In Ovo and Ex Ovo Methods to Study Avian Inner Ear Development
Summary
The chick is a cost-effective, accessible, and widely available model organism for a variety of studies. Here, a series of protocols is detailed to understand the molecular mechanisms underlying avian inner ear development and regeneration.
Abstract
The inner ear perceives sound and maintains balance using the cochlea and vestibule. It does this by using a dedicated mechanosensory cell type known as the hair cell. Basic research in the inner ear has led to a deep understanding of how the hair cell functions, and how dysregulation can lead to hearing loss and vertigo. For this research, the mouse has been the pre-eminent model system. However, mice, like all mammals, have lost the ability to replace hair cells. Thus, when trying to understand cellular therapies for restoring inner ear function, complementary studies in other vertebrate species could provide further insights. The auditory epithelium of birds, the basilar papilla (BP), is a sheet of epithelium composed of mechanosensory hair cells (HCs) intercalated by supporting cells (SCs). Although the anatomical architecture of the basilar papilla and the mammalian cochlea differ, the molecular mechanisms of inner ear development and hearing are similar. This makes the basilar papilla a useful system for not only comparative studies but also to understand regeneration. Here, we describe dissection and manipulation techniques for the chicken inner ear. The technique shows genetic and small molecule inhibition methods, which offer a potent tool for studying the molecular mechanisms of inner ear development. In this paper, we discuss in ovo electroporation techniques to genetically perturb the basilar papilla using CRIPSR-Cas9 deletions, followed by dissection of the basilar papilla. We also demonstrate the BP organ culture and optimal use of culture matrices, to observe the development of the epithelium and the hair cells.
Introduction
The inner ear of all vertebrates is derived from a simple epithelium known as the otic placode1,2. This will give rise to all the structural elements and the cell types necessary to transduce the mechanosensory information associated with hearing and balance perception. Hair cells (HCs), the ciliated sensor of the inner ear, are surrounded by supporting cells (SCs). HCs relay information to the auditory hindbrain through the neurons of the eighth cranial nerve. These are also generated from the otic placode3. The primary transduction of sound is achieved at the apical surface of the auditory HC, through a mechanically sensitive hair bundle4. This is mediated through modified actin-based protrusions called stereocilia, which are arranged in a graded, staircase pattern5. In addition, a modified primary cilium, called the kinocilium, organizes hair bundle formation and is adjacent to the tallest row of stereocilia6,7,8. The architecture of stereocilia is critical for this role in converting mechanical stimuli derived from acoustic energy to electrical neural signals9. Damage to the auditory HC through ageing, infection, otoacoustic trauma, or ototoxic shock can result in partial or complete hearing loss that, in mammals, is irreversible10.
Cellular replacement therapies have been proposed that might repair such damage11,12. The approach of this research has been to understand the normal development of the mammalian hair cell and ask if development programs can be reinitiated in progenitor-like cells that may exist within the inner ear13. A second approach has been to look outside of mammals, to non-mammalian vertebrates in which robust regeneration of auditory hair cells takes place, such as birds14,15. In birds, hair cell regeneration occurs predominantly through the de-differentiation of a supporting cell to a progenitor-like state, followed by asymmetric mitotic division to generate a hair cell and supporting cell16. In addition, direct differentiation of a supporting cell to generate a hair cell has also been observed17.
While the mechanisms of avian auditory development do show significant similarities with that of mammals, there are differences18. HC and SC differentiation in the chick BP is apparent from embryonic day (E) 7, becoming more distinct over time. By E12, a well-patterned and well-polarized basilar papilla (BP) can be visualized, and by E17 well-developed hair cells can be seen19. These time points provide windows into the mechanisms of differentiation, patterning, and polarity, as well as hair cell maturation. Understanding whether such mechanisms are conserved or divergent is important, as they provide insights into the deep homology of the origins of mechanosensory hair cells.
Here, we demonstrate an array of techniques performed at early and late embryonic stages to study cellular processes such as proliferation, fate specification, differentiation, patterning, and maintenance throughout the development of the inner ear organ. This complements other protocols on understanding inner ear development in explant culture20,21,22. We first discuss the introduction of exogenous DNA or RNA into BP precursors within the E3.5 otocyst using in ovo electroporation. Although genetic manipulations can provide valuable insights, the phenotypes thus generated can be pleiotropic and consequently confounding. This is particularly true during later inner ear development, where fundamental processes such as cytoskeletal remodeling play multiple roles in cell division, tissue morphogenesis, and cellular specialization. We present protocols for pharmacological inhibition in cultured explants, which offer advantages in controlling dosage and treatment timing and duration, offering precise spatiotemporal manipulation of developmental mechanisms.
Different organ culture methods can be utilized depending on the treatment duration of small inhibitors. Here we demonstrate two methods of organ culture that allow insights into epithelial morphogenesis and cellular specialization. A method for 3D culture using collagen as a matrix to culture the cochlear duct enables robust culturing and live visualization of the developing BP. For understanding the formation of stereocilia, we present a membrane culture method such that epithelial tissue is cultured on a stiff matrix enabling actin protrusions to grow freely. Both methods allow downstream processing such as live-cell imaging, immunohistochemistry, scanning electron microscopy (SEM), cell recording, etc. These techniques provide a roadmap for the effective use of the chick as a model system to understand and manipulate the development, maturation, and regeneration of the avian auditory epithelium.
Protocol
Protocols involving the procurement, culture, and use of fertilized chicken eggs and unhatched embryos were approved by the Institutional Animal Ethics Committee of the National Centre for Biological Sciences, Bengaluru, Karnataka.
1. In ovo electroporation of chick auditory precursors
- sgRNA design and cloning for CRISPR/Cas9 gene knockout
- For creating gene knockouts, design guide RNAs to disrupt the exon regions of the gene, preferably closer to the 5' end of the coding region.
- Select the potential guide RNAs using a web tool CRISPOR23. Set the browser data to Gallus gallus and the protospacer adjacent motif (PAM) sequence to 5'- NGG -3'. The program determines guide RNAs from the input sequence and assigns different scores based on on-target and off-target activity. Select the top four guides for further studies.
- For the designing of template specific oligos for guide (g)RNA production, remove the PAM sequence (5′- NGG -3′) from the gRNA design tool output. This is not needed for targeting, but contains the Cas9 cleavage recognition sequence. Synthesize two HPLC purified complementary oligos with BsmBI restriction site at both ends, for each potential gRNA.
- Clone the guide sequence in frame with a tracrRNA scaffold of a vector of choice (here, pcU6_1sgRNA vector was used24).
- Dissolve the sgRNA oligonucleotides at a concentration of 100 µM in DNase/RNase-free water. Perform annealing of the two sense and antisense oligo guides using a thermocycler with the following parameters: 95 °C for 3 min ,then 37 °C for 15 min, and then decrease to 4 °C.
- Using standard molecular biology techniques as described in25, setup restriction digestion of the annealed oligos and pcU6_1sgRNA cloning vector with BsmBI enzyme overnight. Setup ligation with the gel-purified linear BsmBI-digested pcU6_1sgRNA vector and digested sgRNA oligos. Transform into a DH5-alpha competent cell and sequence confirm the obtained clone.
- Egg handling and windowing
- Procure freshly laid eggs and clean them by wiping them with 70% ethanol to prevent contamination. Incubate at 37-38 °C, with 45% humidity for 3.5-4 days.
- After incubation, place the egg on its side for at least 5 min before opening. This allows the embryo to reposition to the top of the yolk. Use forceps to make small holes at the top and blunt end of the egg such that a 21G needle can pass through.
- To prevent damage during windowing, remove albumin to lower the embryo away from the shell. To do this, use a 5 mL syringe and a 21G needle to carefully draw 2 mL of albumin from a hole at the blunt end of the egg. Use clear tape to cover the hole at the blunt end.
- To make the egg window, affix clear tape to the top of the eggshell. Cut open a window, around 2 cm long and 1.5 cm wide, using spring bow scissors and expose the embryo. Use forceps to open the chorionic membranes overlying the embryo, allowing access to the embryo.
- Microinjecting plasmids
- For the gene knockout experiment, prepare two solutions: the knock-down mix containing SpCas9 protein and guide plasmid – pcU6_1sgRNA, and the tracer plasmid mix of T2K-eGFP (this is a chick ß-actin promoter driving GFP cassette surrounded by the transposon sites of Tol2) together with the T2TP (the Tol2 transposase cloned in Tol2 construct26,27). The tracer is used to track electroporation efficiency.
- When electroporating multiple plasmids, ensure that the final concentration of DNA is at least 4 µg/µL. Mix the three constructs, guide plasmid – pcU6_1sgRNA, T2K-eGFP, and T2TP, in a 1:1:1 ratio with 1 µg of SpCas9 protein, 30% sucrose, and 0.1% Fast Green dye in a final volume of 10 µL.
- Pull needles for microinjection from standard glass capillaries (length 3 in, OD 1.0 mm) using a vertical pipette puller. Use fine forceps to break off the capillary tip after pulling to obtain a tip diameter of approximately 50 µm with a tapered end.
- Lay the embryos at E3.5 on their left side with the head facing right. This way only the right otic vesicle is accessible for micro-injection. Micro-inject the knock-down mix into the otic vesicle. Inject around a 200 nL volume of DNA solution mix to fill the otic vesicle.
- Determine the guide efficiency using a T7 endonuclease assay28.
- Electroporation
- Add a few drops of 0.719% saline on top of the embryo prior to electroporation to lower the electric resistance and prevent overheating of the embryo.
- Place the positive electrode through the hole made at the blunt side of the egg when the albumin was removed. Maneuver the electrode so that it is under the yolk. Place the negative electrode over the filled otocyst.
- Use electroporation to transfect plasmids into the cells of the embryo. Use a square pulse generator and apply five pulses of 25 V and 100 ms duration each, 50 ms apart. Determine the conditions empirically based on an individual electroporation setup.
- Hydrate the embryo after electroporation by adding a few drops of 0.719% saline. To clean denatured albumin from the surface of the electrodes, thoroughly rinse it with distilled water. Reseal the egg with clear tape and return to the humidified incubator at 37-38 °C for further incubation.
NOTE: Embryos can be cultured until hatching after electroporation, however there is a steep reduction in viability.
2. Basilar papilla dissection
- Use 70% ethanol to disinfect the surgical table, microscope stage, and surrounding area. Heat or alcohol sterilize microdissection equipment which include minimally spring-bow scissors, micro-curette, and two pairs of fine forceps.
- Prepare the following dissection plates: a glass Petri dish with a black silicon base, a 90 mm plastic Petri dish, and a 60 mm Petri dish. Chill either phosphate-buffered saline (PBS) or Hanks' Balanced Salt Solution (HBSS) for dissections.
- Gently crack the egg into the 90 mm Petri dish. Identify the outer ear of the chick. Decapitate the embryo by cutting the neck just below the level of the lower jaw using scissors. Transfer the head to a 60 mm Petri dish filled with ice-cold PBS.
- Orient the embryo with the top of the beak facing toward the experimenter and hold the beak using one of the #5 forceps. Scoop out the eyes using the second #5 forceps. Going from rostral to caudal, cut the skull along its midline. Scoop out the brain.
- Add more ice-cold PBS or HBSS and locate the two shiny structures, close to the level of the pinna. These are the otoliths of the lagena at the end of the cochlear duct and are found close to the midline.
- Cut roughly between the two lagenas, and well above and below this region to isolate two inner ears. Under oblique lighting, visualize the outline of the inner ear. Remove extraneous tissue and the vestibule.
- Transfer the isolated cochlea to a black silicone base plate with ice cold PBS. Using #5 forceps, peel away the cartilaginous cochlear capsule to obtain the cochlear duct. Locate the undulated layer (tegmentum) of the cochlear duct and remove using #55 forceps to expose the BP. Using #55 forceps, remove the tectorial membrane to expose HCs and SCs.
3. Culture of basilar papilla explants
- Membrane culture of the BP
- Take a six-well tissue culture plate and arrange one culture membrane insert per well.
- Collect the dissected basilar papilla (explant) in a 200 µL pipette with 1x HBSS buffer and transfer it onto a membrane. To prevent the tissue from sticking to the wall of the pipette, draw up some media before aspirating the tissue.
- Orient the explant such that the basilar papilla is facing upward, so that the hair and support cells are visible from the top29. Once the explant is positioned, aspirate the HBSS buffer slowly from the culture membrane surface. In this process, the explant will attach to the culture membrane.
- Add 1.2 mL of Dulbecco's modified Eagle medium (DMEM) culture media between the membrane insert and the well wall to fill up the wells of the six-well plate. Up to six explants can be cultured on a single 30 mm culture membrane.
- Collagen culture of the cochlear duct
- Prepare collagen mixture by adding 400 µL of 3 mg/mL rat tail collagen, 50 µL of 10x DMEM, 30 µL of 7.5% NaHCO3, and 5 µL of HEPES in a tissue culture hood.
- Take a four-well plate and add three drops of collagen mixture into each well. Transfer dissected cochlear duct to each collagen drop. Incubate the plate for 10 min at 37 °C, 5% CO2 to cure the collagen matrix.
- Small molecule treatment of cultures
- Prepare culture media (DMEM, N-2 supplement, Penicillin) with either the pharmacological modulator or its solvent alone (for use as a control). Replace the culture media with 700 µL of media supplemented with the inhibitor. Culture the explants in an incubator at 37 °C, 5% CO2.
- Replace 50% of the culture media every day. After appropriate incubation time, remove the culture media and use the explants for downstream assays.
4. Imaging and analysis
- Immunofluorescent analysis
- Remove culture media from wells and wash the explants twice with 1x HBSS. Using a pipette, add 1 mL of 4% paraformaldehyde (PFA) to the well and incubate for 20 min at room temperature.
- Remove PFA and wash the explants three times with 1x PBS at room temperature. To collect the explants from the culture membrane, make a small cut around the membrane containing the piece of the tissue using small spring bow scissors and transfer the tissue along with the membrane to a 48-well culture plate using forceps.
NOTE: If the tissues are floating after the fixation, then aspirate with a 200 µL pipette and transfer it to 48-well plates for further processing. Avoid forceful detachment of the tissue from the membrane. - For collagen droplet culture, use forceps to transfer the entire collagen drop to a silicon plate. Add 200 µL of 1x PBS and remove collagen with the help of forceps, and then transfer the tissue to a 48-well culture plate.
- Permeabilize the explants with 1 mL of 1x PBS supplemented with 0.3% Tween-20 (PBST) for 30 min at room temperature. Incubate the explants with 200 µL of blocking buffer (10% goat serum + 1% bovine serum albumin in PBST) for 1 h at room temperature.
- Incubate the explants with 200 µL of primary antibody solution (1:300, in blocking buffer) overnight at 4 °C. Remove the primary antibody solution and wash the explants thoroughly 5 x 20 min with PBST.
- Incubate the explants with 200 µL of phalloidin and secondary antibody solution (in blocking buffer) for 1 h at room temperature in the dark. Remove the phalloidin and secondary antibody solution and wash the explants thoroughly 5 x 20 min with PBST.
NOTE: Secondary antibody concentration and wash length must be determined for each individual imaging application. For imaging using super resolution microscopy, we typically increase the concentration of the secondary antibody from 1:500 to 1:200 and increase the concentration of phalloidin from 1:400 to 1:200. - Incubate the explants with a solution of DAPI (1:1000 in PBST) for 15 min at room temperature. Remove the DAPI solution and wash the explants 3 x 5 min with PBST.
- Use the mounting media to mount explants on a slide with BP facing in an upward direction (against the coverslip). For confocal imaging use antifade mounting media. Let the mounting media dry overnight at room temperature in the dark. Either image directly or store the slides at 4 °C until imaging.
- OTOTO fixation for SEM analysis
- Prepare all solutions fresh and handle according to local safety guidelines. Fix membrane-cultured explants (from step 4.1.4) in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.3) with 3 mM CaCl2. Perform fixation at 4 °C for 24 to 72 h with a change of fixative every 24 h.
- Remove the fixative and wash the tissue with 0.1 M sodium cacodylate 3 x 5 min at room temperature. Secondary fix with 1% OsO4 (diluted from a 4% OsO4 stock using 0.1 M sodium cacodylate buffer) for 1 h at room temperature. Perform this and the subsequent steps until dehydration in a fume hood.
- Rinse using 0.1 M sodium cacodylate buffer 3 x 5 min at room temperature. Then rinse in double distilled or ultrapure water 3 x 5 min at room temperature.
- Prepare a 0.5% solution of thiocarbohydrazide (TCH) in ultrapure water. Stir at 75 °C for 10 min and filter the solution when it has cooled to room temperature.
NOTE: TCH is extremely hazardous. Usage must be approved by the local safety committee, and care must be taken when handling. - Remove ultrapure water from the sample and add 0.5% TCH drop by drop. If the solution turns brown, stop and rinse the sample with ultrapure water, before carefully adding the 0.5% TCH solution. Once the solution is clear, replace it with 0.5% TCH. Incubate at room temperature for 20 min30,31.
- Rinse the sample with ultrapure water 3 x 5 min at room temperature. Repeat steps 4.2.2, 4.2.3, and 4.2.5 two more times ending with OsO4 incubation followed by rinsing.
- Dehydrate the samples in an ethanol series to 100% anhydrous ethanol (EtOH). Incubate first in 25% ETOH for 10 min, then 50% ETOH for 10 min, 75% EtOH for 10 min, 95% ETOH for 10 min, before incubating for a total of 3x for 10 min each in 100% ETOH.
- Perform critical point drying using liquid CO2. Immediately mount the samples on a SEM stub with double-sided carbon adhesive tape. Proceed to sputter coat to provide 5-10 nm of coating. If not imaging immediately, store the samples in a desiccator under vacuum.
Representative Results
In the electroporation setup, electrode positioning can play a role in the domain of transfection. The positive electrode is placed under the yolk, and the negative above the embryo (Figure 1A). This results in higher GFP expression in much of the inner ear and both vestibular organs (Figure 1B), and auditory basilar papilla (Figure 1C,D), confirming transfection.
In assessing the phenotype of CRISPR-knockdowns, we designed guide RNAs to the hair cell transcription factor Atonal homolog 1 (Atoh1). Mouse mutants of Atoh1 are unable to form hair cells32; after electroporation of Atoh1 guide RNAs and incubation until E10, we find that HC development is impaired when compared to control, empty plasmid control (Figure 2). Although electroporation is mosaic (Figure 2E,F), control electroporated cells are able to form hair cells. In Atoh1 gRNA electroporated samples, GFP positive cells never show the markers of HC development (Figure 2B).
Organ culture of the BP provides accessibility to the tissue. Cultures in a 3D matrix, such as collagen, provide excellent preservation of tissue morphology for up to 5 days. The organization of HC and SC is maintained in these culture conditions (Figure 3).
Organ cultures on a membrane are preferable for imaging stereocilia. Such cultures can be cultured for up to 5 days while maintaining the hair bundle integrity. This can be seen by the localization of the tip-link protein, protocadherin 1533 (Pcdh15) (Figure 4). For interrogating the development of the hair bundle, higher resolution imaging is necessary, and such approaches, using either super-resolution microscopy (Figure 4D) or scanning electron microscopy (Figure 4E), provide more complete information.
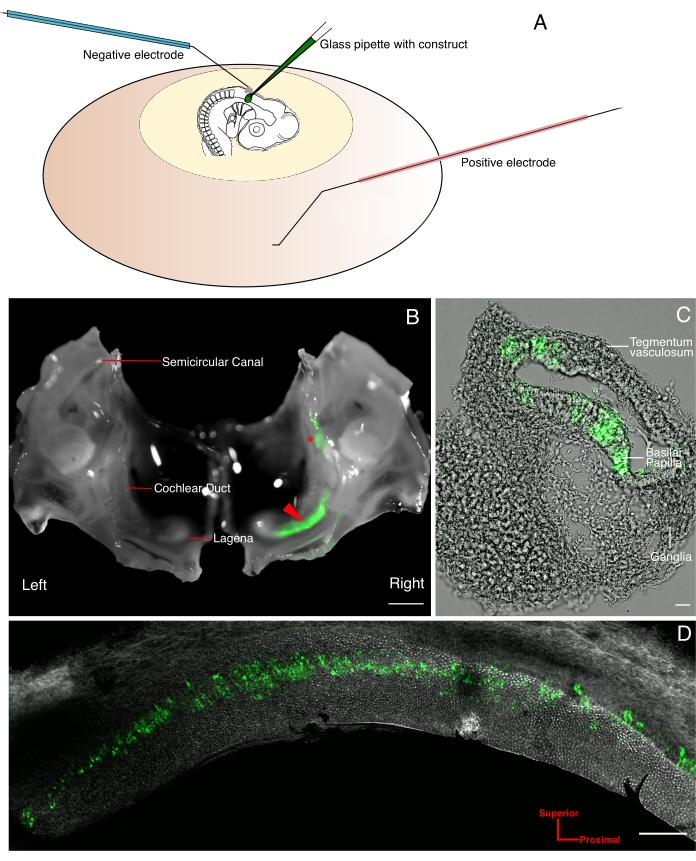
Figure 1. GFP expression is visible at E10 after in ovo electroporation at E4. (A) Schematic diagram illustrating in ovo microinjection and electroporation in chick otic vesicle at E4. The injection pipette is filled with Tol2-eGFP (T2K-eGFP) and Tol2-transposases (T2TP) plasmids, together with fast green dye for visualization. (B) Image of the electroporated inner ear at E10 using a 0.63x air objective lens on a stereomicroscope. The left inner ear is the internal control. The red arrow marks the GFP expression in the cochlear duct of the right inner ear, and the red asterisk shows GFP expression in vestibular organs; scale bar is 2 cm. (C) Widefield fluorescence image of a cross-section of right cochlear duct using a 20x air objective of 0.5 NA. GFP expression is mostly confined to the sensory epithelium; scale bar is 10 µm. (D) Confocal image of whole mount basilar papilla imaged using 10x air objective of 0.5 NA stained with phalloidin conjugated with Alexa 647 fluorophore. GFP expression is observed from proximal to distal end on the neural side of the BP; scale bar is 100 µm. Please click here to view a larger version of this figure.
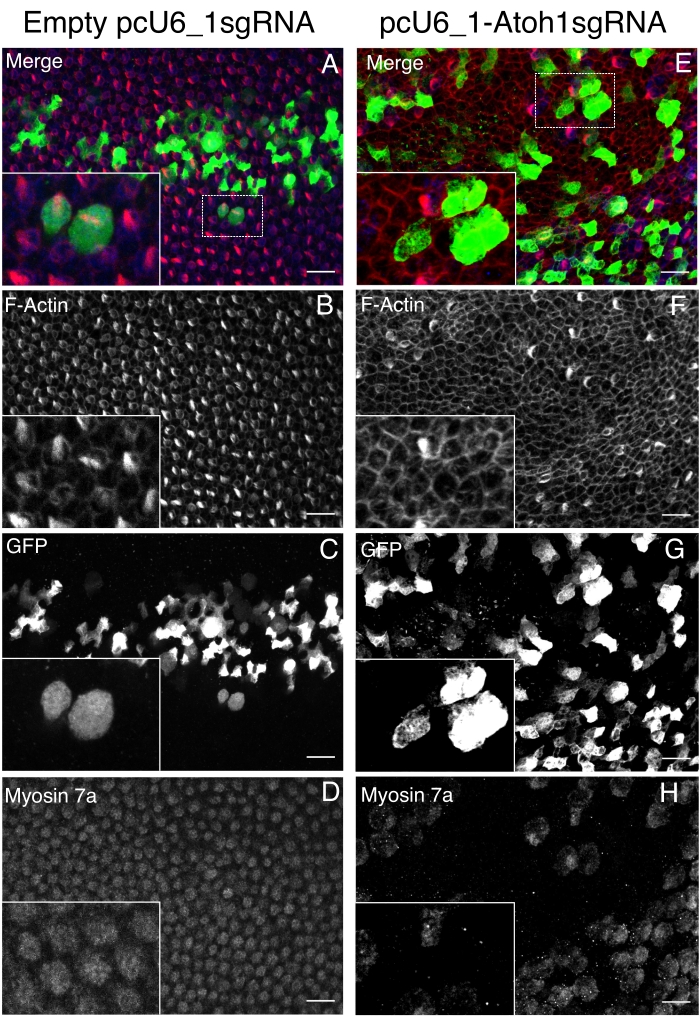
Figure 2. CRISPR/Cas9-mediated Atoh1 gene knockout viain ovo electroporation results in loss of hair cells (HCs). Atoh1 gene guide plasmid pcU6_1-Atoh1sgRNA and tracer plasmids Tol2-eGFP (T2K-eGFP) and Tol2-transposases (T2TP) with SpCas9 protein are microinjected and electroporated in chick otic vesicle at E4. All the images are from chick E10 basilar papilla with a 10 µm scale bar, captured with a 60x oil immersion objective of 1.42 NA using a laser confocal microscope. Zoomed in insets are provided for all images. (A,B,C,D) The left-hand panel contains BP with hair cells (HCs), electroporated with empty pcU6_1sgRNA and T2K-eGFP, and T2TP with SpCas9 protein. (E,F,G,H) The right-hand side panel contains BP with loss of HCs, electroporated with Atoh1 guide (pcU6_1-Atoh1sgRNA) and T2K-eGFP, and T2TP with SpCas9 protein. (A,E) Merged image showing hair cells (HCs) in the basilar papilla. These are immunoreactive for Myosin 7a (blue); F-actin stained with phalloidin conjugated with Alexa 647 (red); GFP expression from Tol2-eGFP plasmids is detected using an anti-GFP antibody (green). The loss of HCs is evident from Myosin 7a immunoreactivity when comparing treatments with empty pcU6_1sgRNA (D) and pcU6_1-Atoh1sgRNA (H). F-actin imaging from both treatments highlights the hair bundle of the HC (B,F). (C,G) T2K-eGFP and T2TP are used to measure transfection location and efficiency. Please click here to view a larger version of this figure.
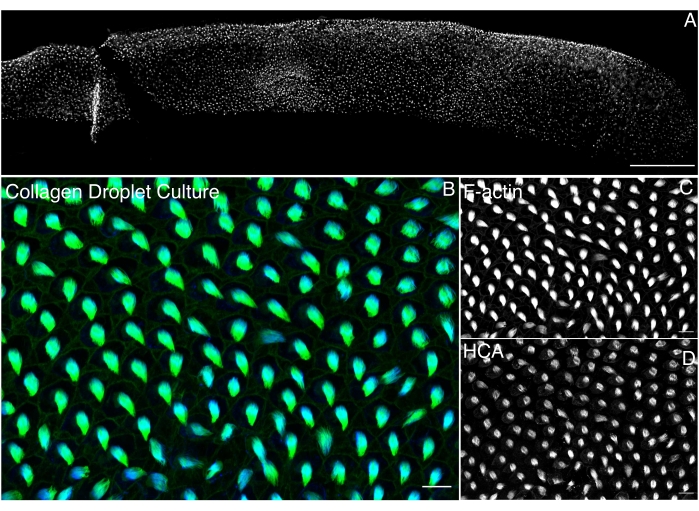
Figure 3. Organ culture of cochlear duct in 3D-collagen droplet culture maintains the organization of sensory epithelium. The BP at E10 is cultured in a 3D collagen droplet culture for 1 day and imaged using 60x oil immersion objective of 1.42 NA using a laser confocal microscope. (A) Image of whole BP of E10 cultured for 1 day in a collagen droplet and stained with antibodies against hair cell antigen (HCA)34. Scale bar is 100 µm. (B) Merged image showing the preserved organization of sensory epithelium from the distal side of BP stained with (C) phalloidin (green) and (D) HCA (blue); scale bar is 10 µm. Please click here to view a larger version of this figure.
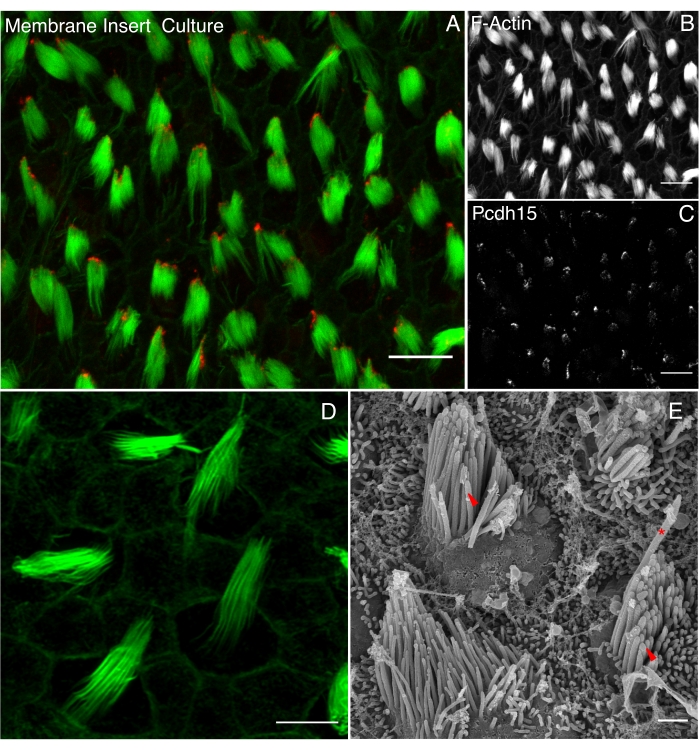
Figure 4. Stereociliary bundles of basilar papilla under electron and light microscope. 0.1% dimethylsulphoxide (DMSO)-treated BP were explanted at E10 and cultured for 3 days in vitro (DIV) on membrane culture inserts. (A) Explant is imaged using a 60x oil immersion objective of 1.42 NA with a laser scanning confocal microscope. The merged image shows the expression of the protocadherin 15 (Pcdh15) and stereocilia marked by phalloidin conjugated with Alexa 488 (green). Single-channel images of F-actin (B) and Pcdh15 (C) are shown. Scale bar is 10 µm. (D) Super-resolution image of stereocilia stained with phalloidin conjugated with Alexa 488 staining in green. The image was obtained using a 63x oil immersion objective of 1.42 NA in Airyscan mode of laser scanning confocal microscope. Scale bar is 5 µm. (E) SEM image was taken at 16340x magnification using 7 kV electron high tension (EHT) voltage. Red asterisk marks kinocilia and red wedge marks stereocilia. Brightness and contrast were adjusted to auto and the image was sharpened using FIJI. Scale bar is 1 µm. Please click here to view a larger version of this figure.
Discussion
The chick is a cost-effective and convenient addition to the model organisms that a lab may use to research the inner ear. The methods described here are routinely used in our lab and complement ongoing research in the mammalian inner ear. In ovo electroporation is used to introduce genetic manipulations into the chick genome. Electroporation can also be used to introduce constructs that encode fluorescent proteins targeted to particular organelles or subcellular structures35,36. While this is a simple procedure, manipulation of embryos in ovo does impact viability. For efficient electroporation, eggs should be procured fresh (soon after laying). A rise in mortality and/or abnormal development is frequently observed in eggs that have been stored for over 5 days. The use of sterile technique, ensuring that the egg is properly sealed so that it does not lose moisture, and minimizing the manipulations performed on the embryo enhance viability.
We found that targeting the otocyst at E3.5 to E4 reduces the trauma that the embryo faces, and using carefully positioned electrodes allows good targeting of the sensory precursors of the BP. However, by this stage, the precursors of the acoustic-vestibular ganglion have already migrated away from the inner ear37,38. To target these cells, earlier timepoints for otocyst electroporation must be selected, and accommodations made for the corresponding decrease in viability. A further note of caution is the possibility of mosaicism in electroporated embryos. Not all cells take up all the plasmid, and thus mosaicism can confound the interpretation. The use of tracer plasmids helps in data interpretation and provides some control over possible mosaic effects. Use of multiple repeats, careful analyses, and statistical methods will aid in the assessment of mosaic phenotypes. An alternative approach is to use genetically modified quails39. Currently, these numbers are limited and are not always available to many labs. However, availability will increase, and quail embryos, which constitutively express sub-cellularly targeted fluorescent proteins, are an attractive proposition for imaging experiments.
In this method, we electroporate protein and DNA for CRISPR mediated knockdowns through non-homologous end joining (NHEJ)24. CRISPR/Cas9 mediated generation of fusion constructs through homology-directed repair (HDR) remains inefficient in this particular paradigm (Singh et al., unpublished observations). The electroporation method can be adapted for use with other types of DNA constructs (these could be constructs encoding fusion proteins), as well as for RNA and protein. It should be noted that during development (and cell division) DNA based expression constructs will become diluted unless the vector incorporates sites that mediate insertion of the exogenous DNA into the genome of these cells (e.g., Tol2 or PiggyBac). This allows more stable expression of the construct.
After electroporation, the embryos are usually cultured in ovo until E10 stage for hair cell differentiation and development studies. But if required, in ovo culture can be continued till the eggs hatch; however, with increasing lengths of incubation there is a corresponding drop in viability. To circumvent this, in ovo electroporation can be combined with BP dissections followed by long-term ex ovo culture. Culturing explants within a 3D matrix allows good preservation of tissue morphology. This method can be used to study changes in tissue patterning, polarity, and differentiation. The culture of the BP on a membrane allows visualization of the apical side of the tissue27, particularly high-resolution imaging of the stereocilia.
In some cases, the limitations of genetic modification can be overcome by using different small molecule-based treatments. The pharmacologically active compounds act as inhibitors or agonists for signaling pathways or cell biological processes. Their application is particularly useful in dissecting the temporal requirement. However, exact modes of delivery and optimal concentrations do need to be empirically determined, as in some cases standardization performed in cell lines is not comparable to the dosage required in tissues. Delivery within the embryo can be problematic, and the amount needed combined with the impact on other organ systems may compromise viability significantly. A ready alternative is organ culture methods20,21,22. However, the exact culture method does depend on the types of study and analyses that are intended. While collagen culture preserves tissue morphology of the BP, membrane culture is better suited for studying the apical hair bundles of the HC. The tissue can simply be placed on top of the membrane to visualize the apical surface using scanning electron microscopy or super-resolution microscopy. Dissections for organ culture do require good practice. These include maintaining a sterile workspace and using sharpened instruments. Such microdissection tools are sensitive, and we find the use of silicon dishes important in preserving the integrity of microsurgical tools.
Together, the techniques represent valuable approaches to further the understanding of inner ear development. The insights in comparative biology and regeneration that the chick embryos offer can provide significant insights into hair cell development and function.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We gratefully acknowledge support from NCBS, TIFR, Infosys-TIFR Leading Edge Research Grant, DST-SERB, and the Royal National Institute for the Deaf. We would like to thank Central Poultry Development Organization and Training Institute, Hesaraghatta, Bengaluru. We are grateful to CIFF and EM facility and lab support at NCBS. We thank Yoshiko Takahashi and Koichi Kawakami for the Tol2-eGFP and T2TP constructs, and Guy Richardson for HCA and G19 Pcdh15 antibody. We are grateful to Earlab members for their constant support and valuable feedback on the protocol.
Materials
| Alexa Fluor 488 Phalloidin | Thermo Fisher Scientific | A12379 | |
| Alexa Fluor 647 Phalloidin | Thermo Fisher Scientific | A22287 | |
| Alt-R S.p. HiFi Cas9 Nuclease V3 | Integrated DNA Technologies | 1081061 | High fidelity Cas9 protein |
| Anti-GFP antibody | Abcam | ab290 | Rabbit polyclonal to GFP |
| Bovine Serum Albumin | Sigma-Aldrich | A9647 | |
| Calcium Chloride Dihydrate | Thermo Fisher Scientific | Q12135 | |
| Collagen I, rat tail | Thermo Fisher Scientific | A1048301 | |
| Critical Point Dryer Leica EM CPD300 | Leica | ||
| CUY-21 Electroporator | Nepagene | ||
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D8418 | |
| DM5000B Widefield Microscope | Leica | ||
| DMEM, high glucose, GlutaMAX Supplement, pyruvate | Thermo Fisher Scientific | 10569010 | |
| Dumont #5 Forceps | Fine Science Tools | 11251-20 | |
| Dumont #55 Forceps | Fine Science Tools | 11255-20 | |
| Fast Green FCF | Sigma-Aldrich | F7252 | |
| Fluoroshield | Sigma-Aldrich | F6182 | |
| FLUOVIEW 3000 Laser Scanning Microscope | Olympus | ||
| Glutaraldehyde (25 %) | Sigma-Aldrich | 340855 | |
| Goat anti-Mouse IgG Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | A-11001 | |
| Goat anti-Mouse IgG Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | A-11032 | |
| Goat anti-Rabbit IgG Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | A-11008 | |
| Goat Serum Sterile filtered | HiMedia | RM10701 | Heat inactivated |
| Hanks' Balanced Salt Solution (HBSS) | Thermo Fisher Scientific | 14025092 | |
| LSM980 Airyscan Microscope | Zeiss | ||
| Millicell Cell Culture Insert, 30 mm, hydrophilic PTFE, 0.4 µm | Sigma-Aldrich | PICM03050 | |
| MVX10 Stereo Microscope | Olympus | ||
| MYO7A antibody | DSHB | 138-1 | Mouse monoclonal to Unconventional myosin-VIIa |
| MZ16 Dissecting microscope | Leica | ||
| N-2 Supplement (100X) | Thermo Fisher Scientific | 17502048 | |
| Noyes Scissors, 14cm (5.5'') | World Precision Instruments | 501237 | |
| Osmium tetroxide (4%) | Sigma-Aldrich | 75632 | |
| Paraformaldehyde | Sigma-Aldrich | 158127 | |
| PC-10 Puller | Narishige | ||
| pcU6_1sgRNA | Addgene | 92395 | Mini vector with modified chicken U6 promoter |
| Penicillin G sodium salt | Sigma-Aldrich | P3032 | |
| Phosphate Buffered Saline (PBS) | Thermo Fisher Scientific | 10010023 | |
| ProLong Gold Antifade Mountant | Thermo Fisher Scientific | P36934 | |
| SMZ1500 Dissecting microscope | Nikon | ||
| Sodium Cacodylate Buffer, 0.2M | Electron Microscopy Sciences | 11652 | |
| Sodium chloride | HiMedia | GRM853 | |
| Sputtre Coater K550X | Emitech | ||
| Standard Glass Capillaries 3 in, OD 1.0 mm, No Filament | World Precision Instruments | 1B100-3 | |
| Sucrose | Sigma-Aldrich | 84097 | |
| The MERLIN Compact VP | Zeiss | ||
| Thiocarbohydrazide | Alfa Aesar | L01205 | |
| TWEEN 20 | Sigma-Aldrich | P1379 |
References
- Sai, X., Ladher, R. K. Early steps in inner ear development: induction and morphogenesis of the otic placode. Frontiers in Pharmacology. 6, 19 (2015).
- Groves, A. K., Fekete, D. M. Shaping sound in space: the regulation of inner ear patterning. Development. 139 (2), 245-257 (2012).
- Driver, E. C., Kelley, M. W. Development of the cochlea. Development. 147 (12), (2020).
- Richardson, G. P., Petit, C. Hair-bundle links: genetics as the gateway to function. Cold Spring Harbor Perspectives in Medicine. 9 (12), 033142 (2019).
- Tilney, L. G., Cotanche, D. A., Tilney, M. S. Actin filaments, stereocilia and hair cells of the bird cochlea. VI. How the number and arrangement of stereocilia are determined. Development. 116 (1), 213-226 (1992).
- Jones, C., et al. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nature Genetics. 40 (1), 69-77 (2008).
- Sipe, C. W., Lu, X. Kif3a regulates planar polarization of auditory hair cells through both ciliary and non-ciliary mechanisms. Development. 138 (16), 3441-3449 (2011).
- May-Simera, H. L., Kelley, M. W. Cilia, Wnt signaling, and the cytoskeleton. Cilia. 1 (1), 1 (2012).
- Ebrahim, S., et al. Stereocilia-staircase spacing is influenced by myosin III motors and their cargos espin-1 and espin-like. Nature Communications. 7, 10833 (2016).
- Corwin, J. T., Cotanche, D. A. Regeneration of sensory hair cells after acoustic trauma. Science. 240 (4860), 1772-1774 (1988).
- Collado, M. S., Burns, J. C., Hu, Z., Corwin, J. T. Recent advances in hair cell regeneration research. Current Opinion in Otolaryngology & Head and Neck Surgery. 16 (5), 465-471 (2008).
- Edge, A. S., Chen, Z. Y. Hair cell regeneration. Current Opinion in Neurobiology. 18 (4), 377-382 (2008).
- Atkinson, P. J., Huarcaya Najarro, E., Sayyid, Z. N., Cheng, A. G. Sensory hair cell development and regeneration: similarities and differences. Development. 142 (9), 1561-1571 (2015).
- Brignull, H. R., Raible, D. W., Stone, J. S. Feathers and fins: non-mammalian models for hair cell regeneration. Brain Research. 1277, 12-23 (2009).
- Rubel, E. W., Furrer, S. A., Stone, J. S. A brief history of hair cell regeneration research and speculations on the future. Hearing Research. 297, 42-51 (2013).
- Stone, J. S., Cotanche, D. A. Hair cell regeneration in the avian auditory epithelium. The International Journal of Developmental Biology. 51 (6-7), 633-647 (2007).
- Roberson, D. W., Alosi, J. A., Cotanche, D. A. Direct transdifferentiation gives rise to the earliest new hair cells in regenerating avian auditory epithelium. Journal of Neuroscience Research. 78 (4), 461-471 (2004).
- Fritzsch, B., Beisel, K. W., Pauley, S., Soukup, G. Molecular evolution of the vertebrate mechanosensory cell and ear. The International Journal of Developmental Biology. 51 (6-7), 663-678 (2007).
- Tilney, L. G., DeRosier, D. J. Actin filaments, stereocilia, and hair cells of the bird cochlea. IV. How the actin filaments become organized in developing stereocilia and in the cuticular plate. Developmental Biology. 116 (1), 119-129 (1986).
- Oesterle, E. C., Tsue, T. T., Reh, T. A., Rubel, E. W. Hair-cell regeneration in organ cultures of the postnatal chicken inner ear. Hearing Research. 70 (1), 85-108 (1993).
- Honda, A., Freeman, S. D., Sai, X., Ladher, R. K., O’Neill, P. From placode to labyrinth: culture of the chicken inner ear. Methods. 66 (3), 447-453 (2014).
- Matsunaga, M., et al. Initiation of supporting cell activation for hair cell regeneration in the avian auditory epithelium: an explant culture model. Frontiers in Cellular Neuroscience. 14, 583994 (2020).
- Concordet, J. P., Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research. 46, 242-245 (2018).
- Gandhi, S., Piacentino, M. L., Vieceli, F. M., Bronner, M. E. Optimization of CRISPR/Cas9 genome editing for loss-of-function in the early chick embryo. Developmental Biology. 432 (1), 86-97 (2017).
- Green, M. R., Sambrook, J. Cloning and transformation with plasmid vectors. Cold Spring Harbor Protocols. 2021 (11), (2021).
- Sato, Y., et al. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Developmental Biology. 305 (2), 616-624 (2007).
- Takahashi, Y., Watanabe, T., Nakagawa, S., Kawakami, K., Sato, Y. Transposon-mediated stable integration and tetracycline-inducible expression of electroporated transgenes in chicken embryos. Methods in Cell Biology. 87, 271-280 (2008).
- Mashal, R. D., Koontz, J., Sklar, J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nature Genetics. 9 (2), 177-183 (1995).
- Ogier, J. M., Burt, R. A., Drury, H. R., Lim, R., Nayagam, B. A. Organotypic culture of neonatal murine inner ear explants. Frontiers in Cellular Neuroscience. 13, 170 (2019).
- Davies, S., Forge, A. Preparation of the mammalian organ of Corti for scanning electron microscopy. Journal of Microscopy. 147, 89-101 (1987).
- Parker, A., Chessum, L., Mburu, P., Sanderson, J., Bowl, M. R. Light and electron microscopy methods for examination of cochlear morphology in mouse models of deafness. Current Protocols in Mouse Biology. 6 (3), 272-306 (2016).
- Bermingham, N. A., et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 284 (5421), 1837-1841 (1999).
- Goodyear, R. J., Forge, A., Legan, P. K., Richardson, G. P. Asymmetric distribution of cadherin 23 and protocadherin 15 in the kinocilial links of avian sensory hair cells. The Journal of Comparative Neurology. 518 (21), 4288-4297 (2010).
- Bartolami, S., Goodyear, R., Richardson, G. Appearance and distribution of the 275 kD hair-cell antigen during development of the avian inner ear. The Journal of Comparative Neurology. 314 (4), 777-788 (1991).
- Funahashi, J., Nakamura, H. Electroporation in avian embryos. Methods in Molecular Biology. 461, 377-382 (2008).
- Nakamura, H., Funahashi, J. Electroporation: past, present and future. Development, Growth & Differentiation. 55 (1), 15-19 (2013).
- Olaya-Sanchez, D., et al. Fgf3 and Fgf16 expression patterns define spatial and temporal domains in the developing chick inner ear. Brain Structure & Function. 222 (1), 131-149 (2017).
- Jones, J. M., Warchol, M. E. Expression of the Gata3 transcription factor in the acoustic ganglion of the developing avian inner ear. The Journal of Comparative Neurology. 516 (6), 507-518 (2009).
- Serralbo, O., et al. Transgenesis and web resources in quail. Elife. 9, 56312 (2020).

