353 | Material removed from column and used to pack nano capillaries (pulledto integrate tip used directly in line with instrument inlet) |
| CAE-Ti-IMAC, 100% | J&K Scientific | 2749380-1G | Material used for dual-functional Ti(IV)-IMAC; can also be used for conventional IMAC/conventional phosphopeptide enrichment |
| Cellcrusher kit | Cellcrusher | n/a | Used for grinding tissue samples into powder before extraction |
| Eppendorf 5424R Microcentrifuge | Fisher Scientific | 05-401-205 | For temperature-controlled centrifugation |
| cOmplete protease inhibitor cocktail tablets | Sigma | 11697498001 | |
| DTT, Molecular Grade (DL-Dithiothreitol) | Promega | V3151 | Protein reducing agent |
| Ethanol, 200 proof (100%), USP | Fisher | 22-032-601 | |
| Fisherbrand Analog Vortex Mixer | Fisher Scientific | 02-215-414 | |
| Fisherbrand Low-Retention Microcentrifuge Tubes (1.5 mL) | Fisher Scientific | 02-681-320 | |
| Fisherbrand Low-Retention Microcentrifuge Tubes (2 mL) | Fisher Scientific | 02-681-321 | |
| Fisherbrand Model 120 Sonic Dismembrator | Fisher Scientific | FB120110 | For sample lysis using ultrasonication |
| Formic Acid, 99.0+%, Optima LC/MS Grade | Fisher Scientific | A117-50 | |
| Fused silica capillary (75 μm inner diameter, 360 μm outer diameter) | Polymicro Technologies LLC | 100 m TSP075375 | For in-house pulled and packed columns with integrated emitter |
| Hydrofluoric acid (48 wt. % in H2O) | Sigma-Aldrich | 339261-100ML | Used for opening emitter of pulled capillary column |
| Iodoacetamide, BioUltra | Sigma | I1149-5G | Protein reducing reagent |
| MaxQuant software | n/a | n/a | Free software used for phosphoproteomic analysis (https://www.maxquant.org/) |
| Multi-therm Shaker with heating and cooling | Benchmark Scientific | H5000-HC | Heating block |
| Oasis HLB 1 cc Vac Cartridge, 10 mg Sorbent per Cartridge, 30 µm, 100/pk | Waters | 186000383 | Larger-scale cartridge desalting for tryptic digests (loading capacity approximately up to 1 mg each) |
| OMIX C18 pipette tips, 100 µL tip, 10 – 100 μL elution volume, 1 x 96 tips | Agilent | A57003100 | Smaller-scale packed pipette tip for desalting for enrichment elutions |
| P-2000 Micropipette Puller | Sutter Instrument Co. | P-2000/F | For pulling nano-capillary columns for LC-MS |
| PhosSTOP phosphatase inhibitor tablets | Sigma | 4906845001 | |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 | |
| Pierce Quantitative Colorimetric Peptide Assay | Thermo Fisher Scientific | 23275 | |
| PolySAX LP (12 μm, pore size 300 Å) | PolyLC | BMSX1203 | Material for strong anion-exchange chromatography used for ERLIC/conventional glycopeptide enrichment |
| Potassium Phosphate Monobasic (Crystalline/Certified ACS) | Fisher Scientific | P285-500 | |
| Pressure injection cell with integrated magnetic stirplate | Next Advance | PC77-MAG | For packing nano-capillary columns with stationary phase up to 2500 psi limit |
| Proteome Discoverer software | Thermo Fisher Scientific | n/a | Commercial software for proteomics anaysis (with integrated database searching software nodes) and data visualization (https://www.thermofisher.com/us/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html) |
| SpeedVac SC110 Vacuum Concentrator Model SC110-120 | Savant | n/a | Centrifugal vacuum concentrator for drying samples (under heat) |
| SDS Solution, 10% Sodium Dodecyl Sulfate Solution, Molecular Biology/Electrophoresis | Fisher Scientific | BP2436200 | |
| Sequencing Grade Modified Trypsin | Promega | V5111 | |
| Sodium Chloride (Crystalline/Certified ACS) | Fisher Scientific | S271-500 | |
| TopTip, Empty, 10-200 µL, Pack of 96 | Glygen Corporation | TT2EMT.96 | Empty pipette tip with micron-sized hole used that can be used to pack chromatographic materials for enrichments, bundled with tube adapters |
| Triethylammonium bicarbonate buffer (TEAB, 1 M, pH 8.5 (volatile)) | Sigma | 90360-100ML | |
| Trifluoroacetic acid, Reagent Grade, 99% | Fisher Scientific | 60-017-61 | |
| Tris Base (White Crystals or Crystalline Powder/Molecular Biology) | Fisher Scientific | BP152-500 | |
| Trypsin/Lys-C Mix, Mass Spec Grade | Promega | V5071 | |
| Urea (Certified ACS) | Fisher Scientific | U15-500 | |
| Water, Optima LC/MS Grade | Fisher Scientific | W64 |
References
- Smith, L. M., Kelleher, N. L. Proteoform: a single term describing protein complexity. Nature Methods. 10 (3), 186-187 (2013).
- Pan, S., Brentnall, T. A., Chen, R. Glycoproteins and glycoproteomics in pancreatic cancer. World Journal of Gastroenterology. 22 (42), 9288-9299 (2016).
- Tabang, D. N., Ford, M., Li, L. Recent advances in mass spectrometry-based glycomic and glycoproteomic studies of pancreatic diseases. Frontiers in Chemistry. 9, 707387 (2021).
- Khoury, G. A., Baliban, R. C., Floudas, C. A. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Scientific Reports. 1, 90 (2011).
- Alpert, A. J. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. Journal of Chromatography A. 499, 177-196 (1990).
- Riley, N. M., Bertozzi, C. R., Pitteri, S. J. A pragmatic guide to enrichment strategies for mass spectrometry-based glycoproteomics. Molecular & Cellular Proteomics. 20, 100029 (2020).
- Low, T. Y., et al. Widening the bottleneck of phosphoproteomics: Evolving strategies for phosphopeptide enrichment. Mass Spectrometry Reviews. 40 (4), 309-333 (2021).
- Cho, K. C., Chen, L., Hu, Y., Schnaubelt, M., Zhang, H. Developing workflow for simultaneous analyses of phosphopeptides and glycopeptides. ACS Chemical Biology. 14 (1), 58-66 (2019).
- Zhou, Y., et al. An integrated workflow for global, glyco-, and phospho-proteomic analysis of tumor tissues. Analytical Chemistry. 92 (2), 1842-1849 (2020).
- Tang, R., et al. Facile preparation of bifunctional adsorbents for efficiently enriching N-glycopeptides and phosphopeptides. Analytica Chimica Acta. 1144, 111-120 (2021).
- Wang, Z., Wang, J., Sun, N., Deng, C. A promising nanoprobe based on hydrophilic interaction liquid chromatography and immobilized metal affinity chromatography for capture of glycopeptides and phosphopeptides. Analytica Chimica Acta. 1067, 1-10 (2019).
- Glover, M. S., et al. Characterization of intact sialylated glycopeptides and phosphorylated glycopeptides from IMAC enriched samples by EThcD fragmentation: Toward combining phosphoproteomics and glycoproteomics. International Journal of Mass Spectrometry. 427, 35-42 (2018).
- Alpert, A. J. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Analytical Chemistry. 80 (1), 62-76 (2008).
- Cui, Y., et al. Counterion optimization dramatically improves selectivity for phosphopeptides and glycopeptides in electrostatic repulsion-hydrophilic interaction chromatography. Analytical Chemistry. 93 (22), 7908-7916 (2021).
- Cui, Y., et al. Finding the sweet spot in ERLIC mobile phase for simultaneous enrichment of N-Glyco and phosphopeptides. Journal of the American Society for Mass Spectrometry. 30 (12), 2491-2501 (2019).
- Tabang, D. N., et al. Analysis of pancreatic extracellular matrix protein post-translational modifications via electrostatic repulsion-hydrophilic interaction chromatography coupled with mass spectrometry. Molecular Omics. 17 (5), 652-664 (2021).
- Huang, J., et al. Dual-functional Titanium(IV) immobilized metal affinity chromatography approach for enabling large-scale profiling of protein Mannose-6-Phosphate glycosylation and revealing its predominant substrates. Analytical Chemistry. 91 (18), 11589-11597 (2019).
- Huang, J., et al. Dual-functional Ti(IV)-IMAC material enables simultaneous enrichment and separation of diverse glycopeptides and phosphopeptides. Analytical Chemistry. 93 (24), 8568-8576 (2021).
- Jami-Alahmadi, Y., Pandey, V., Mayank, A. K., Wohlschlegel, J. A. A robust method for packing high resolution C18 RP-nano-HPLC columns. Journal of Visualized Experiments: JoVE. (171), e62380 (2021).
- Bern, M., Kil, Y. J., Becker, C. Byonic: advanced peptide and protein identification software. Current Protocols in Bioinformatics. , (2012).
- Tyanova, S., Temu, T., Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols. 11 (12), 2301-2319 (2016).
- Perez-Riverol, Y., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research. 47, 442-450 (2019).
- Zhou, Y., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications. 10 (1), 1523 (2019).
- Zacharias, L. G., et al. HILIC and ERLIC enrichment of glycopeptides derived from breast and brain cancer cells. Journal of Proteome Research. 15 (10), 3624-3634 (2016).
- Yang, W., et al. Comparison of enrichment methods for intact N- and O-linked glycopeptides using strong anion exchange and hydrophilic interaction liquid chromatography. Analytical Chemistry. 89 (21), 11193-11197 (2017).
- Toghi Eshghi, S., Shah, P., Yang, W., Li, X., Zhang, H. GPQuest: A spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides. Analytical Chemistry. 87 (10), 5181-5188 (2015).
- Liu, M. -. Q., et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nature Communications. 8 (1), 438 (2017).
- Lu, L., Riley, N. M., Shortreed, M. R., Bertozzi, C. R., Smith, L. M. O-Pair Search with MetaMorpheus for O-glycopeptide characterization. Nature Methods. 17 (11), 1133-1138 (2020).
- Caval, T., Heck, A. J. R., Reiding, K. R. Meta-heterogeneity: Evaluating and describing the diversity in glycosylation between sites on the same glycoprotein. Molecular & Cellular Proteomics. 20, 100010 (2021).
- Lee, J. S., Smith, E., Shilatifard, A. The language of histone crosstalk. Cell. 142 (5), 682-685 (2010).
- Leutert, M., Entwisle, S. W., Villén, J. Decoding post-translational modification crosstalk with proteomics. Molecular & Cellular Proteomics. 20, 100129 (2021).
- Hart, G. W., Slawson, C., Ramirez-Correa, G., Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annual Review of Biochemistry. 80 (1), 825-858 (2011).

