Simplified, High-throughput Analysis of Single-cell Contractility using Micropatterned Elastomers
Summary
This work presents a flexible protocol for utilizing fluorescently labeled elastomeric contractible surfaces (FLECS) Technology in microwell format for simplified, hands-off quantification of single-cell contractile forces based on visualized displacements of fluorescent protein micropatterns.
Abstract
Cellular contractile force generation is a fundamental trait shared by virtually all cells. These contractile forces are crucial to proper development, function at both the cellular and tissue levels,and regulate the mechanical systems in the body. Numerous biological processes are force-dependent, including motility, adhesion, and division of single-cells, as well as contraction and relaxation of organs such as the heart, bladder, lungs, intestines, and uterus. Given its importance in maintaining proper physiological function, cellular contractility can also drive disease processes when exaggerated or disrupted. Asthma, hypertension, preterm labor, fibrotic scarring, and underactive bladder are all examples of mechanically driven disease processes that could potentially be alleviated with proper control of cellular contractile force. Here, we present a comprehensive protocol for utilizing a novel microplate-based contractility assay technology known as fluorescently labeled elastomeric contractible surfaces (FLECS), that provides simplified and intuitive analysis of single-cell contractility in a massively scaled manner. Herein, we provide a step-wise protocol for obtaining two six-point dose-response curves describing the effects of two contractile inhibitors on the contraction of primary human bladder smooth muscle cells in a simple procedure utilizing just a single FLECS assay microplate, to demonstrate proper technique to users of the method. Using FLECS Technology, all researchers with basic biological laboratories and fluorescent microscopy systems gain access to studying this fundamental but difficult-to-quantify functional cell phenotype, effectively lowering the entry barrier into the field of force biology and phenotypic screening of contractile cell force.
Introduction
Cell-generated mechanical forces are essential to proper function in various organs throughout the body such as the intestines, bladder, heart, and others. These organs must generate stable patterns of cell contraction and relaxation to maintain the internal homeostatic state. Abnormal smooth muscle cell (SMC) contraction can lead to the onset of various disorders, including, for example, intestinal dysmotility, characterized by abnormal patterns of intestinal smooth muscle contraction1, as well as the urologic conditions of overactive2 or underactive bladder3. Within the airways, SMCs that exhibit irregular contraction patterns can trigger asthmatic hyperresponsiveness4, potentially tightening the airways and decreasing airflow of oxygen into the lungs. Another widespread physical condition, hypertension, is caused by fluctuations in the smooth muscle contraction within blood vessels5. Clearly, contractile mechanisms within cells and tissues can lead to diseases that require treatment options. As these conditions unmistakably stem from the dysfunctional contractile behaviors of cells, it becomes logical and necessary to measure the cell contractile function itself, when screening potential drug candidates.
Recognizing the need for tools to study cellular contractile force, several quantitative contraction assay methods have been developed by academic researchers including traction force microscopy (TFM)6, micropatterned TFM7, floating gel assays8, and elastomeric micropost assays9. These technologies have been utilized in single-dish format as well as multi-well-plate format in numerous studies and have even been proposed for three-dimensional force measurements10,11,12,13,14. While these technologies have enabled pioneering research within the expansive field of cell force biology, they have all been largely limited to labs possessing specific capabilities and resources, in particular: ability to fabricate TFM substrates, ability to properly apply complex and non-intuitive algorithms to solve TFM displacement maps, and relatively precise microscopy systems that can register images taken before and after sample removal from the stage (for cell dissociation). Thus, for an untrained researcher, the entry barrier to use these methods can be quite high given the extensive set of requirements to apply these technologies. In addition, the imaging resolution required for many existing technologies (40x objectives or greater) can significantly limit experimental throughput, while bulk measurement technologies could mask contributions from outlier cells and prevent discovery of milder contractile differences. Of note, as far as the authors are aware, only the low-throughput and semi-quantitative floating gel assay approach has matured sufficiently to become available to researchers (see Figure 1).
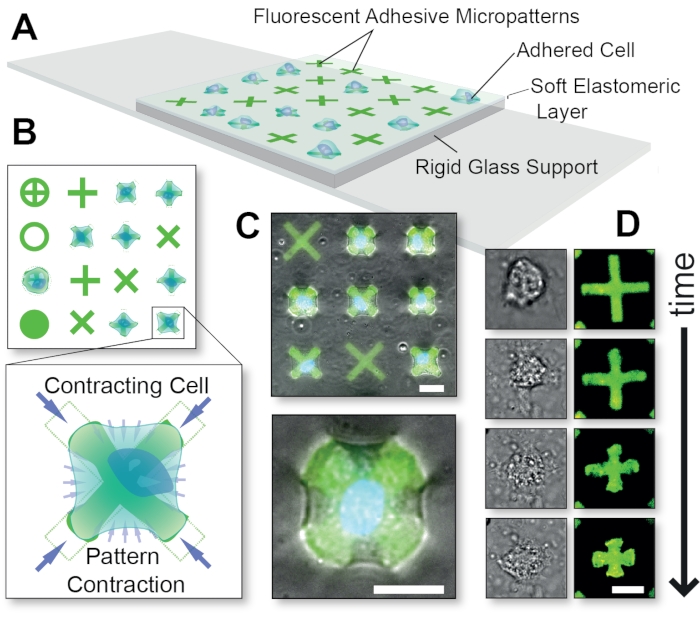
Figure 1: Overall schematic of the FLECS Technology method. (A) Cells are adhered to adhesive protein micropatterns that are covalently embedded into a thin elastomeric layer supported by glass. (B) Top-view of various possible micropattern shapes and a blow-up of a cell contracting an 'X'-shaped micropattern. (C) Overlay of fluorescent micropatterns and phase contrast images of a contracting cell. (D) Time-course images of a single contracting cell. Scale bars = 25 µm. This figure was adapted with permission from Pushkarsky et al15. Please click here to view a larger version of this figure.
Following recent advances in microtechnology, the authors developed a microplate-based technology enabling quantitative measurements of single-cell contraction in hundreds of thousands of cells called FLECS (fluorescently labelled elastomeric contractible surfaces)15,16,17,18,19,20, as an alternative to TFM. In this approach, fluorescent protein micropatterns are embedded into soft films which deform and shrink when cells apply traction forces to them, in an intuitive and measurable manner. Importantly, the protein micropatterns constrain cell position, shape, and spread area, leading to uniform test conditions. These allow simple measurements based only on their dimensional changes, which are highly resolved spatially even in 4x magnification images. The method includes a browser-based image analysis module and enables straightforward analysis of contractile cell force without requiring delicate handling procedures or registration of fiduciary markers, such that it should be operable by any researcher with a basic cell culture facility and simple fluorescent microscope with low magnification (Figure 2). This technology, which is shelf-ready and commercially available, was designed with the end-user in mind and aims to reduce the entry barrier for any laboratory scientist to study cellular force biology.
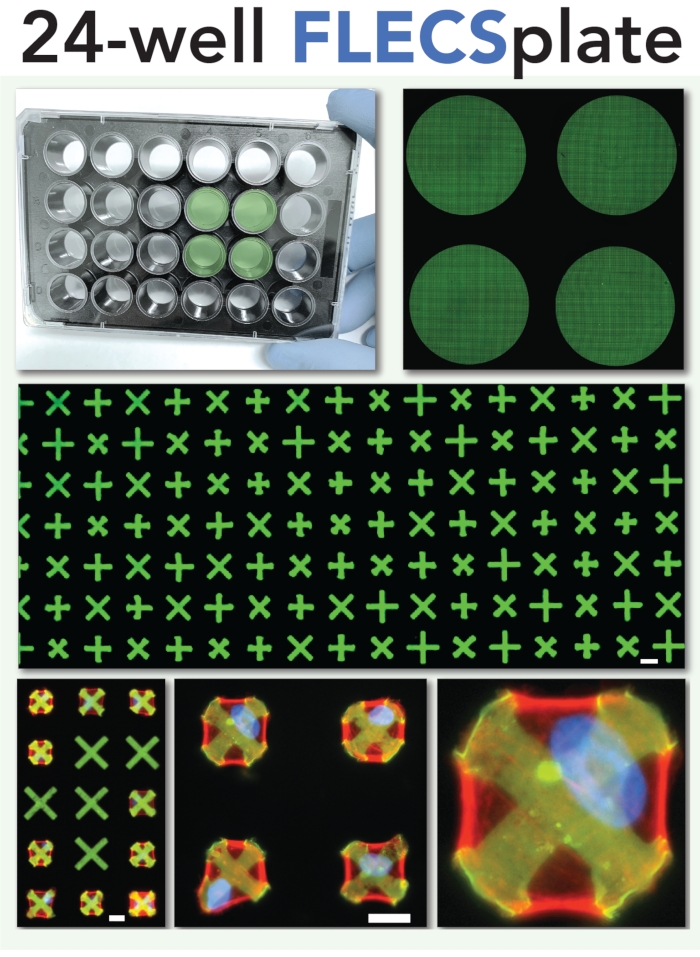
Figure 2: Schematic of the 24-well plate format for the single-cell contractility assay. This format was used in the experiments described herein and depicted in the video portion of the article. Scale bars = 25 µm. Please click here to view a larger version of this figure.
In this work, we present a protocol for applying the 24 well plate format of the FLECS Technology platform to quantify the effects of force-modulating drugs on cellular contractility in primary bladder smooth muscle cells. This general-purpose protocol can be adapted and modified as needed to account for various other timescales, cell-types, and treatment conditions of interest, and scaled to answer other questions in force biology.
Protocol
1. Day 1: Preparation of the 24 well plate
- Begin the procedure by adding 20 mL of the cell culture medium into a 50 mL conical tube. In this experiment, F12 Ham's based medium supplemented with 10% fetal bovine serum (FBS) is used.
- Obtain a 24 well plate designed to assay cell contractility. Set the pipette to 500 µL and obtain a cell strainer for cell passaging.
NOTE: The plate is available from the authors upon request. - Lift and hold the plate in one hand and proceed to gently peel the plastic film from the top of the plate. Then carefully set the plate back down.
- Turn on a vacuum aspirator and aspirate the top layer of phosphate-buffered saline (PBS) from the wells to prevent spilling. Remove the PBS one row at a time. Once the wells are no longer completely full, hold the plate in one hand and carefully remove the rest of the PBS from the wells. Quickly fill with 500 µL of cell culture medium. Take care to avoid contact between the aspirator and the bottom of the well.
- Shake the plate gently and tap on the side to ensure the entire bottom of the well is covered with solution. Once all wells are filled with medium, set the plate to the side.
2. Day 1: Cell seeding
- Retrieve the culture flask with passage 7 or lower primary human bladder smooth muscle (BSM) cells from the 37 °C incubator. Under a microscope, check cell morphology and ensure cells have grown to at least 90% confluency but less than 100% confluency.
- Conduct the cell dissociation protocol. Within a sterile biosafety cabinet, trypsinize the cells for 2.5 min until cells are detached before quenching them with serum-supplemented medium (10% FBS).
- Once cells are dissociated, use a hemocytometer to count the cells and dilute the cell suspension to approximately 50,000 cells/mL in serum-supplemented medium. Importantly, cells require at least 2% serum to attach to the micropatterns.
- Prior to seeding, rapidly strain the cell suspension through a 40 or 100 µm cell strainer into a 50 mL conical tube with a serological pipette to break up clumps of cells into single cells.
- Carefully add 500 µL of the 50,000 cells/mL cell suspension into each of the 24 wells on the plate using a P1000 pipette by dispensing the solution dropwise across different positions in the wells.
- After cell seeding, let the plate sit at room temperature for 1 h to let the cells settle directly onto the micropatterns without becoming influenced by microcurrents generated by evaporation. After 1 h, place the plate into a 37 °C incubator overnight. The cells will self-assemble and spread over the adhesive micropatterns during this time and begin exerting basal contraction levels.
3. Day 2: Addition of test drug
NOTE: The final concentration of dimethyl sulfoxide (DMSO) in the wells containing adhered cells cannot exceed 1% and drug/DMSO cannot be added directly to the cells but should first be diluted and mixed into an intermediate solution of cell medium.
- Create a six-step, eight-fold drug dilution series by transferring 30 µL of the stock drug into consecutive 210 µL volumes of DMSO and thoroughly mixing between each transfer step. In this work, a six-step, eight-fold dilution series of blebbistatin is prepared in doses ranging from 40 µM to 1 nM, in DMSO.
- For each stock drug solution (from Step 3.1), mix 30 µL of drug into 470 µL of cell culture medium. The intermediate solution yields a 16.7-fold dilution of DMSO.
- Transfer 200 µL of each intermediate solution to the appropriate well on the 24 well plate (which already contains 100 µL of medium in each well). This yields an additional six-fold dilution of DMSO.
- Steps 3.2 and 3.3 collectively yield a 1% final concentration of DMSO in the wells.
- Place the treated plate into a 37 °C incubator for the appropriate duration. For this experiment, a 30 min incubation is used.
- Immediately prior to imaging, add Hoechst 33342 live nuclear stain solution to each well (1:10,000 final dilution). Allow it to incubate for an additional 15 min to label the cell nuclei.
4. Day 2: Imaging the well-plate
- Access a microscope that is equipped to image the channels for both the cell nuclei (DAPI) and the micropatterns (TRITC).
- First image the cells with DMSO only.
- Then focus on and image both the micropatterns and the labeled cell nuclei in order to identify single cells and ensure both channels are perfectly aligned to enable automated image analysis.
- Repeat at multiple positions in each of the 24 wells on the plate. Images may be taken with 4x objective (or optionally higher to expedite imaging and acquire maximal data points per image).
- Export the images as TIF files and open them on a web-connected computer with ImageJ to analyze the data.
5. Post-experiment: Image analysis
NOTE: Image analysis was performed using Biodock.ai portal and imaging software.
- Upload the acquired images to a computer.
- Ensure that corresponding pairs of micropattern and nuclear images are named properly.
- Ensure that micropattern image names all take the form "sharedCoreName_pt.tif".
- Ensure that nuclei image names all take the form "sharedCoreName_dapi.tif".
- Convert images from TIF to PNG using ImageJ.
- Once ImageJ is opened, load in one channel at a time. For this experiment, first load micropattern images as a stack into ImageJ.
- Using Image > Adjust > Brightness/Contrast, adjust the image brightness to emphasize the micropatterns and reduce the background to black. In addition to this, smooth out the images.
- Using Image > Type > 8-bit, convert the images to 8-bit.
- Then export the image to PNG type and check the box slice level as file name. Now create a new folder PNGs and save the PNG files there with the same name.
- Repeat this process for nuclei images.
- Upload PNG image pairs into the image processing software for the analysis.
- Create and validate the account. The authors have an account to enable open access to academic users.
- Validate the account by contacting the authors.
- Log into the software.
- Under the Data tab on the left, click Upload Batch.
- Import the images by dragging and dropping image pairs into the window that pops up and name the batch. Click OK.
- Check the box next to the batch name and click Analyze.
- On the next screen, scroll down and check the box next to Contractility Analysis and click Select. Lower down the page, select 10x as the magnification that was used for imaging from a drop-down menu.
- Click 提交稿件. Once the data analysis reads Completed, click on the batch name. On the next screen, click Download Data on the right side of the page.
NOTE: The downloaded files will contain images that were analyzed, summary results reporting average contraction in each image, and a detailed analysis of every micropattern that was detected in any image, reporting their sizes, positions, number of cells adhered, and contraction. - Plot contraction values against drug concentrations to generate a concentration-response curve and determine the relative potencies of different treatments.
- Click 提交稿件. Once the data analysis reads Completed, click on the batch name. On the next screen, click Download Data on the right side of the page.
Representative Results
Regions of the images acquired from wells that were treated with DMSO only and those that were treated with 40 µM blebbistatin are shown side-by-side in Figure 3. It can be clearly observed that DMSO-only treated cells exhibit a significant level of contraction based on the very prominent deformations of the micropatterns adhered to by bladder smooth muscle cells (BSMCs) in that well. Conversely, in the image of the well treated with 40 µM blebbistatin, significant cell relaxation is observed7 as the micropatterns adhered to by BSMCs are nearly indistinguishable in size from the micropatterns that are not adhered to by cells, indicating minimal contraction. These images demonstrate the intuitive and clear visual representation of single-cell contractility offered by the fluorescent micropatterning method. Unlike TFM-based methods, where the omnidirectional movement of numerous fluorescent particles randomly distributed beneath a dense cell monolayer is meant to convey relative contractile force, here, the uniform and marked contracted geometries of the micropatterns provide immediate and easily interpretable qualitative information about contraction of individual cells. These can be directly quantified by applying standard binary object operations on the images.
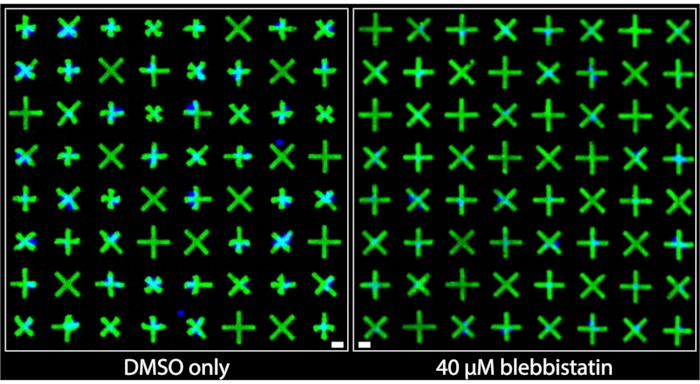
Figure 3: Side-by-side comparisons of images taken of wells containing either only 1% DMSO treatment (left) or containing 40 µM of blebbistatin (right). It can be clearly observed that treatment with blebbistatin significantly reduces the contractility of the single cells as indicated by the larger, un-contracted micropatterns. Blue nuclei indicate which micropatterns are bound by cells. Scale bar = 25 µm. Please click here to view a larger version of this figure.
By applying the browser-based analysis module to analyze the acquired image pairs of micropatterns and cell nuclei, single-cell contractility distributions are obtained for each population as shown in Figure 4. Described in detail in a prior report on the FLECS methodology15, the analysis works by locating the positions and orientations of each "X" shaped micropattern, counting the number of nuclei adhered directly over the center of each micropattern, computing the mean length of each micropattern, and calculating the pixel distance of the contraction of each micropattern with respect to the mean length of empty micropatterns (zero contraction reference). Therefore, the empty micropatterns serve an important purpose for normalizing the contraction data. Importantly, cells that do not bind to the micropatterns will accumulate on the well boundaries due to microcurrents where they will not affect image analysis. As seen in these plots, the unperturbed contractility of the cell population treated only with DMSO controls spans a large range as high as 20 pixels, with a center found at around 10 pixels. Meanwhile, cells treated with blebbistatin contract significantly less and their distribution is pushed down to a center of just over 6 pixels. Importantly, every single micropattern found in the image that binds exactly on cell is represented in these distributions. This demonstrates the ability of the method to convey differential cellular responses to drug treatments.
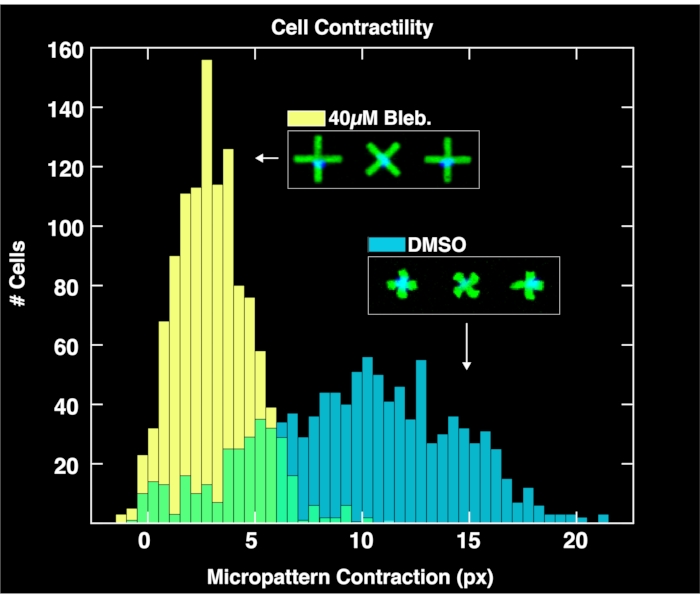
Figure 4: Histograms depicting single-cell contractility data obtained from the analysis of images taken of wells containing 1% DMSO only (blue) or 40µM blebbistatin. The distribution of DMSO-treated cells is broad and centered at a much larger contraction value (~10 pixels) than the blebbistatin treated distribution, demonstrating the quantitative effects of treatment of cells with blebbistatin. Please click here to view a larger version of this figure.
By utilizing all of the wells on a single 24-well plate and imaging at least 3 sites per well, six-point dose-response curves are simultaneously generated for two drug compounds. Figure 5 shows the concentration-response data for BSMCs treated with the same range of doses of blebbistatin or cytochalasin D (both known contractility inhibitors). As evident from the concentration-response profiles, cytochalasin D is the more potent inhibitor of tonic contraction in these cells. By fitting a sigmoidal curve to the data points, IC50 values can be calculated for each drug. Our experiments indicate that the IC50 are 7.9 µM and 100 nM for blebbistatin and cytochalasin D, respectively, following ~30 min of exposure to the drugs. Importantly, overall, these values are consistent with prior reports, validating the quantitative accuracy of the method to determine potency of contraction inhibitors7,21.
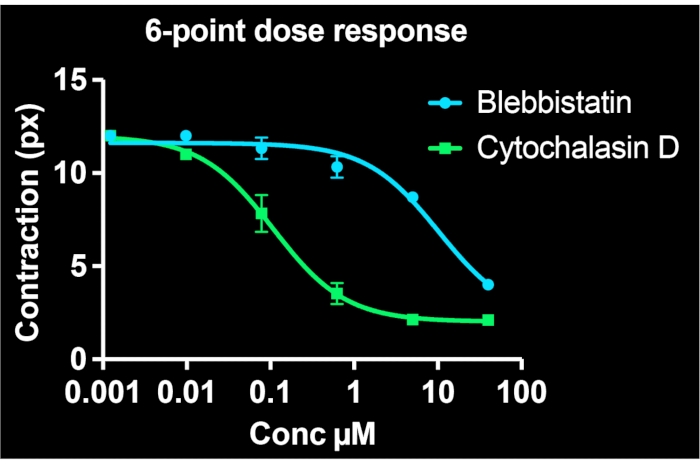
Figure 5: Concentration-response curves depicting the effects of blebbistatin and cytochalasin D on cellular contractility in single-cells. Each data point comprises three images for that condition. A sigmoidal curve was fit to each set of data. The results indicate that cytochalasin D is more potent, having a lower IC50 value. This data is collectible from a single 24-well FLECS plate. Please click here to view a larger version of this figure.
Discussion
This simplified method to quantitatively measure contraction in hundreds of thousands of cells at a time under different treatment conditions and using only standard microscopy instruments provides an accessible alternative to traditional TFM for researchers to study cellular force biology. Because the presented technology provides a visual display of cell contraction by analyzing changes in regularly shaped fluorescent micropatterns, the magnitude of the contraction produced by any given cell is intuitively understood – the smaller the micropattern, the larger the contractile force exerted by the cell.
Notably, by offering control over factors such as shape, spread-area, and adhesion molecule comprising the micropatterns (all factors known to regulate cell contractility22,23,24), the presented technology systematically eliminates additional variables that may confound interpretations of cell contraction studies.
In this experiment, 10 kPa stiffness was used in the gel and a 70 µm (diagonal length) micropattern was used comprised of type IV collagen. Besides these parameters, the adhesive molecule can be replaced with various collagens, fibronectin, gelatin, and other extracellular matrix (ECM). The stiffness of the gel can be tuned down to 0.1 kPa, and up into the MPa range. The micropattern geometry can be designed de novo to be any shape with minimal feature size of ~5 µm. These parameters are decoupled and can be independently optimized for a particular biological context.
This technology has been extensively validated to be compatible with highly adhesive and contractile cell types of a mesenchymal phenotype including various smooth muscle cell types (primary human bladder, intestinal, tracheal, bronchial, uterine, aortic, and arterial), mesenchymal stem cells and their differentiated progeny, various fibroblasts (pulmonary, dermal, and cardiac), myofibroblasts, and endothelial cells. In addition, monocyte-derived macrophages will also produce large measurable phagocytic force on the micropatterns, particularly if the micropattern consists of a known opsonin. Various cancer lines can also be assayed using the method.
The method may pose some challenges for the use with cells that are either relatively small such as T-cells and neutrophils, or cell types with a predominantly epithelial phenotype. The main reason for this is that the method relies on strong adhesion and complete spreading of cells over the micropattern in order to generate the measurable contractile signal. Cells that bind weakly, bind to each other, or do not completely spread will not produce measurable contractile signals. These behaviors, which are relatively rare, can be mitigated by adjusting the micropattern size to be smaller, or by using alternative adhesive molecules within the micropatterns that will better promote adhesion and spreading in those cells.
Users of the technology must carefully evaluate different possible cell culture medium formulations for their particular cell type of interest, as different components, growth factors, serum levels, and pH sensitivities may drive variable behaviors in different cells. Optimization of the protocol should precede scaling of any experimental workflows, and media components should always be fresh, sterile, and consistent with prior batches.
Ultimately, if single-cell resolution is not necessary for a user's goals, or if the target cell type has minimal spreading capacity, then traditional TFM may be equally or more suitable for such experiments. The authors' aim and hope is that this tool provides an additional avenue for cell biologists to study cellular contraction, particularly in the context of automated high-throughput phenotypic drug screens.
Specific to future uses in drug screens, higher throughput plates such as a 384-well FLECS plate may be used. In such plates, 4x objectives on many microscopes can capture an entire single well in their field of view, ensuring that all cellular contractile responses are captured. By using a high-throughput imaging system, an entire 384-well plate can be imaged in approximately 5 min, making this system dramatically faster than other options, and, therefore, suitable for high-throughput phenotypic drug discovery. Indeed, the authors routinely run weekly drug screens on ~50 384-wellplates (totaling more than 19,000 wells) using automation.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The laboratory work was conducted with the support of the UCLA Molecular Shared Screening Resource (MSSR) where Forcyte is sponsoring research activities, and the Magnify Incubator at the California NanoSystems Institute (CNSI), where Forcyte Biotechnologies, Inc. is a resident company. The authors will grant access to the Biodock.ai FLECS analysis module to all academic researchers upon request. L.H. and I.P. contributed equally to this work.
Materials
| Bladder smooth muscle cell culture | Sciencell | #4310 | |
| Blebbistatin | Sigma-Aldrich | B0560 | |
| Cell culture media | Thermofisher | 11765054 | Ham's F12 medium supplemented with 10% FBS and 1% p/s |
| Cell strainer | Fisher Scientific | 7201432 | |
| Conical Tube | Fisher Scientific | 05-539-13 | |
| Culture flask | Fisher Scientific | FB012941 | |
| Cytochalasin D | Sigma-Aldrich | C8273 | |
| DMSO (Dimethyl sulfoxide) | Fisher Scientific | D1284 | |
| Eppendorf tubes | Fisher Scientific | 05-402-31 | |
| Fluorescent microscope | Molecular Devices | ImageXpress Confocal | |
| Forcyte-manufactured 24-well plate | Forcyte Biotechnologies | 24-HC4R-X1-QB12 | |
| Hoescht 3342 Live Nuclear Stain | Thermofisher | 62249 | |
| Phosphate Buffered Saline (PBS) | Fisher Scientific | BP39920 |
References
- Ohama, T., Hori, M., Ozaki, H. Mechanism of abnormal intestinal motility in inflammatory bowel disease: How smooth muscle contraction is reduced. Journal of Smooth Muscle Research. 43 (2), 43-54 (2007).
- Peyronnet, B., et al. A comprehensive review of overactive bladder pathophysiology: On the way to tailored treatment. European Urology. 75 (6), 988-1000 (2019).
- Aldamanhori, R., Osman, N. I., Chapple, C. R. Underactive bladder: Pathophysiology and clinical significance. Asian Journal of Urology. 5 (1), 17-21 (2018).
- Sanderson, M. J., Delmotte, P., Bai, Y., Perez-Zogbhi, J. F. Regulation of airway smooth muscle cell contractility by Ca2+ signaling and sensitivity. Proceedings of the American Thoracic Society. 5 (1), 23-31 (2008).
- Brozovich, F. V., et al. Mechanisms of vascular smooth muscle contraction and the basis for pharmacologic treatment of smooth muscle disorders. Pharmacological Reviews. 68 (2), 476-532 (2016).
- Munevar, S., Wang, Y., Dembo, M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophysical Journal. 80 (4), 1744-1757 (2001).
- Tseng, Q., et al. A new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab on a Chip. 11 (13), 2231-2240 (2011).
- Bell, E., Ivarsson, B., Merrill, C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proceedings of the National Academy of Sciences. 76 (3), 1274-1278 (1979).
- Tan, J. L., et al. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proceedings of the National Academy of Sciences. 100 (4), 1484-1489 (2003).
- Rokhzan, R., et al. high-throughput measurements of cell contraction and endothelial barrier function. Laboratory Investigation. 99 (1), 138-145 (2019).
- Park, C. Y., et al. High-throughput screening for modulators of cellular contractile force. Integrative Biology. 7 (10), 1318-1324 (2015).
- Kaylan, K. B., Kourouklis, A. P., Underhill, G. H. A high-throughput cell microarray platform for correlative analysis of cell differentiation and traction forces. Journal of Visualized Experiments. (121), e55362 (2017).
- Huang, Y., et al. Traction force microscopy with optimized regularization and automated Bayesian parameter selection for comparing cells. Scientific Reports. 9, 539 (2019).
- Franck, C., Maskarinec, S. A., Tirrell, D. A., Ravichandran, G. Three-dimensional traction force microscopy: A new tool for quantifying cell-matrix interactions. PLOS ONE. 6, 17833 (2011).
- Pushkarsky, I., et al. Elastomeric sensor surfaces for high-throughput single-cell force cytometry. Nature Biomedical Engineering. 2 (2), 124-137 (2018).
- Pushkarsky, I. FLECS technology for high-throughput single-cell force biology and screening. ASSAY and Drug Development Technologies. 16 (1), 7-11 (2017).
- Koziol-White, C. J., et al. Inhibition of PI3K promotes dilation of human small airways in a rho kinase-dependent manner. British Journal of Pharmacology. 173 (18), 2726-2738 (2016).
- Orfanos, S., et al. Obesity increases airway smooth muscle responses to contractile agonists. American Journal of Physiology-Lung Cellular and Molecular Physiology. 315 (5), 673-681 (2018).
- Tseng, P., Pushkarsky, I., Carlo, D. D. Metallization and biopatterning on ultra-flexible substrates via dextran sacrificial layers. PLOS ONE. 9, 106091 (2014).
- Yoo, E. J., et al. Gα12 facilitates shortening in human airway smooth muscle by modulating phosphoinositide 3-kinase-mediated activation in a RhoA-dependent manner. British Journal of Pharmacology. 174 (4), 4383-4395 (2017).
- MacGlashan, D., Vilariño, N. Polymerization of actin does not regulate desensitization in human basophils. Journal of Leukocyte Biology. 85 (4), 627-637 (2009).
- Hocking, D. C., Sottile, J., Langenbach, K. J. Stimulation of integrin-mediated cell contractility by fibronectin polymerization. Journal of Biological Chemistry. 275 (14), 10673-10682 (2000).
- Tolić-Nørrelykke, I. M., Wang, N. Traction in smooth muscle cells varies with cell spreading. Journal of Biomechanics. 38 (7), 1405-1412 (2005).
- Ye, G. J. C., et al. The contractile strength of vascular smooth muscle myocytes is shape dependent. Integrative Biology. 6 (2), 152-163 (2014).

