Biochemical Purification and Proteomic Characterization of Amyloid Fibril Cores from the Brain
Summary
This biochemical purification method with mass spectrometry-based proteomic analysis facilitates the robust characterization of amyloid fibril cores, which may accelerate the identification of targets for preventing Alzheimer’s disease.
Abstract
Proteinaceous fibrillar inclusions are key pathological hallmarks of multiple neurodegenerative diseases. In the early stages of Alzheimer's disease (AD), amyloid-beta peptides form protofibrils in the extracellular space, which act as seeds that gradually grow and mature into large amyloid plaques. Despite this basic understanding, current knowledge of the amyloid fibril structure, composition, and deposition patterns in the brain is limited. One major barrier has been the inability to isolate highly purified amyloid fibrils from brain extracts. Affinity purification and laser capture microdissection-based approaches have been previously used to isolate amyloids but are limited by the small quantity of material that can be recovered. This novel, robust protocol describes the biochemical purification of amyloid plaque cores using sodium dodecyl sulfate (SDS) solubilization with sucrose density gradient ultracentrifugation and ultrasonication and yields highly pure fibrils from AD patients and AD model brain tissues. Mass spectrometry (MS)-based bottom-up proteomic analysis of the purified material represents a robust strategy to identify nearly all the primary protein components of amyloid fibrils. Previous proteomic studies of proteins in the amyloid coronae have revealed an unexpectedly large and functionally diverse collection of proteins. Notably, after refining the purification strategy, the number of co-purifying proteins was reduced by more than 10-fold, indicating the high purity of the isolated SDS insoluble material. Negative staining and immuno-gold electron microscopy allowed confirmation of the purity of these preparations. Further studies are required to understand the spatial and biological attributes that contribute to the deposition of these proteins into amyloid inclusions. Taken together, this analytical strategy is well-positioned to increase the understanding of amyloid biology.
Introduction
Amyloid is an extremely stable supramolecular arrangement that is found in a diverse panel of proteins, some of which lead to pathological changes1. The accumulation of intra- or extracellular amyloid aggregates is observed in several neurodegenerative diseases2. Amyloid aggregates are heterogeneous and are enriched with a large number of proteins and lipids3. In recent years, interest in the amyloid proteome has generated substantial interest among basic and translational neuroscientists. Several methods have been developed to extract and purify amyloid aggregates from mouse and post-mortem human brain tissues. Laser-capture microdissection, immunoprecipitation, decellularization, and biochemical isolation of amyloid aggregates are widely used methods to extract and purify amyloid plaques, fibrils, and oligomers4,5,6,7. Many of these studies have focused on determining the protein composition of these tightly packed fibrillar deposits using semi-quantitative MS. However, the available results are inconsistent, and the surprisingly large number of co-purifying proteins previously reported are challenging to interpret.
The primary limitation of the existing literature describing the amyloid core proteome in AD and AD mouse model brains is that the purified material contains an unmanageable number of co-purifying proteins. The overall goal of this method is to overcome this limitation and develop a robust biochemical purification for isolating amyloid fibril cores. This strategy employs a previously described sucrose density gradient ultracentrifugation-based biochemical method for the isolation of SDS insoluble enriched amyloid fractions from post-mortem AD human and mouse brain tissues8,9. This method builds on the existing literature but goes further with ultrasonication and SDS washes to remove most of the loosely bound amyloid-associated proteins, leading to the isolation of highly purified amyloid fibrils (Figure 1). The fibrils purified by this protocol overcome several existing challenges frequently encountered in structural studies of amyloid fibrils isolated from brain extracts. Visualization of these fibrils with transmission electron microscopy (TEM) confirms the integrity and purity of the purified material (Figure 2). In this study, the isolated fibrils are solubilized and digested to peptides with trypsin, and label-free MS analysis can readily reveal the identity of the proteins forming the fibril core. Notably, some of these proteins have an inherent tendency to form supramolecular assemblies in non-membrane-bound organelles. In addition, many of the proteins identified in the analysis of amyloid-beta (Aβ) fibrils are also associated with other neurodegenerative diseases, suggesting that these proteins may play a key role in multiple proteinopathies.
This SDS/ultrasonication method is unlikely to alter or disrupt the structure of the fibril cores. The purified material is also suitable for a wide range of top-down and bottom-up proteomic analysis approaches and additional MS-based structural analysis strategies, such as chemical crosslinking or hydrogen-deuterium exchange. The overall recovery using this method is relatively high and, thus, is suitable for detailed structural studies, which require micrograms to milligrams of the purified material. The purified material is also suitable for structural studies using cryoEM and atomic force microscopy. This protocol, in combination with the stable isotopic labeling of mammals, can facilitate solid-state nuclear magnetic resonance (NMR) studies of amyloid structure10.
Protocol
This protocol involves the use of human or vertebrate brain tissues. All the research was performed in compliance with the approved institutional guidelines of the Northwestern University. The current workflow is standardized using APP-knock in (AppNL-G-F/NL-G-F) mouse brain cortical and hippocampal brain region extracts11. This protocol has been optimized for brain extracts from mice at 6-9 months of age, and it can effectively purify amyloids from both male and female animals.
NOTE: For a better understanding of the overall experimental procedure, see Figure 1 for a schematic of the workflow.
1. Tissue harvesting and amyloid purification
NOTE: Ideally, amyloid fibrils should be isolated from freshly dissected brain regions. However, this method also works well with snap- or flash-frozen brain tissues. Below is a brief outline of snap-freezing brain tissues for storage for use at a later time.
- Brain tissue harvesting and storage: Dissect mouse brains and quickly harvest the amyloid-laden regions (i.e., cortex and hippocampus), followed by snap freezing in liquid nitrogen or a dry ice alcohol bath.
NOTE: Snap freezing helps to preserve the structural attributes of the tissue's constituents. The mice are euthanized by isoflurane and cervical dislocation12,13. It is advised to avoid using CO2 since that can compromise brain proteome integrity. - Post-dissection, cut and store fresh tissues in small blocks (e.g., 5 mm x 5 mm), as this facilitates more efficient homogenization before amyloid extraction. Transfer the tissue to the sterile cryogenic vial, tighten its cap, and rapidly submerge.
- Keep tissues frozen in ultracold conditions (i.e., -80 °C or liquid nitrogen). If extraction is to be performed a few days later, store the tissues at -20 °C and avoid freeze and thaw cycles. Thaw the frozen tissues on wet ice when ready for extraction and purification.
NOTE: Direct contact of the tissues with liquid nitrogen can cause tissue and protein damage. Note that an alcohol bath can remove permanent markers from the tube labels.
2. Enrichment of SDS insoluble material
NOTE: Perform all the steps on ice and centrifuge at 4 °C, unless stated otherwise. Details of all the buffers and solutions used in this protocol are provided in Supplementary File 1. Manufacturers and catalog numbers of chemicals and instruments are provided in the Table of Materials.
- Start with freshly dissected or snap-frozen brain tissue regions (thawed on ice) placed into a 2 mL tube containing 6-8 ceramic beads and freshly prepared ice-cold homogenization buffer (1 mL for 0.25-1 g of wet tissue mass).
- Transfer the tubes to the bead mill homogenizer and grind the tissue contents at 4000 rpm, with two cycles of 30 s each and an interval of 30 s in between.
NOTE: Alternatively, motorized stirrer or homogenizer systems can be used to grind and homogenize the tissues. - Add 9 mL of ice-cold homogenization buffer to the 1 mL of brain whole tissue homogenate in 15 mL tubes, seal with laboratory wax film strips, and rotate end-to-end overnight at 4 °C to ensure robust solubilization.
NOTE: To remove unwanted large cellular debris and lipids, on the next morning, spin the tubes at 800 x g at 4 °C for 10 min and collect the supernatant in a fresh tube. Resuspend the remaining pellet in 2 mL of ice-cold homogenization buffer, mix well, spin again for 10 min at 4 °C, and combine the two supernatants. - Slowly add solid sucrose to the tissue extract suspension to a final concentration of 1.2 M, mix well, and centrifuge at 250,000 x g for 45 min at 4 °C.
- Carefully remove and discard the supernatant using a pipette. Resuspend the pellet in 2 mL of homogenization buffer containing 1.9 M sucrose by triturating, followed by centrifugation at 125,000 x g for 45 min at 4 °C.
NOTE: The buffer volume can be adjusted based on the amount of material recovered from the previous step. In general, 10 volumes of homogenization buffer (V/V) is ideal for this step. - Collect the top white solid layer using a pipette, transfer it to a fresh tube, and solubilize in 1 mL of ice-cold wash buffer by pipetting up and down several times, without introducing air bubbles.
- Apart from the top layer, the pellet is also enriched with amyloid fibrils. For a higher yield, combine the two fractions and proceed. Carefully remove the middle aqueous layer using a pipette and discard.
- Centrifuge the combined fractions at 8000 x g for 20 min at 4 °C. Discard the supernatant.
- Add 1 mL of ice-cold digestion buffer to the washed pellet, resuspend, and incubate at room temperature (RT) for 3 h.
- Centrifuge at 8000 x g for 20 min at 4 °C and remove the supernatant using a pipette.
- Resuspend and wash (same as Step 2.8.) the digested pellet two times in 1 mL of ice-cold Tris buffer.
- Resuspend the washed pellet in 1 mL of solubilization buffer containing 1% SDS and 1.3 M sucrose by pipetting up and down. Centrifuge quickly at 200,000 x g for 60 min at 4 °C.
NOTE: Although the amyloid fibrils are highly resistant to detergents and denaturants, very long exposures to the detergent (1% SDS) may affect the integrity of the fibrils or remove the tightly bound proteins, which will compromise subsequent analyses. Therefore, immediately after resuspending the pellets, proceed to centrifugation. - Save the pellet and increase the volume of the remaining supernatant by adding 50 mM Tris buffer (in ratio 1:0.3) to reduce the sucrose concentration from 1.3 to 1 M and centrifuge one more time at 200,000 x g for 45 min at 4 °C.
- Dissolve the two pellets in 100 µL of Tris buffer containing 0.5% SDS and pool for amyloid purification. Visible pellets are small and should appear opaque and off-white (see Figure 2A).
3. Amyloid purification
NOTE: Combine the two pellets, solubilize by pipetting until obtaining a uniform solution and proceed with the following steps of amyloid purification.
- For complete solubilization of the amyloid-rich material present in pellets, subject the tube to mechanical shearing produced by ultrasound waves in a bath sonication device running for 30 s ON and 30 s OFF at a medium-range frequency for 20 cycles.
- Immediately centrifuge the material at 20,000 x g for 30 min at 4 °C and resuspend the pellet in 500 µL of 0.5% SDS Tris buffer.
- Repeat Step 3.1. and Step 3.2. four times for a total of five washes. The number of washes can be increased to improve the purity.
NOTE: Sonication and washing steps are optimized at 0.5% SDS as higher concentrations may affect the structure, integrity, or protein composition of the fibrils. Increasing the SDS concentration at this step is not recommended. - After the final centrifugation step, wash the pellet in 200 µL of ultra-pure water and centrifuge at 20,000 x g for 30 min at 4 °C to remove any remaining detergent. The washed pellet containing purified amyloid fibrils appears semi-transparent in color and is difficult to see sometimes.
- Dissolve the final pellet containing purified amyloid fibrils in 100 µL of ultrapure water. Proceed to the next step immediately for downstream processing or save the fibril suspension at -20 °C for further analysis. If frozen, thaw on ice before starting precipitation.
4. Methanol chloroform precipitation
NOTE: If the final goal is to perform protein analysis, it is recommended to desalt and remove additional non-proteinaceous impurities.
- Add 400 µL of methanol to the purified 100 µL of amyloid fibrils in a 1.5 mL tube and vortex well.
- Add 100 µL of chloroform to the tube and vortex well.
- Add 300 µL of ultrapure water and vortex thoroughly. This turns the mixture cloudy due to protein precipitation.
- Centrifuge at 12,000 x g for 2 min at RT.
- Carefully remove the aqueous layer (i.e., top) without disturbing the interface layer containing the protein flake.
- Add the same volume of methanol again and centrifuge at 12,000 x g for 2 min at RT.
- Discard the supernatant using a pipette and air-dry the pellet at RT. Avoid over-drying; amyloid fraction is difficult to redissolve if over-dried.
5. Trypsin digestion
- Dissolve the pellet in 50 µL of guanidine hydrochloride (GuHCl) buffer. Sonicate in an ice-cold water bath and heat at 95 °C for 5 min, if required.
- Vortex thoroughly at RT for 45 min to 1 h to dissolve it completely. The pellet will not be visible after this step, and the solution looks clear in appearance.
- Add the same volume of 0.2% surfactant solution. Solubilize at RT with vortexing for 60 min. This step enhances trypsin enzymatic activity.
- Add 1 µL of tris-2-carboxyethylphosphine (TCEP) from the 500 mM stock solution. Incubate for 60 min.
- Add 2 µL of 500 mM iodoacetamide (IAA) and incubate in the dark for 20 min.
- Quench the IAA with 5 µL of TCEP solution for 15 min.
- Add the required volume of 50 mM ammonium bicarbonate solution to the tube to reduce the guanidine concentration to 1.5 M.
- Add 1% surfactant solution to the tube (1 µL/50 µg of protein).
- Add trypsin (1 µg/100 µg of protein) and leave the tube mixing at 37 °C overnight.
6. Peptide cleanup
- Next morning, acidify the digested peptide solution by lowering the pH to 2.0 by adding formic acid.
- Activate the C18 (n-octadecyl) spin column in a 2 mL receiver tube by adding 200 µL of 50% methanol solution and spin at 1500 x g for 2 min at RT. Repeat the activation step.
- Equilibrate the C18 column resin beds by adding 200 µL of equilibration buffer and spinning for 2 min with the same conditions. Repeat this step.
- Load the acidified peptide solution on the C18 column and centrifuge at 1500 x g for 2 min at RT. Collect the flow-through and reload it one more time. Discard the second flow-through.
- Wash the peptides bound to the C18 resin with the wash buffer 2x, as done in Step 6.3. Use the same equilibration buffer for washing the column.
- Elute the peptides by adding 40 µL of elution buffer followed by centrifugation at 1500 x g, for 2 min at RT. Repeat the elution step three times to increase the yield of the recovered peptides.
- Dry the peptides in a speed vacuum concentrator by evaporating the aqueous solution. Dry pellets can be saved at -20 °C for a few weeks before the MS analysis.
7. Setting up mass spectrometer for peptide analysis
NOTE: For MS parameters, see Supplementary File 1 (adapted from a previous publication from the lab)14.
- Before loading the peptide samples for the MS analysis, dissolve the dry peptide pellets in 20 µL of sample loading buffer and perform micro BCA to quantify the concentration of recovered peptides.
- Transfer the dissolved peptides in a glass vial and load 3 µg (quantified from micro BCA) of peptides into the UPLC system autosampler.
- Load the samples onto a vented trap column (C18 HPLC column, 0.075 mm x 20 mm) with a flow rate of 250 nL/min.
- Arrange the trap column in line with an analytical column (0.075 µm x 500 mm) and assemble an emitter tip to the electrospray ionization (ESI) source subjected to a spray voltage of 2000 V.
NOTE: In this study, ESI is performed, in which the mobile phase is subjected to high-voltage ionization into the gas phase. The spray created by this is directed to the vacuum chamber of the MS through a heated capillary. While under vacuum, the de-solvation of droplets and ejection of ions occurs in the presence of heat and voltage. Thereafter, the ions are accelerated toward the mass analyzer in the presence of a high voltage environment. - Analyze the samples with 2 h acquisition runs. This study is performed using data-dependent acquisition with the most intense top 20 precursor ion selection paradigm.
NOTE: In data-dependent acquisition, a limited number of precursor peptides are detected in the MS1 scan and subjected to fragmentation for MS2 analysis. However, the dynamic exclusion is problematic here since highly abundant Aβ peptides will be omitted for fragmentation and result in an underestimate of their quantity. - For absolute quantification of Aβ peptides, run the same samples one more time using the targeted MS/MS method. For this study, an exhaustive list of all m/z ratios for various possible tryptic Aβ peptides for the APP knock-in mouse models is prepared (see Supplementary Table 1). Other groups should prepare a similar list in accordance with the available mouse model and the possible peptides generated from the protein of interest (e.g., Aβ for AD).
NOTE: To address the exclusion of highly abundant peptides, use a targeted approach by providing a list of selected m/z values corresponding to Aβ tryptic peptides. This approach addresses the problem of data-dependent exclusion of Aβ peptides and can quantify the absolute quantities of different monomers (Aβ38, 40, and 42 peptides) with high resolution. - The mass spectrometer generates MS spectra of the peptide samples and saves raw data files on the target directory. Use this file to perform spectral matching using well-established statistical and bioinformatics tools. Multiple online and offline database search and analysis tools are available.
8. MS data analysis
- Extract the MS1 and MS2 files using the offline Rawconverter tool (http://www.fields.scripps.edu/rawconv/).
- Perform identification, quantification, and detailed analyses of the data on a web-based MS data search engine. In this study, Integrated Proteomics Pipeline – IP2 (Bruker: http://www.integratedproteomics.com/) was used.
NOTE: Several other online and offline MS data analysis tools are available and can also be used, such as MaxQuant (https://www.maxquant.org/). - After uploading the MS1 and MS2 files, select an updated mouse proteome database. Here, an updated mouse database containing additional App knock-in specific mutations is selected for the identification of peptides using ProLuCId and SEQUEST algorithms11.
- In the IP2 analysis system, select the default parameters and modify them according to the experimental requirements. In this study, a peptide mass tolerance of 50 ppm for precursor and 600 ppm for fragments is used (refer to Supplementary File 1 for other parameters used in IP2).
NOTE: Researchers can modify the search parameters in accordance with the experimental objectives. For example, for identifying post-translational modifications, add differential modification values for various PTMs (e.g., ubiquitination, SUMOylation, phosphorylation).
Representative Results
Here, a detailed method for the isolation and purification of amyloid fibrils using a modified sucrose density gradient ultracentrifugation purification method is summarized (see Figure 1). The innovation in this method is the inclusion of steps of ultrasonication-based washing using a water bath sonication system followed by SDS solubilization, which removes many loosely associated proteins from the amyloid fibrils that co-purify with the highly dense and clean fibrils. The ultrasonication step generates a high shearing force and agitates the fibrils, loosening the hydrophobic forces and releasing the SDS soluble loosely associated proteins into the SDS wash buffer. In turn, small quantities of highly pure amyloid fibril cores are recovered. As shown in Figure 2A, a visible pellet, which initially appears opaque (possibly due to impurities), can be seen after enrichment; however, following the ultrasonication and multiple SDS washes, the pellet turns semi-transparent and is hardly visible. The representative Congo red staining of purified amyloids as compared to the SDS soluble fraction documents the enrichment of the amyloid fibrils (Figure 2B). Congo red staining can be used to confirm the amyloid material in different fractions and can be visualized using a bright field microscope. As shown in Figure 2C, the SDS soluble fraction does not stain with the Congo red dye.
The structure of the purified fibrils with negative staining transmission electron microscopy analysis confirmed the presence of nearly pure amyloid fibrils (Figure 2D). Additionally, immunogold labeling using a combination of Aβ42 (6E10 and 4G8) antibodies confirmed the presence of Aβ42 peptides (Figure 2E). To investigate the composition and structural features of the purified material, we used immunoblot techniques for Aβ peptides and hallmark structural signatures (e.g., fibrils). Representative dot blot analysis of the fractions collected during amyloid isolation showed a relative abundance of Aβ42 peptides and fibrils using anti-Aβ42 and anti-fibril (LOC) antibodies (Figure 3A). Similarly, the western blot of the representative fractions also showed enrichment of Aβ42-containing fibrils in high molecular weight proteins trapped in the wells of the SDS PAGE gel (Figure 3B). To understand the composition of these high molecular weight fibrils, purified amyloid fractions were subjected to MS-based proteomic analysis. These semi-quantitative results revealed the presence of approximately 250 proteins, while the fraction collected before ultrasonication and SDS washes contained more than 2500 proteins (Figure 3C). This indicates the effectiveness of these two crucial steps that are included in this purification protocol. Taken together, multiple independent results indicate the high abundance of similar protein classes in fibril cores.
Gene Ontology (GO) cellular component analysis for one representative MS dataset in Figure 4 revealed that a large number of proteins present in the fibril cores are associated with non-membrane-bound organelle and supramolecular complexes. This observation is likely due to the inherent tendency of many proteins to aggregate themselves or co-aggregate with other proteins in proteinaceous inclusions that are in close proximity. Physical forces play crucial roles in these interactions. The other cellular organelle and components primarily represented by these proteins are mitochondria, cytoskeleton, cell membrane, and myelin sheath. Many of these proteins interact with Aβ peptides, oligomers, or protofibrils at different stages of amyloid formation. They might interact close to the plasma membrane, where Aβ peptides are released. The interaction may also happen while some of these proteins are released directly or via vesicular transport into the extracellular space. Secretion or exocytosis of protein aggregates is one among many strategies that cells use to cope and reduce the burden associated with protein aggregates15. This leaves another opportunity where some of the intracellular proteins can bind to Aβ peptides. The protocol provides a framework for further optimization depending on the experimental goals. For example, the purity and yield, by altering the number of ultrasonication and SDS washes, can be fine-tuned accordingly.
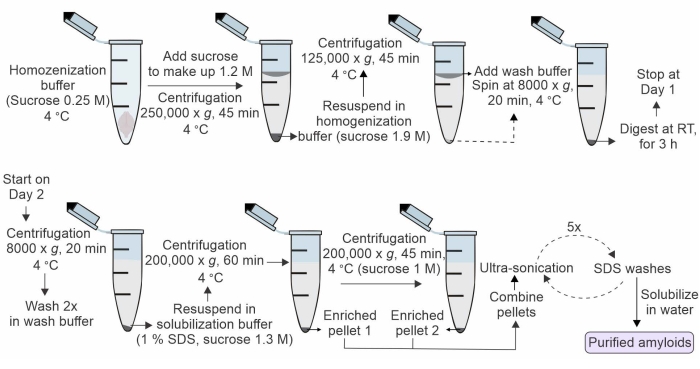
Figure 1: A diagrammatic overview of the workflow for isolation of amyloid fibrils core from AD post-mortem human or model animal brain tissues. Please click here to view a larger version of this figure.
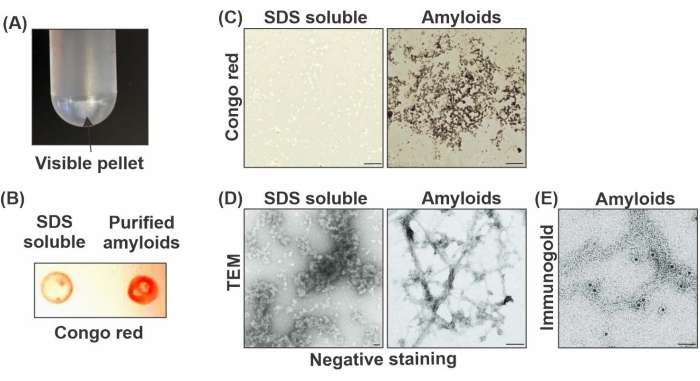
Figure 2: Confirmation of amyloid extraction using biochemical staining and imaging of amyloid fibrils. (A) Enriched amyloid-containing SDS insoluble pellet appears opaque off-white in color. (B) Congo red staining of SDS soluble supernatant and SDS insoluble pellet containing purified amyloid blotted on 0.45 µm nitrocellulose membrane; BCA readings were used for normalization of the loading amount of the proteins. BCA assay was performed as per the manufacturer's instructions. (C) Bright-field images of SDS soluble and amyloid material following Congo red staining (scale bar: 100 µm). (D) Visualization of SDS soluble fraction and purified amyloid fibrils using negative staining under the electron microscope (scale bar: 100 nm). (E) Confirmation of Aβ42 peptides abundance in purified amyloid fibrils by immunogold electron microscopy using Aβ42 (6E10 and 4G8) antibodies (scale bar: 50 nm). Please click here to view a larger version of this figure.
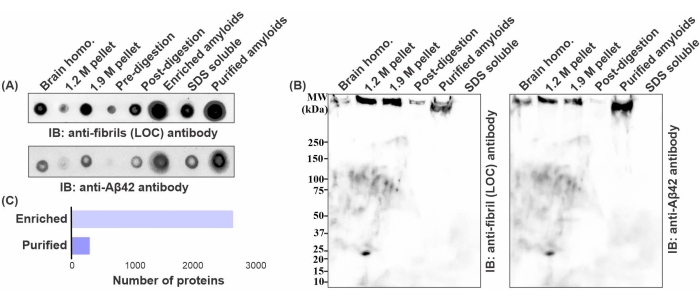
Figure 3: Validation of amyloid purification using immunoblot and MS analysis. (A) Dot blot and (B) Western blot analysis of several representative fractions collected during the amyloid purification process using anti-fibril LOC and anti-Aβ42 antibody; BCA assay readings were used for normalization of the loading amount of the proteins. (C) Number of proteins recovered in label-free mass spectrometry analysis of enriched and purified amyloid fractions; micro BCA assay was used for loading 3 µg of digested peptides for each MS analysis. BCA and micro BCA assays were performed as per the manufacturer's instructions. Please click here to view a larger version of this figure.
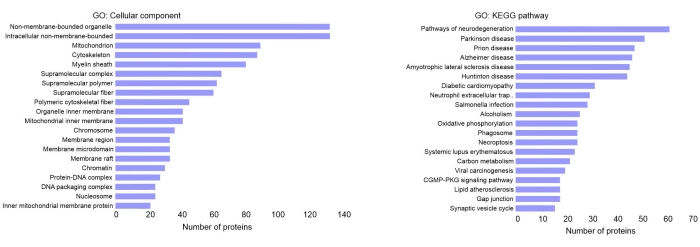
Figure 4: Gene Ontology analysis of proteins abundant in purified amyloid fractions. (A) Cellular components and (B) KEGG pathways. Please click here to view a larger version of this figure.
Supplementary File 1: Buffers and solutions, mass spectrometry parameters and ProLuCID search parameters for identification of peptides. Please click here to download this File.
Supplementary Table 1- A representative list of m/z ratios identified in MS run for amyloid beta peptides of APP knock in mouse models. Please click here to download this Table.
Discussion
Developing a clear understanding of amyloid structure and composition is challenging for structural biologists and biochemists due to the biological complexities and experimental limitations in extracting purified fibrils from AD brain tissues16,17. Amyloid fibrils are polymorphic at the molecular level, showing a heterogeneous population of varying lengths and complexities18,19. To better understand their biological features and pathological relevance, an exhaustive characterization of the composition of polymorphic amyloid fibrils obtained from post-mortem human and AD mouse brain tissues is required20,21. A large subset of proteins directly interacts with Aβ42, while others may have a tendency to form large fibrillar structures or protein complexes22,23,24. It is more challenging to determine the roles played by the proteins interacting with Aβ42 in amyloid formation, stabilization, and elongation as it relates to AD pathology. In recent years, several proteomic studies have elucidated the differences and similarities in the protein composition of various supramolecular arrangements, membrane-less organelles, inclusion bodies, and protein aggregates25,26,27. In the early stages of AD, multiple cellular proteins (e.g., Aβ42 and microtubule-associated tau protein and apolipoprotein E) misfold and co-aggregate in multiple brain regions28,29. Amyloidogenic proteinaceous formations are one major pathological hallmark of AD and likely contribute to pathogenesis3.
The purification of amyloid fibrils from diseased human brains is a tedious and challenging task and has multiple limitations. One major drawback of the existing methods is the low purity of extracted material, which often restricts their detailed structural analysis using imaging methods, such as cryoEM. Likewise, NMR studies require a few milligrams of purified material, which also needs to be labeled with heavy isotopes (i.e., 15N)30. Post-mortem human brains can meet the first requirement, as the starting material can be increased up to a few grams of human tissues; however, labeling human brain tissues with heavy isotopes is not possible. On the other hand, labeling the AD mouse models with 15N isotopes is possible (although costly) and is now increasingly common31,32. Our lab has used pulse-chase labeling to study the synaptic protein turnover dynamics during disease and aging brains14. We have also used whole-animal heavy isotope labeling to identify long-lived proteins and understand their physiological relevance in elaborate biological structures33,34. However, for the mouse brain, large quantities of starting material are required to obtain a workable quantity of the fibril cores. This method successfully addresses these issues by improving the yield and purity of the extracted amyloid fibrils by modifying the existing biochemical isolation principles. Therefore, this robust protocol for amyloid fibrils extraction from brain tissues can be readily utilized for cryoEM and NMR-based structural studies.
This method utilizes the existing sucrose density gradient-based subcellular fractionation paradigm and removes nonspecific co-purifying proteins in successive steps. After the removal of cell debris, myelin, DNA, and cellular lipids, the amyloid-rich SDS insoluble pellets are isolated. The incorporation of additional steps of multiple ultrasonication coupled SDS washes helps to reduce the numbers of co-purifying cellular components, loosely bound proteins, and smaller SDS soluble polymorphs of amyloids. The final pellet is solubilized in ultrapure water and can be used for many applications, including seeding experiments, biochemical or pharmacological studies, and structural analysis. The purified amyloid fibrils from AD brain tissues are also used to understand the proteomic composition and structural features of fibril cores using MS-based proteomics. This analysis confirmed the presence of a subset of cellular proteins (represented in Figure 4) in the core of the fibrils, which may indicate the possible roles of more than one protein in the formation, elongation, and stabilization of amyloid fibrils over a long course of time. Some of the proteins identified in this analysis are known for their association with more than one neurodegenerative disorder, for example, Adam22, APP, ApoE, β-catenin neurofilament proteins, 14-3-3 proteins, and others.
There are possibilities that some contaminating proteins may appear in proteomic analysis owing to the fact that, following homogenization, some unwarranted interactions may happen among proteins due to the high hydrophobicity of amyloid fibrils. Some of these proteins stick to the cores and are not removed even after multiple rounds of sonication and SDS washing steps. This is one limitation of this amyloid purification strategy. However, it could be addressed using suitable negative controls and performing effective statistical cutoffs in large-scale proteomic studies. Another limitation that we encountered relates to the underestimation of Aβ peptide abundance following trypsin digestion. This has been addressed in this workflow by a targeted MS/MS analysis strategy. GO analysis for KEGG pathways indicates the abundance of proteins belonging to pathways involved in many neurodegenerative diseases, for example, Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis (ALS), and Prion diseases. These proteins are important players in multiple pathological pathways and, thus, have known involvement in disease initiation and progression. Interestingly, some of these proteins require further analysis to determine their possible involvement in the pathology of AD and other neurodegenerative diseases.
Future studies on the pure amyloid material from other disease models may provide an in-depth understanding of the structural patterns and composition of core fibrils and may help in identifying key therapeutic targets.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the NIH grant R01AG061865 to R.J.V. and J.N.S. The authors thank Vassar and Savas research group members at Northwestern University for their thoughtful discussions. We also sincerely thank Dr(s). Ansgar Seimer and Ralf Langen at the University of South California for their crucial input. We thank Dr. Farida Korabova for sample preparation and negative staining electron microscopy imaging at Northwestern University Center for Advanced Microscopy.
Materials
| Acclaim PepMap 100 C18 HPLC column 0.075 mm x 20 mm | Thermo Scientific | 164535 | Alternative instruments, chemicals and antibodies from other manufacturers can be used |
| Ammonium bicarbonate | Sigma-Aldrich | 9830 | |
| anti-amyloid beta (1-16) 6E10 antibody | Biolegend | 803001 | |
| anti-amyloid beta (17-24) 4G8 antibody | Biolegend | 800701 | |
| anti-amyloid beta (N terminus 82E1) antibody | IBL America | 10323 | |
| anti-amyloid fibril LOC antibody | EMD Millipore | AB2287 | |
| BCA kit | Thermo Fisher Scientific | 23225 | |
| Bioruptor Pico Plus | Diagenode | B01020001 | |
| Calcium Chloride | Sigma-Aldrich | C1016 | |
| Collagenase | Sigma-Aldrich | C0130 | |
| Complete Protease Inhibitor Cocktail | Sigma-Aldrich | 11697498001 | |
| Dnase I | Thermo Fisher Scientific | EN0521 | |
| EDTA | Sigma-Aldrich | EDS | |
| Guanidine hydrochloride | Sigma-Aldrich | G4505 | |
| HyperSep C18 Cartridges | Thermo Fisher Scientific | 60108-302 | |
| Integrated Proteomics Pipeline – IP2 | http://www.integratedproteomics.com/ | ||
| Iodoacetamide (IAA) | Sigma-Aldrich | I1149 | |
| K54 Tissue Homogenizing System Motor | Cole Parmer | Glas-Col 099C | |
| MaxQuant | https://www.maxquant.org/ | ||
| Micro BCA kit | Thermo Fisher Scientific | 23235 | |
| Nanoviper 75 μm x 50 cm | Thermo Scientific | 164942 | |
| Optima L-90K Ultracentrifuge | Beckman Coulter | BR-8101P-E | |
| Orbitrap Fusion TribridMass Spectrometer | Thermo Scientific | IQLAAEGAAPFADBMBCX | |
| Pierce C18 Spin Columns | Thermo Fisher Scientific | 89870 | |
| Precellys 24 tissue homogenizer | Bertin Instruments | P000062-PEVO0-A | |
| ProteaseMAX(TM) Surfactant Trypsin Enhancer | Promega | V2072 | |
| RawConverter | http://www.fields.scripps.edu/rawconv/ | ||
| Sodium azide | VWR | 97064-646 | |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich | 74255 | |
| Sorvall Legend Micro 21R Microcentrifuge | Thermo Fisher Scientific | 75002446 | |
| Speed Vaccum Concentrator | Labconco | 7315021 | |
| Tris-2-carboxyethylphosphine (TCEP) | Sigma-Aldrich | C4706-2G | |
| Tris-HCl | Thermo Fisher Scientific | 15568025 | |
| Trypsin Gold-Mass spec grade | Promega | V5280 | |
| UltiMate 3000 RSLCnano System | Thermo Scientific | ULTIM3000RSLCNANO |
References
- Willbold, D., Strodel, B., Schröder, G. F., Hoyer, W., Heise, H. Amyloid-type protein aggregation and prion-like properties of amyloids. Chemical Reviews. 121 (13), 8285-8307 (2021).
- Rambaran, R. N., Serpell, L. C. Amyloid fibrils: abnormal protein assembly. Prion. 2 (3), 112-117 (2008).
- Upadhyay, A., et al. Complex inclusion bodies and defective proteome hubs in neurodegenerative disease: New clues, new challenges. The Neuroscientist. , (2021).
- Greiner, E. R., Kelly, J. W., Palhano, F. L. Immunoprecipitation of amyloid fibrils by the use of an antibody that recognizes a generic epitope common to amyloid fibrils. PLOS ONE. 9 (8), 105433 (2014).
- Kourelis, T. V., et al. A proteomic atlas of cardiac amyloid plaques. JACC: CardioOncology. 2 (4), 632-643 (2020).
- Mangione, P. P., et al. Increasing the accuracy of proteomic typing by decellularisation of amyloid tissue biopsies. Journal of Proteomics. 165, 113-118 (2017).
- Rostagno, A., Neubert, T. A., Ghiso, J. Unveiling brain Aβ heterogeneity through targeted proteomic analysis. Methods in Molecular Biology. 1779, 23-43 (2018).
- Roher, A. E., et al. Morphology and toxicity of Aβ-(1-42) dimer derived from neuritic and vascular amyloid deposits of Alzheimer’s disease. Journal of Biological Chemistry. 271 (34), 20631-20635 (1996).
- Lu, J. -. X., et al. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 154 (6), 1257-1268 (2013).
- Tycko, R. Solid-state NMR studies of amyloid fibril structure. Annual Review of Physical Chemistry. 62, 279-299 (2011).
- Saito, T., et al. Single App knock-in mouse models of Alzheimer’s disease. Nature Neuroscience. 17 (5), 661-663 (2014).
- Meyerhoff, J., et al. Microdissection of mouse brain into functionally and anatomically different regions. Journal of Visualized Experiments: JoVE. (168), e61941 (2021).
- Spijker, S., Li, K. Dissection of Rodent Brain Regions. Neuroproteomics. Neuromethods. 57, (2011).
- Hark, T. J., et al. Pulse-chase proteomics of the App knockin mouse models of Alzheimer’s disease reveals that synaptic dysfunction originates in presynaptic terminals. Cell Systems. 12 (2), 141-158 (2021).
- Liu, S., et al. Highly efficient intercellular spreading of protein misfolding mediated by viral ligand-receptor interactions. Nature Communications. 12 (1), 5739 (2021).
- Toyama, B. H., Weissman, J. S. Amyloid structure: conformational diversity and consequences. Annual Review of Biochemistry. 80, 557-585 (2011).
- Sundaria, N., et al. Neurodegeneration & imperfect ageing: Technological limitations and challenges. Mechanisms of Ageing and Development. 200, 111574 (2021).
- Cendrowska, U., et al. Unraveling the complexity of amyloid polymorphism using gold nanoparticles and cryo-EM. Proceedings of the National Academy of Sciences. 117 (12), 6866-6874 (2020).
- Seuring, C., et al. Amyloid fibril polymorphism: almost identical on the atomic level, mesoscopically very different. The Journal of Physical Chemistry B. 121 (8), 1783-1792 (2017).
- Close, W., et al. Physical basis of amyloid fibril polymorphism. Nature Communications. 9 (1), 699 (2018).
- Tycko, R. Amyloid polymorphism: Structural basis and neurobiological relevance. Neuron. 86 (3), 632-645 (2015).
- Konstantoulea, K., et al. Heterotypic Amyloid β interactions facilitate amyloid assembly and modify amyloid structure. The EMBO Journal. 41, 108591 (2022).
- Hondius, D. C., et al. Proteomics analysis identifies new markers associated with capillary cerebral amyloid angiopathy in Alzheimer’s disease. Acta Neuropathologica Communications. 6 (1), 1-19 (2018).
- Luo, J., Wärmländer, S. K., Gräslund, A., Abrahams, J. P. Cross-interactions between the Alzheimer disease amyloid-β peptide and other amyloid proteins: a further aspect of the amyloid cascade hypothesis. Journal of Biological Chemistry. 291 (32), 16485-16493 (2016).
- Hosp, F., et al. Spatiotemporal proteomic profiling of Huntington’s disease inclusions reveals widespread loss of protein function. Cell Reports. 21 (8), 2291-2303 (2017).
- Wallace, E. W. J., et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 162 (6), 1286-1298 (2015).
- Darling, A. L., Liu, Y., Oldfield, C. J., Uversky, V. N. Intrinsically disordered proteome of human membrane-less organelles. Proteomics. 18 (5-6), 1700193 (2018).
- Kepchia, D., et al. Diverse proteins aggregate in mild cognitive impairment and Alzheimer’s disease brain. Alzheimer’s Research & Therapy. 12 (1), 1-20 (2020).
- Espay, A. J., et al. Revisiting protein aggregation as pathogenic in sporadic Parkinson and Alzheimer diseases. Neurology. 92 (7), 329-337 (2019).
- Fändrich, M., Schmidt, M., Grigorieff, N. Recent progress in understanding Alzheimer’s β-amyloid structures. Trends in Biochemical Sciences. 36 (6), 338-345 (2011).
- Bonnin, E. A., Fornasiero, E. F., Lange, F., Turck, C. W., Rizzoli, S. O. NanoSIMS observations of mouse retinal cells reveal strict metabolic controls on nitrogen turnover. BMC Molecular and Cell Biology. 22 (1), 1-10 (2021).
- Michno, W., et al. Following spatial Aβ aggregation dynamics in evolving Alzheimer’s disease pathology by imaging stable isotope labeling kinetics. Science Advances. 7 (25), (2021).
- Toyama, B. H., et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 154 (5), 971-982 (2013).
- Bomba-Warczak, E., Edassery, S. L., Hark, T. J., Savas, J. N. Long-lived mitochondrial cristae proteins in mouse heart and brain. Journal of Cell Biology. 220 (9), 202005193 (2021).

