DNA Sequence Recognition by DNA Primase Using High-Throughput Primase Profiling
Summary
Protein binding microarray (PBM) experiments combined with biochemical assays link the binding and catalytic properties of DNA primase, an enzyme that synthesizes RNA primers on template DNA. This method, designated as high-throughput primase profiling (HTPP), can be used to reveal DNA-binding patterns of a variety of enzymes.
Abstract
DNA primase synthesizes short RNA primers that initiate DNA synthesis of Okazaki fragments on the lagging strand by DNA polymerase during DNA replication. The binding of prokaryotic DnaG-like primases to DNA occurs at a specific trinucleotide recognition sequence. It is a pivotal step in the formation of Okazaki fragments. Conventional biochemical tools that are used to determine the DNA recognition sequence of DNA primase provide only limited information. Using a high-throughput microarray-based binding assay and consecutive biochemical analyses, it has been shown that 1) the specific binding context (flanking sequences of the recognition site) influences the binding strength of the DNA primase to its template DNA, and 2) stronger binding of primase to the DNA yields longer RNA primers, indicating higher processivity of the enzyme. This method combines PBM and primase activity assay and is designated as high-throughput primase profiling (HTPP), and it allows characterization of specific sequence recognition by DNA primase in unprecedented time and scalability.
Introduction
HTPP makes use of DNA binding microarray technology combined with biochemical analysis (Figure 1) to statistically identify specific features of DNA templates that affect the enzymatic activity of DNA primase. Therefore, HTPP provides a technological platform that facilitates a knowledge leap in the field. The classical tools used to determine primase recognition sites do not have the ability to yield massive amount of data, whereas HTPP does.
PBM is a technique routinely used to determine the binding preferences of transcription factors to DNA1,2; however, it is not suitable for detection of weak/transient binding of proteins to DNA. Unlike the universal PBM that provides information about average protein binding specificity to all possible sequences consisting of eight base pairs, HTPP is based on the library of single-stranded DNA templates comprising unique sequence elements. Such DNA sequence elements involve tens of thousands short (few tens of bp) genomic sequences, as well as computationally designed DNA sequences enriched in certain DNA repetitive sequence elements present in the genome, which possess different average GC content. Such a high-throughput approach allows determination of, in a systematic, quantitative, and hypothesis-driven way, the sequence-related properties that are important for primase binding and its enzymatic activity3. In particular, the important link between primase-DNA binding preferences, (modulated by DNA sequences flanking specific tri-nucleotide binding sites) and primase processivity has been identified for this enzymatic system4.
The new technology was applied to revisit our understanding of primase recognition sites even for the T7 DNA primase that has been extensively studied5. Specifically, re-examination of classical concepts, such as DNA recognition sites of T7 DNA primase (which were determined almost four decades ago 6) using protein-DNA binding microarray (PBM) has led to unprecedented insight into features related to the flanking sequence of these recognition sites3. It was expected that the sequences flanking tri-nucleotide recognition site of T7 DNA primase (5'-GTC-3') will be random. Instead, we found that TG-rich flanking sequences increase the chances of T7 DNA primase to synthesize longer RNA primers indicating an increase in processivity.
Other methods that can be used to study DNA-binding properties of proteins in vitro include the electrophoretic mobility shift assay (EMSA)7, DNase I footprinting8, surface-plasmon resonance (SPR)9, and Southwestern blotting10. These are, however, low-throughput methods only applicable to investigating a small number of DNA sequences. In addition, the precision and sensitivity of some of these techniques (e.g., EMSA) is low. On the other hand, in vitro selection11 is a technique that, similarly to PBM, can be used for the identification of numerous binding sequences. However, low affinity sequences are usually excluded in most applications of in vitro selection; therefore, this approach is not suitable for obtaining comparative binding data for all available sequences. The universal PBM1,2 is mainly used to characterize the binding specificities of transcription factors from prokaryotes and eukaryotes as well as specific factors (e.g., presence of certain ligands, cofactors, etc.) that may affect this interaction12.
HTPP expands the PBM application to DNA processing enzymes by combining unprecedented high-throughput statistical power with high precision to provide information on binding sequence context. Such data has not yet been obtained for primases and related enzymes (that have weak/transient binding to DNA) due to aforementioned technical limitations of other available techniques.
Protocol
1. Design of microarray
NOTE: DNA probes represent custom 36-nucleotide sequences, consisting of the recognition site for T7 DNA primase (GTC) located between two variable flanking regions, followed by a constant 24-nucleotide sequence tethered to a glass slide3. We used a 4 x 180,000 microarray format, which enabled spotting of each DNA sequence in six replicates, randomly distributed on the slide.
- Design of DNA library for primases with known recognition sequences
- Ensure that each sequence is composed of 60 nucleotides. Keep the first 24 nucleotides constant (to be attached to the glass slide). Variable regions should have the following general form: (N)16GTC(N)17; where N represents any desired nucleotide.
- Design the flanking region to address a specific scientific question (an example of the design is presented in the following text). The flanking regions may be designed to contain specific features such as the repeat elements or specific symmetry.
NOTE: We have created eight categories of different flanking sequences composed of two or three specific nucleotides: T and G (group 1); T and C (group 2); C and A (group 3); A and G (group 4); G, C and T (group 5); C, T and A (group 6); G, A and T (group 7); G, A and C (group 8). 2,000 different sequences were designed for each group. Each group had subgroups of sequences with different types and different numbers of sequence repeats. - Include a set of negative control sequences (lacking the specific binding site)4. The presence of such sequences allows to validate the occurrence of specific binding in the experiment.
- Design of DNA library for primases with unknown recognition sequences
- If the recognition sequence is unknown, create the DNA library by selecting the sequences (with or without specific features mentioned previously) from the genome of desired organism (bacteria, fungi, etc.).
- Design of microarray slide
- Purchase custom slides (e.g., 4x180K, AMADID #78366) from a commercial supplier (for more details see Table of Materials). Order each sequence in six replicates, where each replicate should be attached to a randomly selected spot on the slide.
2. Primase DNA binding experiment
NOTE: The day before (or at least 2 h before) the PBM, prepare the blocking solution [2% w/v skim milk in phosphate buffer saline (PBS)] and stir it on a magnetic stirrer. Prior to use, filter the solution with a 0.45 µM filter. To detect the primase binding to the DNA strands, several steps should be performed in the following order.
- Blocking procedure
- Pre-wet the microarray slide in a Coplin jar with 0.01% v/v Triton X-100 in PBS (5 min at 125 rpm on a lab rotator).
- Briefly wash the gasket slide (also referred to as coverslip) with water and ethanol and dry using compressed air.
- Assemble bottom part of steel hybridization chamber (PBM chamber) with the gasket slide facing up (commercial label facing up). For more details regarding the assembly, refer to the Table of Materials.
- Pipet blocking solution (2% w/v skim milk in PBS) into each chamber.
- Remove the microarray slide from the Coplin jar, then dry the non-DNA side (the DNA should be on the same side as the company label) and edges using a fine wipe. Slowly place the microarray on the gasket slide, avoiding bubbles (company label facing down). Immediately assemble and tighten steel hybridization chamber apparatus. Incubate for 1 h at room temperature (RT).
- Fill one staining dish (the “PBS” dish) with 800 mL of fresh PBS. Fill a second staining dish (the “WASH” dish) with 800 mL of freshly prepared 0.5% v/v Tween-20 in PBS.
- Unscrew the PBM chamber and remove the microarray slide-coverslip “sandwich”, taking care not to break the seal. Disassemble it underwater in the WASH dish by placing forceps between the slide and coverslip.
- Shake the microarray slide underwater and quickly transfer to a Coplin jar.
- Wash once with 0.1% v/v Tween-20 in PBS (5 min at 125 rpm on a lab rotator).
- Wash once with 0.01% v/v Triton X-100 in PBS (2 min at 125 rpm on a lab rotator).
- Quickly transfer the slide to a Coplin jar containing PBS.
- Protein binding
- Assemble the PBM chamber as previously described (steps 2.1.2–2.1.3).
- Pipette protein binding mixture containing 5 µM T7 DNA primase, 30 mM Tris-HCl pH 7.5, 6.5 mM MgCl2, 30 mM K-glutamate, 6 mM dithiothreitol (DTT), 65 μM ribonucleoside triphosphate (rNTP), 2% w/v skim milk, 100 ng/μL bovine serum albumin (BSA), 50 ng/μL salmon testes DNA, and 0.02% v/v Triton X-100 (TX-100) into each chamber of the gasket slide (without touching the rubber sides and without introducing bubbles).
- Rinse the microarray slide briefly by dipping it in the PBS dish, which removes excess detergent from the surface of the slides. Wipe the non-DNA surfaces of the slide with a fine wipe.
- Slowly lower the microarray slide (facing down) onto gasket slide, being careful to prevent leakage from one chamber to another. Immediately assemble and tighten PBM chamber apparatus without introducing bubbles. If bubbles do form, they can be removed by gently tapping the steel chamber against a hard surface.
- Incubate for 30 min at room temperature (RT).
- Fluorescent antibody attachment
- Unscrew the PBM chamber and remove the microarray slide-coverslip “sandwich”, taking care not to break the seal. Disassemble underwater in the WASH dish using forceps. Shake the slide underwater and quickly transfer to a Coplin jar containing PBS.
- Assemble the PBM chamber with gasket slide as previously described (steps 2.1.2–2.1.3).
- Add Alexa 488-conjugated anti-his antibody (10 ng/μL in binding buffer) to the gasket slide.
- Rinse the slide briefly by dipping it in PBS dish as before, then remove the slide from PBS slowly, wipe the non-DNA surface and place it facing down onto gasket slide.
- Incubate 30 min at RT in the dark (fluorescence probe is light- sensitive) to reduce photo-bleaching.
- Disassemble the PBM chamber and coverslip as before, removing the coverslip underwater in the WASH dish.
- Rinse the slides briefly by dipping them in the PBS dish. Dry the slides with compressed air. Store in the dark in a slide box until ready to scan.
- Scanning the microarray slide
- Scan the chip by using microarray scanner (refer to Table of Materials) with excitation of 495 nm and emission of 519 nm and collect the median fluorescence intensity.
3. Microarray data analysis
NOTE: All data processing was performed using custom written scripts in MATLAB.
- Perform the initial PBM data processing using Wilcoxon rank sum test p-value as described previously4.
- Use the median value of the binding intensity for each DNA sequence for further analysis.
- Next, using the one-way ANOVA p-values, compare statistical significance of the observed differences in primase-DNA binding intensities obtained for different groups of DNA probes, as explained above in the DNA library design section3.
4. Template-directed RNA synthesis catalyzed by T7 DNA primase
- Preparation of denaturing polyacrylamide gel
- To prepare 100 mL of gel mixture, combine 62.5 mL of 40% acrylamide-bisacrylamide (19:1), 42 g of urea, 1.1 g of Tris base (2-Amino-2-(hydroxymethyl)-1,3-propanediol), 0.55 g of boric acid, and 0.4 mL of 0.5 M ethylenediaminetetraacetic acid (EDTA) solution.
- Divide the mixture into 14 mL aliquots (the amount needed for one gel of 16.5 cm x 26 cm x 0.03 cm) and keep them protected from light at 4 °C for up to 1 month.
- To prepare one denaturing polyacrylamide gel, add 4 µL of TEMED and 40 µL of 10% w/v ammonium persulfate to 14 mL of the previously prepared mixture, cast it, and leave it to polymerize for at least 2 h at RT. The gel can be kept for one day at 4 °C.
- Sample preparation
NOTE: Keep the reagents, reaction mixture, and samples on ice.- To prepare the reaction mixture required for ten reactions combine 11 µL of 5x activity buffer (200 mM Tris-HCl, pH = 7.5; 50 mM dithiothreitol, 250 mM potassium glutamate, 50 mM MgCl2), 5.5 µL of 2.5 mM mixture of nucleotide tri-phosphate (NTPs), and 5.5 µL of radiolabeled ribonucleotide [e.g., ATP, (α-32P) 3000 Ci/mmol].
NOTE: All amounts have been increased by 10% due to possible pipetting errors. Radiolabeled ribonucleotides should be diluted according to the strength of radioactive signal (initial dilution is usually 1:10 or more). - Transfer 2 µL of the reaction mixture to ten PCR tubes.
- Add 1 µL of 100 µM DNA template to the corresponding PCR tube and spin down.
- Add 2 µL of 16 µM T7 DNA primase, spin down, mix gently, and spin down again.
- Incubate the reaction at RT for 20 min.
- Stop the reaction by adding 5 µL of quenching solution (95% formamide, 20 mM EDTA, 0.05% w/v xylene cyanol and bromophenol blue), mix, and spin down.
- To prepare the reaction mixture required for ten reactions combine 11 µL of 5x activity buffer (200 mM Tris-HCl, pH = 7.5; 50 mM dithiothreitol, 250 mM potassium glutamate, 50 mM MgCl2), 5.5 µL of 2.5 mM mixture of nucleotide tri-phosphate (NTPs), and 5.5 µL of radiolabeled ribonucleotide [e.g., ATP, (α-32P) 3000 Ci/mmol].
- Separation of RNA products by electrophoresis through denaturing polyacrylamide gel and subsequent signal detection
- Pre-run the gel for 1 h (10 mA, 600 V) in 1x TBE buffer (90 mM Tris-borate, 2 mM EDTA).
- Load up to 2 µL of each sample and perform electrophoresis (20 mA, 1100 V) in 1x TBE buffer (90 mM Tris-borate, 2 mM EDTA).
- Dry the gel for 2 h at 80 °C under a vacuum.
- Expose the phosphor imaging plate to radioactive gel for a few hours to overnight, depending on the strength of radioactive signal.
- Record the signal (photostimulated luminescence) by using phosphorimager (see Table of Materials).
Representative Results
This technological advance for mapping the primase binding sites allows the obtaining of DNA binding properties that are difficult, if not impossible, to observe using classical tools. More importantly, HTPP enables the revisiting of the traditional understanding of primase binding sites. Specifically, HTPP reveals binding specificities in addition to known 5'-GTC-3' recognition sequences, which leads to changes in functional activities of T7 DNA primase. Namely, two groups of sequences were identified: strong-binding DNA sequences that contained T/G in the flanks and weak-binding sequences that contained A/G in the flanks (all thymines in the strong-binding templates were replaced by adenines). No primase binding to DNA templates that were missing 5'-GTC-3' within their sequence was detected.
The primase DNA recognition sites that contained specific features, such as T/G-rich flanks, increased primase-DNA binding up to 10-fold, and surprisingly also increased the length of newly formed RNA (up to threefold) (Figure 2 in previous publication3). Importantly, HTPP allowed us to observe and quantify the variability in primer length in relation to the sequence of the DNA template.
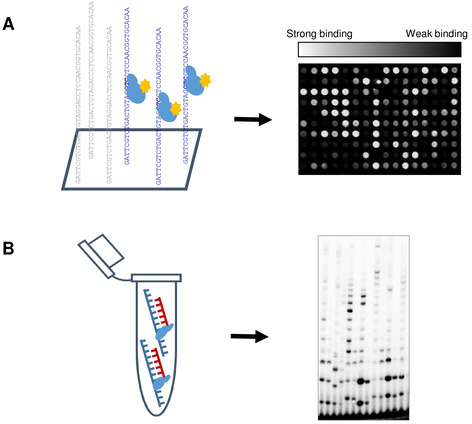
Figure 1: Schematic representation of high-throughput primase profiling (HTPP). (A) The slide was incubated with the primase in activity buffer (40 mM Tris-HCl, pH 7.5; 10 mM MgCl2, 50 mM K-glutamate, 10 mM DTT, 100 µM rNTPs). Next, Alexa 488-conjugated anti-his fluorescent antibody was introduced to label the protein. After the mild washing step, the slide was scanned using microarray scanner. Binding affinity was determined according to the median fluorescence signal and DNA sequences were divided into groups accordingly. (B) Biochemical assays were performed to correlate the DNA-binding results obtained from the microarray experiment, with the functional properties of T7 DNA primase. Please click here to view a larger version of this figure.
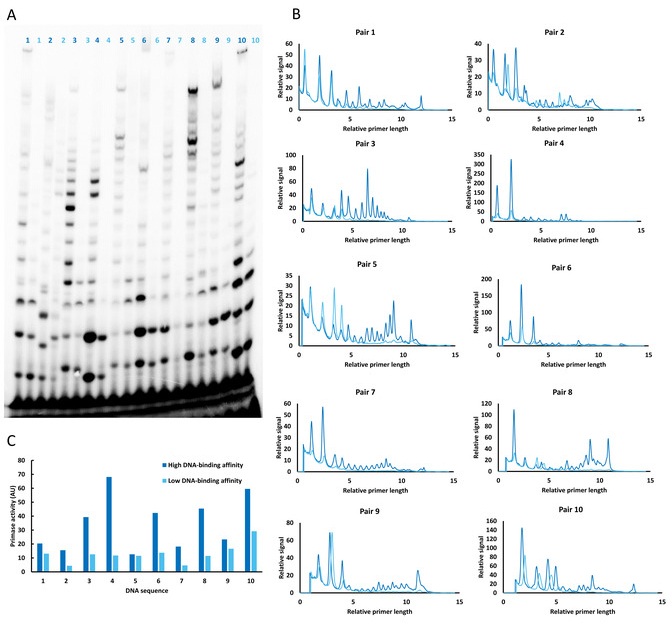
Figure 2: Comparison of catalytic activity of T7 DNA primase on two groups of DNA templates (strong binding with T/G in the flanks and weak binding with A/G in the flanks) obtained from PBM. (A) RNA primer formation catalyzed by the T7 DNA primase. The reactions contained oligonucleotides with the primase recognition sequence (numbered lanes), 32P-α-ATP, ATP, CTP, UTP, and GTP in the standard reaction mixture. One group of DNA oligonucleotides (dark blue) contained T/G in the flanks, whereas all thymines were replaced with adenines in the second group (light blue). After incubation, the radioactive RNA products were separated by electrophoresis through a 25% polyacrylamide gel containing 7M urea and visualized by autoradiography. (B) Relative length (number of ribonucleotides constituting each RNA primer is unknown) and amount of RNA primers synthesized by T7 DNA primase on two groups of DNA templates (panel A). The plots show that longer RNA primers are synthesized (increased processivity) on DNA templates that primase binds with higher affinity (that contain T/G in the flanks) compared to the templates that are bound with lower affinity (that contain A/G in the flanks). (C) Quantification of the amount of synthesized RNA primers on two groups of DNA templates. The results demonstrate a correlation between the DNA-binding affinity and the amount of synthesized RNA by the T7 DNA primase. AU (arbitrary unit) is the measure of intensity of radioactive signal which directly correlates with the amounts of synthesized RNA primers. Please click here to view a larger version of this figure.
Discussion
The PBM method has been widely used to investigate binding properties of transcription factors and can also be applied to DNA processing enzymes, such as DNA primase, that bind to DNA with low affinity. However, certain modifications of experimental procedures are required. The microarray experiment involves several steps: design of the DNA library, preparation of the chip, binding of the protein target, fluorescent labeling, and scanning. Mild washing steps are critical, since the long washes with solutions containing detergents cause dissociation of the protein from the DNA template due to the weak/transient mode of binding. Other critical steps include binding conditions (e.g., buffer composition, cofactors) and incubation times that need to be optimized for each specific enzyme.
The results obtained from microarray need to be validated in biochemical assays. Biochemical assays also provide insight into interesting features of DNA processing enzymes such as the correlation between DNA binding affinity and enzymatic activity/processivity. HTPP enabled us to observe the effect of DNA binding on functional properties of T7 DNA primase. For example, it has been observed that if primase exhibits higher binding affinity for the DNA template, it catalyzes the formation of longer RNA primers (increased processivity).
Overall, the method presented in this article is fast, reliable, and provides the opportunity to simultaneously test binding properties on tens of thousands of diverse DNA sequences in a single experiment in a hypothesis-driven way. Addition of other proteins or metal cofactors to the reaction mixture offers the possibility to investigate their effect on DNA binding properties of primases in a fast, high-throughput manner. On the other hand, the application of this method is limited by relatively high price of microarray slides and the safety precautions required when handling radioactive material for primase activity assays.
It is important to note that different primases require modifications of both microarray binding conditions and buffer conditions for activity assays. For example, primase of Mycobacterium tuberculosis requires substitution of magnesium with divalent manganese in the reaction buffer. Inappropriate metal cofactors or inappropriate reaction buffers may lead to poor primase activity or decreased DNA binding affinity.
As mentioned previously, primases bind to DNA templates with weak affinity; therefore, gentle washing steps are required during microarray binding experiment. Otherwise, they will be washed out from the microarray slide, leading to loss of the florescent signal upon addition of fluorescently labeled antibody.
To summarize, the DNA binding properties of many prokaryotic primases (including the trinucleotide DNA recognition sequence) are still poorly understood, mostly due to the technical limitations of currently available methods. HTPP represents a fast and efficient platform for the discovery of the DNA recognition sites or investigations of other template-related factors (i.e., overall nucleotide composition, GC content, presence of repetitive DNA sequence elements and their symmetry) that affect the binding affinity and activity of ssDNA binding enzymes. In addition, future applications may be directed towards the effects of different proteins or cofactors on DNA binding affinity, recognition patterns, or functional activity of primases and other DNA processing enzymes.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by the ISRAEL SCIENCE FOUNDATION (grant no. 1023/18).
Materials
| 40% acrylamide-bisacrylamide (19:1) solution | Merck | 1006401000 | |
| 95% formamide | Sigma-Aldrich | F9037-100ML | |
| Alexa 488-conjugated anti-his antibody | Qiagen | 35310 | |
| Ammonuium persulfate (APS) | Sigma-Aldrich | A3678-100G | |
| ATP, [α-32P] – 3000 Ci/mmol | Perkin Elmer | NEG003H250UC | |
| Boric acid, granular | Glentham Life Sciences | GE4425 | |
| Bovine Serum Albumin (BSA) | Roche | 10735094001 | |
| Bromophenol blue | Sigma-Aldrich | B0126-25G | |
| Coplin jar | |||
| Dithiothreitol (DTT) | Sigma-Aldrich | D0632-25G | |
| DNA microarray | Agilent | 4x180K (AMADID #78366) https://www.agilent.com |
|
| Ethylenediaminetetraacetic acid (EDTA) | Acros Organics | AC118430010 | |
| Fujifilm FLA-5100 phosphorimager | FUJIFILM Life Science | ||
| Glass slide staining rack | Thermo Scientific | 12869995 | If several slides are used |
| Lab rotator | Thermo Scientific | 88880025 | |
| Magnesium chloride | Sigma-Aldrich | 63064-500G | |
| Microarray Hybridization Chamber | Agilent | G2534A | https://www.agilent.com/cs/library/usermanuals/Public/G2534-90004_HybridizationChamber_User.pdf |
| Microarray scanner (GenePix 4400A) | Molecular Devices | ||
| Phosphate Buffered Saline (PBS) | Sigma-Aldrich | P4417-100TAB | |
| Potassium glutamate | Alfa Aesar | A172232 | |
| Ribonucleotide Solution Mix (rNTPs) | New England BioLabs | N0466S | |
| Salmon testes DNA | Sigma-Aldrich | D1626-1G | |
| Skim milk powder | Sigma-Aldrich | 70166-500G | |
| Staining dish | Thermo Scientific | 12657696 | |
| Tetramethylethylenediamine (TEMED) | Bio-Rad | 1610800 | |
| Tris base (2-Amino-2-(hydroxymethyl)-1,3-propanediol) | Sigma-Aldrich | 93362-500G | |
| Triton X-100 | Sigma-Aldrich | X100-500ML | |
| Tween-20 | Sigma-Aldrich | P9416-50ML | |
| Urea | Sigma-Aldrich | U6504-1KG | |
| Xylene cyanol | Alfa Aesar | B21530 |
References
- Berger, M. F., Bulyk, M. L. Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins. Methods in Molecular Biology. 338, 245-260 (2006).
- Berger, M. F., Bulyk, M. L. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nature Protocols. 4 (3), 393-411 (2009).
- Afek, A., et al. DNA Sequence Context Controls the Binding and Processivity of the T7 DNA Primase. iScience. 2, 141-147 (2018).
- Afek, A., Schipper, J. L., Horton, J., Gordan, R., Lukatsky, D. B. Protein-DNA binding in the absence of specific base-pair recognition. Proceedings of the Nationaly Academy of Sciences of the Unitet States of America. 111 (48), 17140-17145 (2014).
- Frick, D. N., Richardson, C. C. Interaction of bacteriophage T7 gene 4 primase with its template recognition site. Journal of Biological Chemistry. 274 (50), 35889-35898 (1999).
- Tabor, S., Richardson, C. C. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proceedings of the Nationaly Academy of Sciences of the Unitet States of America. 78 (1), 205-209 (1981).
- Fried, M., Crothers, D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Research. 9 (23), 6505-6525 (1981).
- Galas, D. J., Schmitz, A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Research. 5 (9), 3157-3170 (1978).
- Jost, J. P., Munch, O., Andersson, T. Study of protein-DNA interactions by surface plasmon resonance (real time kinetics). Nucleic Acids Research. 19 (10), 2788 (1991).
- Bowen, B., Steinberg, J., Laemmli, U. K., Weintraub, H. The detection of DNA-binding proteins by protein blotting. Nucleic Acids Research. 8 (1), 1-20 (1980).
- Oliphant, A. R., Brandl, C. J., Struhl, K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Molecular Cell Biology. 9 (7), 2944-2949 (1989).
- Marmorstein, R., Fitzgerald, M. X. Modulation of DNA-binding domains for sequence-specific DNA recognition. Gene. 304, 1-12 (2003).

