Epicardial Outgrowth Culture Assay and Ex Vivo Assessment of Epicardial-derived Cell Migration
Summary
Here, we describe methods for isolating primary mouse epicardial cells by an outgrowth culture assay and assessing the functional migration of epicardial-derived cells (EPDC) using an ex vivo heart culture system. These protocols are suitable for identifying genetic and chemical modulators of epicardial epithelial-to-mesenchymal transition (EMT) and motility.
Abstract
A single layer of epicardial cells lines the heart, providing paracrine factors that stimulate cardiomyocyte proliferation and directly contributing cardiovascular progenitors during development and disease. While a number of factors have been implicated in epicardium-derived cell (EPDC) mobilization, the mechanisms governing their subsequent migration and differentiation are poorly understood. Here, we present in vitro and ex vivo strategies to study EPDC motility and differentiation. First, we describe a method of obtaining primary epicardial cells by outgrowth culture from the embryonic mouse heart. We also introduce a detailed protocol to assess three-dimensional migration of labeled EPDC in an organ culture system. We provide evidence using these techniques that genetic deletion of myocardin-related transcription factors in the epicardium attenuates EPDC migration. This approach serves as a platform to evaluate candidate modifiers of EPDC biology and could be used to develop genetic or chemical screens to identify novel regulators of EPDC mobilization that might be useful for cardiac repair.
Introduction
The epicardium is a single layer of mesothelial cells that lines the heart and impacts cardiac development, maturation and repair. Through a highly coordinated exchange of paracrine signals, the intimate dialogue between the myocardium and epicardium is indispensable for cardiac growth and the formation of non-myocyte cardiac lineages1. Epicardial-derived cells (EPDC) emerge from a subset of epicardial cells through an epithelial-to-mesenchymal transition (EMT)2, invade the subepicardial space and underlying myocardium, and differentiate largely into coronary vascular mural cells and cardiac fibroblasts, and to a lesser extent, endothelial cells and cardiomyocytes3-9.
The mechanisms regulating epicardial EMT and EPDC invasion have been attributed to the coordinated action of various molecules including secreted ligands10-13, cell surface receptors and adhesion molecules14,15, regulators of apical-basal polarity16,17, small GTPases18,19, and transcription factors2,20,21. While many of the molecular effectors of EPDC migration have been defined, a better understanding of the physiological signals that stimulate EPDC mobilization in the embryo may accelerate the development of strategies to manipulate this process in the adult for improved cardiac repair.
Studies aimed at identifying novel regulators of EPDC mobilization rely on purification of this cell population or tracing the migration of individual epicardial cells. One or a combination of marker genes, including Wt1, Tcf21, Tbx18, and/or Aldh1a2, is commonly used to identify the fetal epicardium1. However, the use of these markers to track migrating EPDC is not optimal as expression of epicardial markers wanes over the course of heart development and is often lost when epicardial cells undergo transdifferentiation into mesenchymal cells.
The application of the Cre/loxP system, in conjunction with lineage-tracing reporters, has been useful in permanently labeling the epicardium and its descendants during heart development and following ischemic injury in the adult7,22,23. Several epicardial-restricted Cre lines have been generated and are widely used to label EPDC and for conditional gene deletion strategies1. These studies have led to the characterization of various epicardial lineages, and the identification of critical mediators of EPDC motility and differentiation. However, accumulating evidence also suggests the epicardium is a heterogeneous population of progenitor cells4,24,25. Thus, only a subset of epicardial cells will be targeted using a given Cre driver.
In order to label the entire epicardium regardless of Cre specificity, a number of laboratories have utilized outgrowth culture systems or ex vivo organ culture methods to faithfully isolate or label epicardial cells without depending upon the expression of a genetic lineage markers26-28. For ex vivo migration studies, embryonic hearts are isolated prior to epicardial EMT and cultured in medium supplemented with a green fluorescent protein (GFP)-expressing adenovirus (Ad/GFP)9,18. This approach allows for efficient labeling of the entire epicardium rather than a subset of cells by Cre-mediated recombination. Heart cultures are subsequently exposed to known inducers of EMT to stimulate the mobilization of EPDC28,29. Ex vivo and in vivo analyses are complemented by in vitro epicardial explant cultures, which are a particularly useful approach for exploring detailed mechanisms driving EPDC migration.
Here, we describe methods for isolating primary epicardial cells for in vitro studies of epicardial EMT, as well as an organ culture system for ex vivo analyses of EPDC motility. We have recently demonstrated the utility and robustness of this method by genetically modulating the myocardin-related transcription factor (MRTF)/serum response factor (SRF) signaling axis to manipulate actin-based migration of EPDC9. While our findings highlight one signaling pathway necessary for regulating EPDC migration, these methods are suitable for unraveling the mechanisms that collectively orchestrate EPDC migration and differentiation. Furthermore, the explant and ex vivo culture systems could be implemented in functional screens to identify novel regulators of EPDC mobilization for therapeutic applications in cardiac regeneration.
Protocol
NOTE: All experimentation with mice were approved by the University Committee on Animal Resources at the University of Rochester.
1. Epicardial Outgrowth Culture Assay (Figure 1A)
- Preparations
- Prepare 5 ml medium A for primary epicardial cell isolation by supplementing Medium 199 (M199) with 5% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin (Pen/Strep).
- Pre-warm medium A and 100 ml Hanks' Balanced Salt Solution (HBSS) in a 37 °C water bath.
- Allow collagen-coated chamber slides to warm to room temperature for 30 min.
- Add 100 µl medium A to each well of the chamber slide and gently rotate to evenly coat the collagen surface. Repeat for 8-12 wells. Remove media after coating chambers.
- Aliquot pre-warmed HBSS into two 10 cm2 plates containing 50 ml each of HBSS.
- Isolate Embryonic Hearts from Pregnant Dam at 11.5 Days Post Coitum (dpc).30
- Anesthetize mouse with 0.5 ml of 13 mg/ml ketamine/0.88 mg/ml xylazine cocktail in 1x Dulbecco's Phosphate-Buffered Saline (DPBS) via intraperitoneal injection. Confirm proper anesthetization using a toe pinch test. A gentle pinch is sufficient to determine if the mouse is responsive. Any movement or paw withdraw indicates the animal is not sufficiently anesthetized to do surgery. Euthanize by cervical dislocation.
- Lay the mouse facing ventral side up and spray the abdomen with 70% ethanol to reduce contamination by hair.
- Pinch the skin with forceps and cut a lateral incision at the abdomen with surgical scissors. Hold the skin firmly and then pull apart in opposing directions towards the head and tail.
- Pinch the peritoneum with forceps and cut with scissors to expose the abdominal cavity.
- Carefully remove the decidua by cutting along the mesometrium. Transfer the dissected decidua to pre-warmed HBSS in the first 10 cm2 plate.
- Separate the decidua by cutting between embryos.
- Transfer an embryo to the second 10 cm2 plate with pre-warmed HBSS under a dissecting microscope. Carefully remove the extraembryonic tissue and yolk sac, then decapitate the embryo.
- Pinch the thoracic wall with forceps and tear the tissue apart at the midline, exposing the chest cavity.
- Place the forceps behind the heart. Grasp the outflow tract and pull the heart away from the embryo. Separate the pulmonary tract and lungs from the heart, if still attached.
- Transfer the heart to a single well of the collagen-coated slide and place it dorsal side down (Figure 1B). Repeat steps 1.2.7-1.3 for each embryo.
- Once all hearts have been transferred to individual wells, incubate the chamber slide for 30 min at 37 °C, 5% CO2 to allow sufficient adherence to the collagen matrix.
- Carefully add 20-50 µl of pre-warmed explant medium A around each heart. Important: Do not add the medium too quickly or to a level above the heart, otherwise hearts may detach from the collagen substrate.
- Culture the hearts at 37 °C, 5% CO2 for approximately 24 hr to allow epicardial outgrowth. Note: Outgrowths can be observed as early as 3-4 hr of culture (Figure 1C) and primarily consists of mesothelial cells displaying cobblestoned morphology, with minimal mesenchymal cell contamination (Figures 1D, E). Representative results were acquired using a dissection microscope equipped with a camera.
2. Genetic Ablation and Induction of EMT in Epicardial Outgrowths
Caution: Handle adenoviruses with care according to approved biosafety guidelines.
- Prepare 15 ml medium B (M199 supplemented with 1% FBS and 1% Pen/Strep) and pre-warm to 37 °C in a water bath.
- For excision of floxed alleles, prepare explant medium B supplemented with 2.5 x 104 pfu/ml of adenovirus expressing Cre recombinase (Ad/Cre) or control virus.
Note: Adenoviral mediated overexpression of candidate genes can also be carried out to evaluate gain of function phenotypes. Viral titers should be determined by the end user. - Carefully remove epicardium-depleted hearts from outgrowth cultures using forceps. Transfer hearts to 1.5 ml tubes with 800 µl Trizol and store at -80 °C for later RNA isolation and gene expression analysis.
- Wash each well with DPBS to remove blood cells and excess cellular debris.
- Add 500 µl explant medium B supplemented with adenovirus(es) to each outgrowth culture and incubate for 24 hr at 37 °C, 5% CO2.
- For EMT induction, remove media and replenish with 37 °C explant medium B supplemented with recombinant transforming growth factor (TGF)-β1 [10 ng/ml] or vehicle (4 mM hydrochloric acid (HCl) containing 1 mg/ml bovine serum albumin (BSA)). Culture explants for an additional 24 hr at 37 °C, 5% CO2. Proceed with gene/protein expression analyses or immunocytochemistry as detailed in the subsequent protocols.
Note: Induction of epicardial EMT has been stimulated with various concentrations of TGF-β1 ranging from 0.5 ng/ml – 10 ng/ml26,31-33. Higher concentrations of ligand may result in non-specific binding to all TGF-β receptors. The concentration of TGF-β1 should reflect the specific experimental goals.
3. Gene Expression Analysis of Epicardial Outgrowths
- Thaw epicardium-depleted hearts previously stored in Trizol (2.2).
- Aspirate media from EPDC cultures and wash twice with DPBS.
- Add 800 µl room temperature Trizol to explant cultures.
- Incubate epicardium-depleted hearts and EPDC cultures in Trizol for 5 min at room temperature. Pipette up and down ten times to homogenize the lysed sample.
Note: Hearts may require further pipetting to completely homogenize. - Transfer lysates to 1.5 ml tubes and proceed with total RNA isolation per manufacturer's instructions. Add 10 µg glycogen prior to precipitation to increase RNA yield. A given explant will yield approximately 0.5-1.0 µg RNA.
- Remove contaminating DNA with DNase I and reverse transcribe 0.5 µg mRNA per manufacturer's instructions.
- Validate the purity of epicardial explants and/or induction of EMT with quantitative reverse transcription (qRT)-PCR 34 using SYBR green and the markers described in Table 1. Normalize mRNA levels to glyceraldehyde 3-phosphate dehydrogenase (Gapdh) and calculate relative expression using the 2–ΔΔCt method 35. Typical results are shown in Figure 2A.
4. Protein Analysis of Epicardial Outgrowths
- Aspirate media from EPDC cultures and wash twice with DPBS.
- Add 10 µl ice-cold protein lysis buffer (50 mM Tris pH 7.5, 150 mM sodium chloride, 1 mM ethylenediaminetetraacetic acid pH 8.0, 1% Triton X-100, 1x protease inhibitor cocktail) to each chamber well and place the chamber slide on ice. Cut off approximately ¼ of the bottom on a P200 pipette tip and use the remaining piece to scrape the cells off of the slide. Pipette up and down to lyse epicardial cells and transfer to a 1.5 ml tube.
- Sonicate lysed explants on low (160 W) for 5 min in an ice water bath with 30 sec ON and OFF cycles.
- Centrifuge lysates for 10 min at 10,000 x g, 4 °C. Transfer supernatant to a fresh tube and determine the protein concentration using a protein assay36.
Note: A given explant will yield up to 1.5 µg total protein. We have found that pooling explants from two hearts and loading 2 µg protein per well is sufficient to detect smooth muscle α-actin (ACTA2) and GAPDH 9. The detection of other target proteins may require pooling multiple explant samples as determined by the end user.
5. Immunocytochemistry of Epicardial Outgrowths
- Remove media from explant cultures and wash twice with DPBS.
- Gently add 500 µl of 4% (v/v) paraformaldehyde or ice-cold methanol to each well and fix explants for 5 min at room temperature or 10 min at -20 oC, respectively. Fix according to antibody specifications as recommended by the manufacturer.
- Remove fixative and rinse each well with DPBS three times for 5 min.
- Permeabilize the cells with 500 µl of 0.1% (v/v) Triton X-100 in DPBS for 5 min.
- Remove permeabilization solution and rinse with DPBS three times for 5 min.
- Block the cells with 10% serum in DPBS for 30 min at room temperature.
- Proceed with primary antibody staining per manufacturer's recommendations and optimal conditions as determined by the end user.
- Remove antibody solution and rinse with DPBS three times for 5 min.
- Apply secondary antibody per manufacturer's instructions and counterstain with an appropriate nuclear dye.
- Remove antibody solution and rinse with DPBS three times for 5 min.
- Remove chamber slide walls per manufacturer’s instructions and add aqueous mounting medium directly to the cells. Place a coverslip over the cells and seal with nail polish. Typical results are shown in Figure 2B–2D using inverted confocal scanning microscopy.
6. Ex Vivo Culture Assay (Figure 3A)
- Preparations
- Prepare 12 ml ex vivo culture medium using a (1:1) mixture of Dulbecco's Modified Eagle Medium (DMEM):M199 supplemented with 10% FBS and 1% Pen/Strep.
- In a 37 °C water bath, pre-warm ex vivo culture medium and 100 ml HBSS.
- Aliquot 1 ml pre-warmed medium into 8-12 wells of a 24 well plate.
- Transfer pre-warmed HBSS to two 10 cm2 plates, containing 50 ml each.
- Isolate mouse embryonic hearts in HBSS (as described in 1.2) from pregnant dams at 12.5 dpc.
- Transfer each heart to a single well of the 24 well plate containing ex vivo culture medium. Incubate the plate at 37 °C, 5% CO2 with gentle rocking manually or using a rocking platform to prevent adherence. Immediately proceed to step 7.1.
7. Epicardial Labeling and EMT Induction in Ex Vivo Cultures
- To label the epicardium, prepare 12 ml of 37 °C ex vivo culture medium supplemented with 3.0 x 106 pfu/ml of Ad/GFP. For targeted ablation of floxed alleles, transfer 6 ml of Ad/GFP supplemented culture medium to a new 15 ml conical tube. Add Ad/Cre or control adenovirus to a final concentration of 1.5 x 106 pfu/ml.
Note: Gain of function analyses can also be carried out with appropriate adenoviral mediated gene delivery. - Carefully remove the culture medium from each well of the 24 well plate using a P1,000 pipette tip.
- Gently add culture medium supplemented with adenovirus(es) to each well.
- Incubate the plate at 37 °C, 5% CO2 for 24 hr with gentle rocking.
- For EMT induction, prepare 12 ml of 37 °C ex vivo culture medium containing 10 ng/ml TGF-β1 and 20 ng/ml platelet-derived growth factor (PDGF)-BB or vehicle (4 mM HCl containing 1 mg/ml BSA).
- Carefully remove culture medium from each well of the 24 well plate using a P1,000 pipette tip. Gently rinse the hearts with 1 ml DPBS.
- Remove the DPBS and replenish hearts with ex vivo culture medium containing TGF-β1 and PDGF-BB or vehicle.
- Incubate for 24 hr at 37 °C, 5% CO2 with gentle rocking.
8. Cryopreservation and Immunohistochemistry of Ex Vivo Cultures
- Prepare the Hearts for Cryopreservation.
- Remove the culture medium and wash hearts twice with ice-cold DPBS.
- Add 1 ml of 4% (v/v) paraformaldehyde to each heart and fix for 2 hr at 4 °C with gentle rocking.
- Remove the fixative and wash hearts twice with DPBS. Transfer the hearts to fresh wells with 1 ml 10% (w/v) sucrose prepared in DPBS. Incubate the plate overnight at 4 °C with gentle rocking.
- Carefully transfer hearts to new wells with 18% (w/v) sucrose prepared in DPBS. Incubate the plate overnight at 4 °C with gentle rocking.
- Transfer each heart to a cryomold containing tissue freezing medium. Immediately freeze blocks using liquid nitrogen and store at -80 °C.
- Section hearts at 5 µm using a cryostat and place tissue sections on positively charged microscope slides. Store slides at -80 °C until staining.
- Perform immunohistochemistry on sections for detection of the sub-epicardial basement membrane.
- Thaw slides for at least 30 min at room temperature.
- Rehydrate the sections by washing twice in DPBS for 5 min with gentle rocking.
- Permeabilize section with 0.1% (v/v) Triton X-100 in PBS for 5 min at room temperature.
- Wash the slides twice in DPBS for 5 min with gentle rocking.
- Blot off excess DPBS and define the staining region with a hydrophobic labeling pen. Block sections with 10% serum in DPBS for 30 min at room temperature.
- Remove block and apply collagen type IV antibody (ColIV, 1:200) diluted in block. Incubate overnight at 4 °C in a humidity-controlled chamber.
- Wash the slides three times in DPBS for 5 min with gentle rocking.
- Dilute fluorescently conjugated secondary antibody (1:500) and DAPI (1:5,000) in DPBS and apply to the slides. Incubate for 1 hr at room temperature. Choose an appropriate secondary antibody with an emission and excitation wavelength distinct from GFP.
- Wash the slides three times in PBS for 5 min with gentle rocking. Mount slides with mounting medium.
9. Imaging and Quantification of Ex Vivo Cultures
- Have a blinded user image and quantify stained sections. Image at least five non-consecutive tissue sections from a given heart in 4-5 arbitrary locations along the epicardium (edge of the heart) using a fluorescent or confocal microscope at 40X magnification. Typical results are shown in Figure 3B using inverted confocal scanning microscopy.
- Manually count the total number of GFP positive cells in a given image. Identify migrating EPDC as GFP positive cells located within or beyond the ColIV subepicardial basement membrane. Determine the percent of GFP migrating cells as the number of migrating GFP positive cells over the total number of GFP positive cells in a given image. Typical results are shown in Figure 3C-D using inverted confocal scanning microscopy.
Representative Results
The epicardium can be efficiently isolated using an outgrowth culture assay by taking advantage of its exterior location and exploiting the developmental plasticity and intrinsic migratory behavior of EPDC. Murine embryonic hearts are isolated at E11.5 prior to epicardial EMT and cultured dorsal side down on collagen-coated chamber slides26 (Figure 1A, B). Explanted hearts will continue to contract; however, the epicardium will adhere to the collagen matrix and migrate off from the myocardium within 24 hr of culture (Figure 1C). Epicardial cells can be observed emanating away from the heart with cobblestone-like morphology, indicative of mesothelial identity (Figures 1C-E). After 24 hr of culture, epicardium-depleted hearts are removed for gene or protein expression analysis.
In addition to observing epithelial morphology, the purity of epicardial cell cultures can be further validated by the expression of epicardial restricted gene and protein markers. Epicardial cell explants display robust expression of epicardial and mesothelial markers (Aldh1a2, Tcf21, and Wt1) and low expression of cardiomyocyte genes (Nkx2-5 and Myh7) compared to epicardium-depleted hearts (Figure 2A). To further verify identity, epicardial cell cultures are fixed 48 hr after heart removal and co-stained with antibodies against Wilm's tumor 1 (WT1) and zona occludens protein 1 (ZO1) to label mesothelial cells and intercellular tight junctions, respectively37,38 (Figure 2B). This outgrowth culture method yields a relatively high purity of mesothelial cells; however, users may observe a small percentage of cell contamination. Fibroblasts and smooth muscle cells exhibit an elongated mesenchymal phenotype with a notable absence of nuclear WT1 and organized tight junctions (Figure 2C). Troubleshooting techniques to limit the amount of non-epicardial cell contamination are expanded in the discussion.
The explant outgrowth assay is a useful culture system for interrogating epicardial biology in vitro. To demonstrate epicardial cell plasticity, explants were stimulated with TGF-β1 [10 ng/ml] to induce transdifferentiation into mesenchymal cells26,28. After 24 hr of stimulation, EPDC adopt a mesenchymal phenotype with reduced ZO1 staining and robust de novo formation of smooth muscle α-actin positive stress fibers (ACTA2 or SMA), particularly at the periphery of the culture39 (Figure 2D, red). In contrast to stress fiber assembly at the periphery of explants, confluent epicardial cells with more defined intercellular contacts (ZO1, green) at the center of a monolayer display ACTA2 protein expression in cortical actin.
In addition to undergoing EMT, EPDC will invade the myocardium and differentiate, constituting a substantial proportion of non-cardiomyocyte populations. The motility of EPDC can be assessed using a previously established ex vivo organ culture system whereby the exterior of E12.5 hearts are labeled with an adenovirus expressing GFP9,18 (Figure 3A). Labeled hearts are further cultured for three days in the presence of TGF-β1 [10 ng/ml] and PDGF-BB [20 ng/ml] to promote EPDC migration. Organ cultures are periodically rotated to prevent adhesion of ex vivo hearts to the surface of culture plates. This system allows for efficient labeling of the entire epicardium and assessment of EPDC motility in a three-dimensional model as opposed to directional migration with in vitro scratch assays. The migration of EPDC is observed as the invasion of GFP positive epicardial cells into or beyond the ColIV (red) subepicardial basement membrane16 (Figures 3B, C). To illustrate the utility of this system, we have recently shown that manipulation of the MRTF/SRF signaling axis can influence the motility of EPDC9. Deletion of Mrtfa and Mrtfb by Cre-mediated excision of floxed alleles (MRTFdKO) attenuates the migration of EPDC into the subepicardial space. Conversely, forced expression of MRTF-A (Ad/MRTF-A) in C57BL/6 hearts results in increased EPDC migration and disruption of the ColIV basement membrane (Figures 3C, D).
| Gene | Forward | Reverse | NCBI Accession |
| Aldh1a2 | GGCACTGTGTGGATCAACTG | TCACTTCTGTGTACGCCTGC | NM_009656 |
| Gapdh | CGTGCCGCCTGGAGAAAC | TGGGAGTTGCTGTTGAAGTCG | NM_001289726 |
| Myh7 | GTGGCTCCGAGAAAGGAAG | GAGCCTTGGATTCTCAAACG | NM_080728 |
| Nkx2-5 | GACGTAGCCTGGTGTCTCG | GTGTGGAATCCGTCGAAAGT | NM_008700 |
| Tcf21 | CATTCACCCAGTCAACCTGA | CCACTTCCTTCAGGTCATTCTC | NM_011545 |
| Wt1 | ATCCGCAACCAAGGATACAG | GGTCCTCGTGTTTGAAGGAA | NM_144783 |
Table 1: Sequences of Forward and Reverse Primers Used for Validation of Epicardial Outgrowth Culture Purity by Analysis of Epicardium and Myocardium Restricted Gene Expression. Gene expression is determined by the qRT-PCR with the following parameters: denature at 95 °C for 10 sec; anneal at 60 °C for 30 sec; repeat 50x cycles.
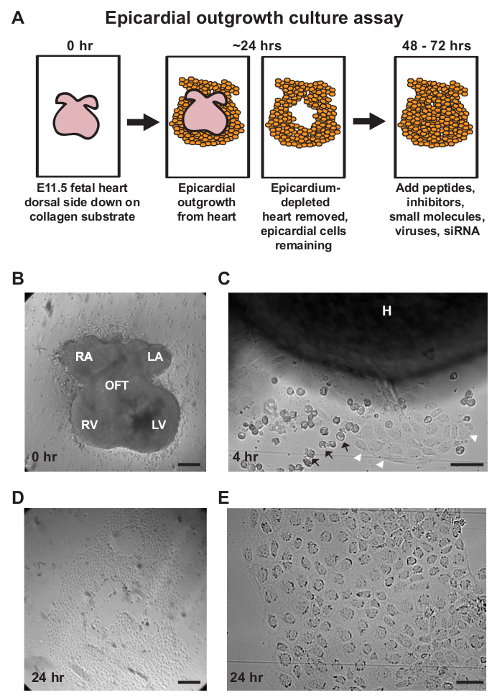
Figure 1: Isolation of Primary Epicardial Cells Using an Outgrowth Culture Assay. (A) A schematic representation of the epicardial outgrowth explant assay with a recommended timeline for epicardial modulation and analysis. (B–E) Representative brightfield images of various time points during the explant assay. (B) Embryonic day E11.5 murine hearts are placed dorsal side down on a collagen substrate. (C) Epicardial cells (white arrowheads) will begin to migrate off the heart within 3-4 hr of culture. Blood cells (black arrows) may accumulate nearby or cover explants during epicardial outgrowth. (D, E) After approximately 24 hr of culture, hearts are removed and epicardial monolayers remain adhered to the chamber slide. Cellular outgrowths display epithelial morphology with a cobblestone-like arrangement of cells. Scale bars = 250 µm (B-D) or 50 µm (C, E). H = heart, LA = left atrium, LV = left ventricle, OFT = outflow tract, RA = right atrium, RV = right ventricle. Please click here to view a larger version of this figure.
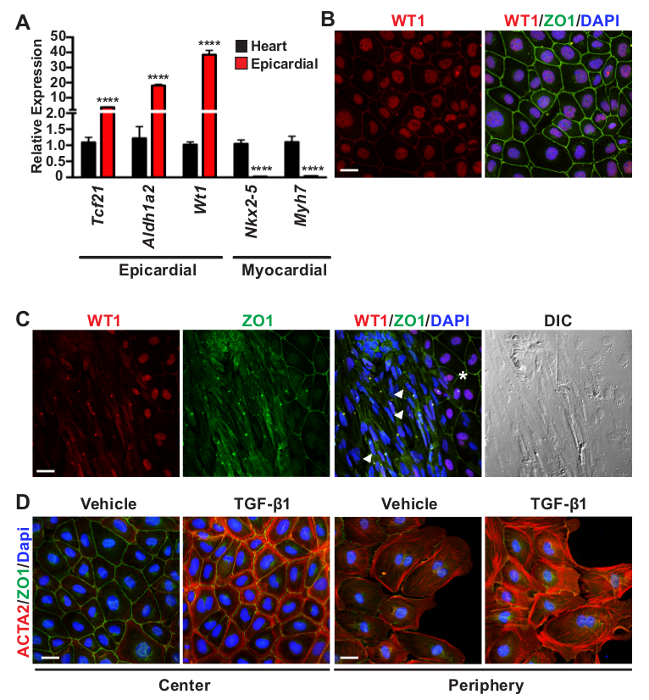
Figure 2: Evaluation of Epicardial Cell Outgrowth Purity. (A) qRT-PCR validation of epicardial restricted markers (Tcf21, Aldh1a2, and Wt1) in outgrowth cultures compared to myocardial gene expression (Nkx2-5 and Myh7) in epicardium-depleted E11.5 hearts. Data represent the mean ± SEM (n = 8 per group). Asterisks denote statistical significance using a Student T-test with ****P <0.0001. (B) Representative co-immunofluorescent staining of unstimulated outgrowths for epicardial expression (WT1, red), intercellular tight junctions (ZO1, green), and nuclei (DAPI, blue). (C) An example of cell contamination in an outgrowth culture. Epicardial cells (asterisk) are observed to the right of the panel with characteristic mesothelial identity (nuclear WT1, red) and organized epithelial tight junctions (ZO1, green). The adjacent cells in the center (white arrowheads) exhibit an elongated mesenchymal morphology, shown by differential interference contrast (DIC) imaging, with minimal nuclear WT1 protein and unorganized ZO1 staining. (D) Epicardial cell cultures were stimulated with vehicle (4 mM HCl in 1mg/ml BSA) or TGF-β1 [10 ng/ml] to induce epicardial EMT. TGF-β1 stimulation promotes robust expression of ACTA2 (red) in cortical regions of confluent cells at the center of explants or in stress fibers of cells migrating at the edge or periphery of the monolayer. Cell adhesions are labeled by ZO1 (green). Scale bars = 25 µm (B-D). Please click here to view a larger version of this figure.
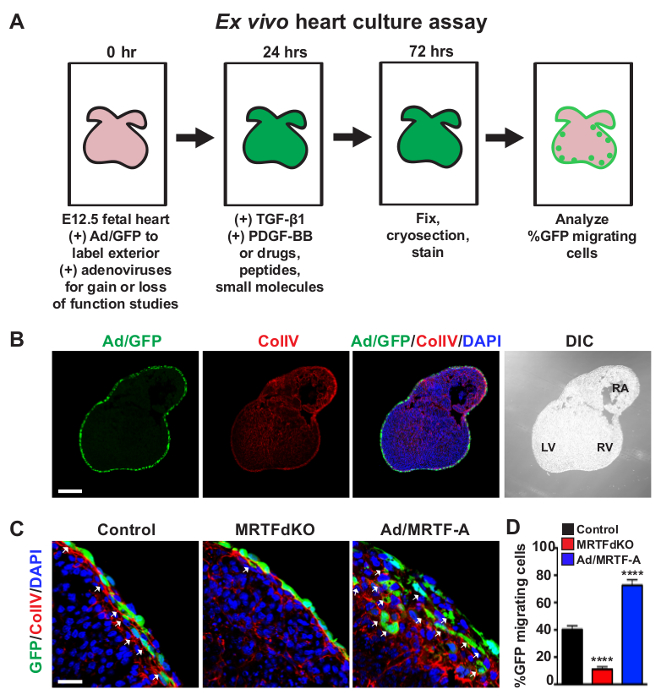
Figure 3: Ex Vivo Assessment of EPDC Motility. (A) A schematic timeline of the ex vivo heart culture assay. (B) A representative 5 µm section of a C57BL/6 E12.5 heart transduced with Ad/GFP (green) for 24 hr to label the exterior of the heart. Ex vivo hearts were subsequently cultured for an additional 48 hr with TGF-β1 [10 ng/ml] and PDGF-BB [20 ng/ml] to stimulate EPDC migration. Sections were co-stained for ColIV (red) to label the subepicardial basement membrane and nuclei (DAPI, blue). Ex vivo heart culture morphology is shown by DIC. LV = left ventricle, RV = right ventricle, RA = right atrium. (C, D) An example application of the ex vivo migration assay by modulating the MRTF/SRF regulatory axis. This figure has been modified from [9]. E12.5 hearts were isolated from Mrtfa−/−; Mrtfbfl/fl embryos and transduced with Ad/GFP and a control virus or an adenovirus expressing Cre recombinase for 24 hr. Ex vivo cultures were then stimulated with TGF-β1 [10 ng/ml] and PDGF-BB [20 ng/ml] for an additional 48 hr. EPDC motility (white arrows) is observed as the migration of GFP positive cells (green) into or beyond the ColIV (red). Cre-mediated deletion of Mrtfb in the Mrtfa-/- background (MRTFdKO) results in attenuation of EPDC migration compared to control hearts. Conversely, forced expression of MRTF-A (Ad/MRTF-A) in E12.5 C57BL/6 hearts enhances EPDC migration. (D) Quantification of %GFP positive cell migration. Data are presented as the mean ± SEM (n = 4 hearts for each condition). Data was analyzed using a one-way ANOVA with asterisks denoting statistical significance by Tukey post-hoc tests with ****P <0.0001. Scale bars = 200 µm (B) or 25 µm (C). All images were acquired using inverted confocal laser scanning microscopy. Please click here to view a larger version of this figure.
Discussion
Here we outline detailed methods to isolate primary epicardial cell by outgrowth and track EPDC migration in ex vivo heart cultures. For outgrowth cultures, the appropriate time to remove the epicardium-depleted heart varies slightly between experiments. Periodic monitoring of the explants after an overnight incubation is recommended to gauge the extent of epicardial outgrowth and to extract the heart before fibroblasts appear. To ensure purity, hearts should be removed within 24 hr of explanting to prevent accumulation of non-epicardial lineages. Trimming away the outflow tract and both atria prior to transferring the heart to a chamber slide can also reduce contaminating cell types27,40. Since fetal hearts are isolated from a mouse litter during a narrow window of time and cultured under the same conditions, the time for outgrowths to appear is relatively similar from one heart to another within an experiment. Therefore, all hearts should be removed when the majority of explants exhibit epicardial outgrowths. In some cases, epicardial cells do not migrate out efficiently from a heart that is contracting more vigorously than the others, in which case this explant must be excluded from the experiment.
The accumulation of blood cells can often conceal a flat monolayer of epicardial cells (arrows, Figure 1C), thus making it difficult to visualize outgrowths. To avoid excessive blood cell contribution, hearts can remain in HBSS for a few minutes upon dissection from the embryo. This allows sufficient time for the fetal heart to pump out residual blood in the heart chambers before being transferred to a chamber slide. Alternatively, supplementing the cultures with explant medium A after the first overnight incubation will help to flush away some excess blood and reveal outgrowths; however, the level of media should not rise above the heart as the surface tension can cause the heart to pull away from the substrate. Robust epicardial outgrowths are observed using collagen type I as the culture substrate; however, other substrates have been reported depending on the organism and/or application27,40-42. Using this protocol, primary fetal epicardial cells can thrive for 4 days following heart removal and daily medium replenishment; however, experimental designs requiring longer periods of culture should be determined by the end user. Since epicardial EMT is strongly activated during embryonic development, this outgrowth culture technique is limited to the enrichment of fetal epicardial cells. Other groups have described methods to isolate adult epicardial cells43,44.
For ex vivo migration assays, heart extraction and epicardial cell labeling are recommended at E12.5 for two reasons. First, hearts isolated after E12.5 are larger and hence require more perfusion to support viability. Simple diffusion of nutrients is not sufficient to maintain organ cultures from larger hearts. Second, epicardial EMT and EPDC migration occur around E12.5. Therefore, the epicardium should be labeled with an adenovirus expressing a fluorescent protein immediately after heart extraction to maximize the detection of EPDC. Alternatively, carboxyfluorescein diacetate succinimidyl ester (CFSE) has been previously reported to label the exterior of the heart and trace EPDC migration10,45.
In contrast to outgrowth assays, ex vivo cultures should be rocked periodically to avoid heart attachment and epicardial cell outgrowth. Cultures could also be continuously agitated in an incubator equipped with a rocker on the lowest speed setting. Users will notice changes in cardiac morphology within a few days of organ culture and should avoid analyses beyond 72 hr of heart isolation. Using this assay, EPDC invasion is observed within 48 to 72 hr of culture. While the Cre-loxP system has been used in vivo to map the localization of EPDC to the coronary vasculature, the myocardium interstitium, and the interventricular septum7,22, this ex vivo technique is limited to EPDC migration within a few cell layers of the sub-epicardial space. Thus, this method is more suitable for determining regulators of EPDC motility and invasion rather than defining the final destination of migrating EPDC.
Epicardial cell isolations and ex vivo migration assays, in combination with gain or loss of function approaches, have already proven useful for evaluating candidate genes that might impact EPDC biology9,18,46. These methods also provide a platform for gain-of-function or loss-of-function screens to identify novel regulators of EPDC migration. This strategy, in conjunction with fluorescence activated cell sorting or single cell RNA-sequencing, may also provide an opportunity to probe epicardial heterogeneity and EPDC differentiation in greater detail. Finally, modifications of these procedures could be used to develop screens for small molecules that impact epicardial cell and EPDC biology for the purpose of cardiac regenerative medicine.
Disclosures
The authors have nothing to disclose.
Acknowledgements
E.M.S. was supported by grants from the National Institutes of Health (NIH) [grant number R01HL120919]; the American Heart Association [grant number 10SDG4350046]; University of Rochester CTSA award from the NIH [grant number UL1 TR000042]; and startup funds from the Aab CVRI. M.A.T. was supported in part by a grant to the University of Rochester School of Medicine and Dentistry from the Howard Hughes Medical Institute Med-Into-Grad Initiative; and an Institutional Ruth L. Kirschstein National Research Service Award from the NIH [grant number GM068411].
Materials
| Dulbecco's Modified Eagle Medium | Hyclone | SH3002201 | DMEM |
| Hanks' Balanced Salt Solution | Hyclone | SH3003102 | HBSS |
| Medium 199 | Hyclone | SH3025301 | M199 |
| Dulbecco’s Phosphate-Buffered Saline | Hyclone | SH3002802 | DPBS |
| Fetal Bovine Serum | Gemini | 100-106 | FBS |
| Penicillin/Streptomycin Solution | Hyclone | SV30010 | |
| Corning Biocoat Collagen I, 4-Well Culture Slides | Corning | 354557 | |
| Multiwell 24-well plates | Falcon | 08-772-1 | |
| Dissecting microscope | Leica | M50 | Equipped with Leica KL300 LED might source |
| Confocal miscroscope | Olympus | 1X81 | Equipped with muli-argon laser 405/488/515, FV10-MCPSU laser 405/440/635, 559 laser, and mercury lamp |
| Bioruptor | Diagenode | UCD-200 | |
| Transforming growth factor beta 1 | R&D Systems | 100-B-001 | TGF-β1 |
| Platelet-derived growth factor BB | R&D Systems | 220-BB-010 | PDGF-BB |
| Trizol Reagent | Applied Biosystems | 15596-026 | Caution, hazardous material |
| TURBO DNA-free Kit | Life Technologies | AM1907 | DNaseI |
| iScript cDNA Synthesis Kit | BioRad | 1708891 | |
| UltraPure Glycogen | Life Technologies | 10814-010 | |
| SYBR Green | BioRad | 170-8880 | |
| Protease inhibtor cocktail tablets | Roche | 11836170001 | |
| Alexa Goat-anti-rabbit 488 | Life Technologies | A11008 | Secondary antibody |
| Alexa Goat anti-rabbit 594 | Life Technologies | A-11012 | Secondary antibody |
| Alexa Goat anti-mouse 594 | Life Technologies | A-11005 | Secondary antibody |
| Mouse anti-smooth muscle alpha actin, Cy3-conjugated | Sigma-Aldrich | C6198 | monoclonal antibody, clone 1A4 |
| Mouse anti-Wilms tumor 1 (WT1) | Life Technologies | MA1-46028 | monoclonal antibody, clone 6F-H2 |
| Rabbit anti-ZO1 | Life Technologies | 40-2200 | polyclonal antibody |
| Rabbit anti-Collagen Type 4 | Millipore | AB756P | polyclonal antibody |
| 4',6-Diamidino-2-Phenylindole, Dihydrochloride | Life Technologies | D1306 | DAPI |
| Fluorescent mounting medium | DAKO | S3023 | |
| 16% Paraformaldehyde solution | Electron Microscopy Sciences | 15710 | PFA diluted to 4% in DPBS. Caution, hazardous material |
| Sucrose | Sigma-Aldrich | S0389 | |
| Tissue Freezing Medium | Triangle Biomedical Sciences | TFM-B | |
| Pap pen | DAKO | S2002 | |
| Disposable mictrotome blades | Sakura Finetek | 4689 | |
| Microscope slides | Globe Scientific | 1358W | positively charged, 25 x 75 x 1mm |
| Cryostat | Leica | CM1950 | |
| Forceps | Fine Science Tools | 11295-00 | |
| Surgical Scissors | Fine Science Tools | 91460-11 | |
| Disposable base molds | Fisher Scientific | 22-363-553 | |
| Ketamine | Hospira | NDC 0409-2051-05 | |
| Xylazine | Akorn | NADA 139-236 |
References
- von Gise, A., Pu, W. T. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res. 110, 1628-1645 (2012).
- Martinez-Estrada, O. M., et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet. 42, 89-93 (2010).
- Gittenberger-de Groot, A. C., Vrancken Peeters, M. P., Mentink, M. M., Gourdie, R. G., Poelmann, R. E. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 82, 1043-1052 (1998).
- Katz, T. C., et al. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell. 22, 639-650 (2012).
- Mikawa, T., Gourdie, R. G. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 174, 221-232 (1996).
- Wilm, B., Ipenberg, A., Hastie, N. D., Burch, J. B., Bader, D. M. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 132, 5317-5328 (2005).
- Zhou, B., et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 454, 109-113 (2008).
- Dettman, R. W., Denetclaw, W., Ordahl, C. P., Bristow, J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 193, 169-181 (1998).
- Trembley, M. A., Velasquez, L. S., de Mesy Bentley, K. L., Small, E. M. Myocardin-related transcription factors control the motility of epicardium-derived cells and the maturation of coronary vessels. Development. 142, 21-30 (2015).
- Mellgren, A. M., et al. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ Res. 103, 1393-1401 (2008).
- von Gise, A., et al. WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Dev Biol. 356, 421-431 (2011).
- Lavine, K. J., et al. Endocardial and Epicardial Derived FGF Signals Regulate Myocardial Proliferation and Differentiation In Vivo. Dev Cell. 8, 85-95 (2005).
- Sanchez, N. S., Barnett, J. V. TGFbeta and BMP-2 regulate epicardial cell invasion via TGFbetaR3 activation of the Par6/Smurf1/RhoA pathway. Cell Signal. 24, 539-548 (2012).
- Dettman, R. W., Pae, S. H., Morabito, C., Bristow, J. Inhibition of 4-integrin stimulates epicardial-mesenchymal transformation and alters migration and cell fate of epicardially derived mesenchyme. Dev Biol. 257, 315-328 (2003).
- Rhee, D. Y., et al. Connexin 43 regulates epicardial cell polarity and migration in coronary vascular development. Development. 136, 3185-3193 (2009).
- Wu, M., et al. Epicardial spindle orientation controls cell entry into the myocardium. Dev Cell. 19, 114-125 (2010).
- Hirose, T., et al. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development. 133, 1389-1398 (2006).
- Baek, S. T., Tallquist, M. D. Nf1 limits epicardial derivative expansion by regulating epithelial to mesenchymal transition and proliferation. Development. 139, 2040-2049 (2012).
- Lu, J., et al. Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev Biol. 240, 404-418 (2001).
- Combs, M. D., Braitsch, C. M., Lange, A. W., James, J. F., Yutzey, K. E. NFATC1 promotes epicardium-derived cell invasion into myocardium. Development. 138, 1747-1757 (2011).
- Acharya, A., et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 139, 2139-2149 (2012).
- Zhou, B., von Gise, A., Ma, Q., Hu, Y. W., Pu, W. T. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev Biol. 338, 251-261 (2010).
- Zhou, B., et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 121, 1894-1904 (2011).
- Singh, M., Epstein, J. Epicardial Lineages and Cardiac Repair. Journal of Developmental Biology. 1, 141-158 (2013).
- Braitsch, C. M., Combs, M. D., Quaggin, S. E., Yutzey, K. E. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev Biol. 368, 345-357 (2012).
- Austin, A. F., Compton, L. A., Love, J. D., Brown, C. B., Barnett, J. V. Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFbeta. Developmental dynamics : an official publication of the American Association of Anatomists. 237, 366-376 (2008).
- Kim, J., Rubin, N., Huang, Y., Tuan, T. L., Lien, C. L. In vitro culture of epicardial cells from adult zebrafish heart on a fibrin matrix. Nat Protoc. 7, 247-255 (2012).
- Compton, L. A., Potash, D. A., Mundell, N. A., Barnett, J. V. Transforming growth factor-beta induces loss of epithelial character and smooth muscle cell differentiation in epicardial cells. Developmental dynamics : an official publication of the American Association of Anatomists. 235, 82-93 (2006).
- Smith, C. L., Baek, S. T., Sung, C. Y., Tallquist, M. D. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res. 108, 15-26 (2011).
- Shea, K., Geijsen, N. Dissection of 6.5 dpc Mouse Embryos. JOVE. 2, (2007).
- Witty, A. D., et al. Generation of the epicardial lineage from human pluripotent stem cells. Nature biotechnology. 32, 1026-1035 (2014).
- Iyer, D., et al. Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development. 142, 1528-1541 (2015).
- Takeichi, M., Nimura, K., Mori, M., Nakagami, H., Kaneda, Y. The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bi-directional regulation of Slug in murine primary epicardial cells. PloS one. 8, e57829 (2013).
- Wong, M. L., Medrano, J. F. Real-time PCR for mRNA quantitation. BioTechniques. 39, 75-85 (2005).
- Schmittgen, T. D., Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 3, 1101-1108 (2008).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 72, 248-254 (1976).
- Moore, A. W., McInnes, L., Kreidberg, J., Hastie, N. D., Schedl, A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 126, 1845-1857 (1999).
- Kim, J., et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci U S A. 107, 17206-17210 (2010).
- Kalluri, R., Weinberg, R. A. The basics of epithelial-mesenchymal transition. J Clin Invest. 119, 1420-1428 (2009).
- Dong, X. R., Maguire, C. T., Wu, S. P., Majesky, M. W. Chapter 9 Development of Coronary Vessels. Methods in Enzymology. 445, 209-228 (2008).
- Ruiz-Villalba, A., Ziogas, A., Ehrbar, M., Perez-Pomares, J. M. Characterization of epicardial-derived cardiac interstitial cells: differentiation and mobilization of heart fibroblast progenitors. PLoS One. 8, e53694 (2013).
- Garriock, R. J., Mikawa, T., Yamaguchi, T. P. Isolation and culture of mouse proepicardium using serum-free conditions. Methods. 66, 365-369 (2014).
- Smart, N., Riley, P. Derivation of epicardium-derived progenitor cells (EPDCs) from adult epicardium. Curr Protoc Stem Cell Biol. , (2009).
- Zhou, B., Pu, W. T. Isolation and characterization of embryonic and adult epicardium and epicardium-derived cells. Methods Mol Biol. 843, 155-168 (2012).
- Morabito, C. J., Dettman, R. W., Kattan, J., Collier, J. M., Bristow, J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev Biol. 234, 204-215 (2001).
- Grieskamp, T., Rudat, C., Ludtke, T. H., Norden, J., Kispert, A. Notch signaling regulates smooth muscle differentiation of epicardium-derived cells. Circ Res. 108, 813-823 (2011).

