Making Precise and Accurate Single-Molecule FRET Measurements using the Open-Source smfBox
Summary
This article provides step-by-step instructions for making fully-corrected accurate FRET measurements on individual, freely diffusing biomolecules using the open-source, inexpensive smfBox, from switch on, through alignment and focusing, to data collection and analysis.
Abstract
The smfBox is a recently developed cost-effective, open-source instrument for single-molecule Förster Resonance Energy Transfer (smFRET), which makes measurements on freely diffusing biomolecules more accessible. This overview includes a step-by-step protocol for using this instrument to make measurements of precise FRET efficiencies in duplex DNA samples, including details of the sample preparation, instrument setup and alignment, data acquisition, and complete analysis routines. The presented approach, which includes how to determine all the correction factors required for accurate FRET-derived distance measurements, builds on a large body of recent collaborative work across the FRET Community, which aims to establish standard protocols and analysis approaches. This protocol, which is easily adaptable to a range of biomolecular systems, adds to the growing efforts in democratising smFRET for the wider scientific community.
Introduction
Single-molecule Förster resonance energy transfer (smFRET) is a technique that measures the FRET efficiency between two dyes-a donor and an acceptor-at the level of individual molecules. FRET is a photophysical process arising from the overlapping energy spectra of two dyes: the donor is excited by light of a specific wavelength and transfers energy non-radiatively to the acceptor, resulting in emission from the acceptor. The efficiency of this transfer is inversely proportional to the sixth power of the distance between the two dyes, so the transfer efficiency varies with distance1. Thus, this FRET efficiency can be used to determine spatial information about the molecule(s)2 to which the dyes are attached, within a range of 3-10 nm. This scale, and the fact that changes in FRET efficiency are sensitive to Angstrom molecular movements3, makes the technique well suited to investigating structural information about biomolecules-such as nucleic acids and proteins-without the complications of ensemble averaging4,5,6. While changes in relative FRET efficiencies can be used to monitor biomolecular interactions and conformational dynamics, shedding light on key cellular processes such as protein (un)folding, transcription, and DNA replication and repair, absolute FRET efficiencies have been used to determine precise distances for biomolecular structure determination7,8,9,10,11, overcoming the need for crystallization or freezing as is required for some other structural methods4,12.
smFRET experiments most commonly take two forms, confocal or total internal reflection fluorescence (TIRF) microscopy. Between both approaches the molecular dynamics of biomolecules can typically be investigated on timescales from pico- to millisecond (confocal, freely diffusing molecules) up to millisecond to hours (TIRF, surface immobilized molecules). This is due to the different setups involved in each technique. In TIRF microscopy, molecules are immobilized on the surface of a slide and excited by an evanescent wave (Figure 1A). Here, however, the focus is on confocal microscopy as this is the format of the smfBox. In confocal microscopy, molecules are not immobilized and instead freely diffuse via Brownian motion through the confocal volume (~1 fL), formed by focusing a laser beam through a high numerical aperture lens into a spot at some designated depth within the solution (Figure 1B). The resulting emission is focused back through the same aperture and filtered through a dichroic mirror (Figure 1C for full schematic). It is then focused through a pinhole in order to remove any out-of-focus light and onto an avalanche photodiode (APD). When the APD detects a photon, it outputs a TTL pulse, the timing of which can be recorded with up to picosecond resolution. The observation time of these freely diffusing molecules within the vicinity of the confocal volume is commonly within the order of milliseconds.
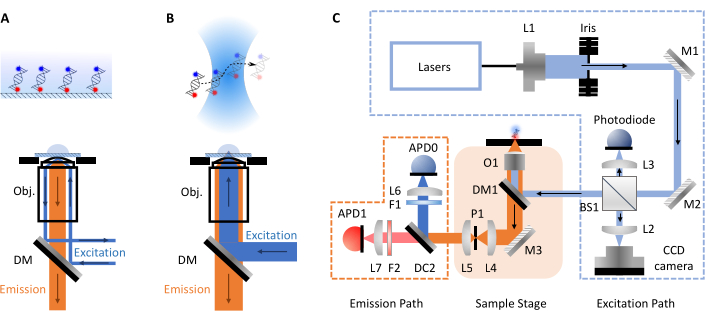
Figure 1: Schematics showing principles of microscopy and the smfBox setup. (A) Total Internal Reflection Fluorescence (TIRF) Microscopy principle: excitation light is directed into the edge of the objective (Obj.) and undergoes total internal reflection at the coverslip-buffer interface generating an exponentially decaying evanescence field to excite surface attached molecules. (B) Confocal Microscopy: Freely diffusing molecules are excited by a near diffraction-limited spot focused into the sample. (C) The smfBox setup used in this protocol, showing all key components: avalanche photodiodes (APD), beam splitter (BS), dichroic mirrors (DM), filters (F), mirrors (M), objective (O) and pinhole (P). Please click here to view a larger version of this figure.
More recently, smFRET techniques incorporated two color excitation, where lasers matching the donor and acceptor excitation wavelengths are alternated5. This can be done in one of two ways, the first by modulating continuous wave lasers on the KHz timescale, which is known as alternating laser excitation (ALEX)13,14. The second method interleaves fast pulses on the MHz timescale; this is nanosecond-ALEX15 or pulsed interleaved excitation (PIE)16. In all these approaches, information from the acceptor laser leads to calculation of the so-called stoichiometry, which can discriminate between molecules with a low FRET efficiency and those lacking an acceptor (either through incomplete labeling or photobleaching). Using PIE/ns-ALEX additionally gives access to fluorescent lifetimes on the single-molecule level, and anisotropies can be measured when coupled with polarizing optics. This combination of measurements is known as multiparameter fluorescence detection (MFD)9.
Despite the many advantages of smFRET, it is not widely used outside of specialist labs due to the high costs of commercial instruments and a lack of simple, self-build alternatives. A growing trend towards development of low-cost opensource microscopy is taking place and other platforms have recently emerged, including Planktonscope17, OpenFlexure Microscope18, Flexiscope19, miCube20, liteTIRF21, and Squid22. Herein the study describes the protocol for using the smfBox, a recently developed cost-effective confocal set-up capable of measuring the FRET efficiency between two dyes on freely diffusing single molecules. Detailed build instructions and all the necessary operational software are freely available at: https://craggslab.github.io/smfBox/23. The optical arrangement of the smfBox is assembled from readily available components purchased from affordable and widely-accessible manufacturers, while the microscope body (responsible for the majority of the expense in a standard confocal set-up) has been replaced by a custom light-tight anodized-aluminium box (allowing measurements to be made under ambient light conditions). This box houses key optical components, including the excitation dichroic, objective, and pinhole, and a mechanical laser interlock, enabling its safe operation as a Class I laser product (see Figure 1C for a full schematic). The smfBox uses ALEX to validate the dye stoichiometry and to determine accurate FRET correction factors. It is operated using custom-written, open-source software (smOTTER), which controls all aspects of the data acquisition and outputs the data in the open-source photon-HDF5 format24, compatible with many third-party analysis tools. The smfBox and the acquisition and data analysis protocols were recently tested against >20 other instruments (both confocal and TIRF) in a multi-lab blind study25. The FRET efficiencies obtained were in excellent agreement with all the other instruments, despite the smfBox costing only a fraction of the price of commercially available setups.
Here, a step-by-step protocol is outlined for acquiring and analyzing accurate, absolute FRET efficiencies on freely diffusing DNA duplexes using the smfBox, all the way from switch on, through alignment and focusing, to data collection and analysis. The samples used here are three duplex DNAs (exhibiting high-, mid- and low-FRET efficiencies, see Table 1) that were assessed in the world-wide blind study25; however, the method is adaptable to many molecular systems, including proteins and other nucleic acids. The hope is that such a detailed protocol, along with the already existing build instructions for the smfBox23, will help to make this powerful technique even more accessible to a wide range of labs.
Protocol
1. Power-on components
- Power on the six plugs (no particular order): Piezoconcept (z-focus), APD0, APD1, green laser, red laser and photodetector.
2. Software 1-Experimental setup
- Launch the laser control center.
- Ensure Continuous Wave – Alternating Constant Current (CW-ACC) mode is selected.
- Power on both the lasers.
- Check/set the laser powers.
NOTE: The laser power will need to be adjusted as measured just before the excitation dichroic, as ND filters and beam splitters in the excitation path will reduce the laser power from the number given on the laser control panel. The numbers given here are power at the excitation dichroic, but the power on the laser control that corresponds to this will need to be worked out.- 220 µW green laser (515 nm, 40 mW)
- µ70 W for red laser (638 nm, 10 mW)
- smOTTER acquisition software
- Connect the lasers, detectors, z-stage, and camera. Configure them correctly (might vary depending on settings of particular NI card setup).
3. Alignment of emission path (not routinely required)
- Setting up alignment
- Pipette 10 µL of free Cy3B dye (~100 nM) onto the microscope and focus as described in steps 4.3-4.5 below.
NOTE: Another donor dye with some leakage into the acceptor channel would also work. - Open the Alignment tab in smOTTER, lower the laser power, and alter the y-axis scale until readout is seen from the detectors.
NOTE: The goal here is to increase the signal, so the scale may need to be changed again after the signal increases. - Unscrew the four screws at the front of the smfBox and remove the front panel.
- Pipette 10 µL of free Cy3B dye (~100 nM) onto the microscope and focus as described in steps 4.3-4.5 below.
- Pinhole alignment
- Turn the x knob on the pinhole positioner while watching the signal on the Alignment tab, trying to increase the signal in green and red.
- Now turn the y knob to align the pinhole in the other direction.
- Return to the x knob to check for any further increase in signal.
- Pinhole-lens alignment
- Turn the lens x knob in one direction, this will decrease the signal.
- Turn the pinhole x knob in the same direction to increase the signal again.
- If the new max signal is higher than before, continue to iteratively move both the pinhole and lens in that direction. If it is lower than before, iteratively move in the opposite direction.
- Repeat above for y.
- APD lens alignment
- Starting with the green APD, move the x knob until the green signal is at a maximum.
- Repeat for the y knob.
- Return to the x knob, move back and forth to find the threshold points where the signal begins to fall, and leave it at a position halfway between these two points.
- Repeat for the x knob.
- Repeat the above steps for the red APD lens, watching the red signal.
- Place the front panel back on the smfBox and replace the screws.
NOTE: Perform the above alignment monthyl, and as a precautionary measure if it is suspected that the microscope has developed an alignment fault between measurements.
4. Measurement-data acquisition
- For the first sample, clean the stage with lens cleaning tissue soaked in methanol; wipe gently across the objective from one end to the other.
- Apply 3-4 drops of immersion oil for microscopy onto the center of the objective; replenish as needed between samples.
- Sample preparation
- Pipette 10 µL of sample (first use Type I ultrapure water) onto the center of a clean cover glass.
- Carefully place the cover glass onto the objective lens lowering it at an angle to the oil to prevent trapping air bubbles between the cover glass and the objective.
NOTE: If necessary, slide cover glass around to push air bubbles in the oil away from the focal point.
- To perform alternating-laser excitation (ALEX) experiments configure the laser in the laser duty cycles panel as follows.
- Donor (green laser) 0 off, 45 on, 55 off
- Acceptor (red laser) 50 off, 45 on, 5 off
- ALEX period (µs): 100
- Click on the Z focus tab in the ribbon; in the acquisition panel, switch the lasers to Live and start the camera; adjust the exposure so that a bright spot appears centrally surrounded by black.
NOTE: The camera captures light reflected by the glass/water interface between the coverslip and the sample and the visible circle of light is at its smallest diameter when the focal point is at this interface. It is therefore necessary to start below this interface, increase the z-height to the interface, and then slightly further to focus above the coverslip and in the sample being measured.- Starting from a low Z position, increase the height until the bright spot reaches its minimal size, then raise the height further by up to 20 µm to focus the laser above the oil and cover glass and in the sample.
- Stop the camera once in focus.
- To confirm that the setup has been performed appropriately, monitor the trace of Type I ultrapure water to see no fluorescence signals; repeat confirmation of purity for the buffers in which samples are prepared.
- Carefully remove the cover glass with rubber-ended tweezers to avoid damaging objective, then place the sample of interest prepared as above onto the objective. Replenish the immersion oil as necessary to maintain a uniform area of contact between cover glass and objective (special attention paid to sample's contact area on cover glass and focal area of objective)
- Concentration bingo
- To ensure single-molecule data is obtained, samples need to be at a concentration at which 1-5 bursts per second are observed in the live trace panel of smOTTER (this minimizes the chance of more than one molecule being present in the detection volume at once). Take the measurements when the appropriate concentration has been determined.
- To prevent evaporation during long experiments, prepare an airtight sample chamber. Press the silicone isolators (8-9 mm diameter hole) down onto the center of a cover glass (colored-in with a pipette tip). Then, carefully pipette a sample into the center, avoiding any contact with the silicone. Then, place a second cover glass on top and press to form a seal.
- Check the live stoichiometry vs FRET efficiency (ES) histogram to see if the sample is behaving as expected, i.e., expected FRET efficiency and reasonable stoichiometry (~0.5).
- When the sample is ready, enter the details for the experiment into the save settings panel. Choose an appropriate directory and filename by clicking on the ellipsis icon (files saved in hdf5/h5 format) in the save settings panel.
- Enter information for sample name, sample details, donor and acceptor labels, buffer, donor and acceptor excitation wavelengths, detection wavelengths and laser power, and user and user affiliation.
- Return to the live trace tab in the ribbon, and in the acquisition panel enter experiment length (in minutes), select an appropriate save interval to mitigate potential data-loss in case of an error. Select Save Laser Powers, if required.
- Press Start to take the data.
NOTE: To enable accurate FRET determination, at least two samples will need to be measured containing the same donor and acceptor dyes, but with sufficiently different FRET efficiencies.
5. Analysis/software 2
- Launch Jupyter notebook with the FRETBursts python package (setup instructions can be found here: https://craggslab.github.io/smfBox/anasoftware.html)
- If correction for absolute FRET efficiencies is not needed (i.e., only relative changes in FRET are sufficient for the measurement) launch Jupyter notebook FRET Analysis 1.4 Uncorrected.ipynb. Use this to export figures or data as csv files for further analysis or plotting in other software.
NOTE: Each notebook contains detailed instructions to guide the user through the analysis procedures. For a more detailed discussion of analysis procedures, including burst search algorithms, background correction, and all correction parameters see23,25. - If correction for absolute FRET efficiencies is needed, begin by calculating the crosstalk parameters alpha (leakage of donor emission into the acceptor channel) and delta (proportion of direct excitation of the acceptor under donor excitation).
- First launch Jupyter notebook Correction Factor Finder Alpha-Delta.ipynb and determine the alpha and delta parameters.
NOTE: If these are inconsistent between samples, the microscope may have developed an alignment problem between measurements or the spectra of the dyes differs between samples. - If the alpha and delta parameters are consistent, launch Jupyter notebook Correction Factor Finder Gamma-Beta.ipynb to determine the gamma and beta parameters (which account for differences in excitation and detection efficiencies between the dyes).
NOTE: If the gamma and beta plot does not fit well, the microscope may have developed an alignment problem between measurements, or the quantum yields, or the extinction coefficients of the dyes differs between samples. - With the four crosstalk parameters determined, these factors may be used in the FRET Analysis 1.4 Corrected.ipynb Jupyter notebook to determine absolute FRET efficiencies.
- First launch Jupyter notebook Correction Factor Finder Alpha-Delta.ipynb and determine the alpha and delta parameters.
6. Troubleshooting
- If all signals are low or counts per burst are lower than expected (this is dye dependant-but for ATTO-550 and ATTO-647N on the smfBox typical values are between 50 and 100 counts per ms during a single-molecule burst), realign the smfBox.
- Bridge between doubly and singly labeled populations on ES histogram.
NOTE: This can be caused either by working at too high a concentration (remedy this by diluting the sample), or by photobleaching, which can be caused by using too high a laser power (remedy this by reducing laser power). - If the burst rate falls throughout the experiment (fluorescently labeled molecules are likely adhering to the glass coverslip), use an increased concentration of BSA to rectify.
- If the burst rate increases throughout the experiment (sample is likely evaporating), prepare an air-tight sample chamber as described above.
- If the signal crashes to zero during alignment (the software is likely being overwhelmed by signal from the detectors), lower the laser power or use a more dilute alignment sample.
- If Z-focus shows concentric rings (can happen if multiple coverslips have been placed on the objective), check for multiple coverslips on the objective.
- If the sample contains bright, long bursts of intermediate stoichiometries (caused by aggregated molecules), use detergents or modify sample purification protocols.
NOTE: Getting a clean buffer can be an issue, as some buffer components will often contain very small amounts of moderately fluorescent contaminants which are enough to present as single color bursts on the time trace. If there is too much of this in the buffer, then it can coincide with sample bursts and change the FRET efficiency or stoichiometry being measured. BSA in particular can often be problematic in this regard, so it is helpful to expose a stock BSA solution to a strong light source to photobleach the contaminants.
Representative Results
The protocol necessitates critical assessment of experimental conditions during setup (see protocol step 4.8). The first results acquired which determine success or failure of the experiment are achieved at this stage. A positive result would be to have between five and one bursts per second (see Figure 2B,C). A negative result would be having too many (Figure 2A) or too few bursts (Figure 2D) within that time frame. It remains possible at this stage to rectify these errors: a sample with too high a concentration needs simply to be diluted; if the concentration is too low, however, a new sample may need to be prepared (the determinant being whether it remains possible at this low concentration to collect data in a reasonable time frame).
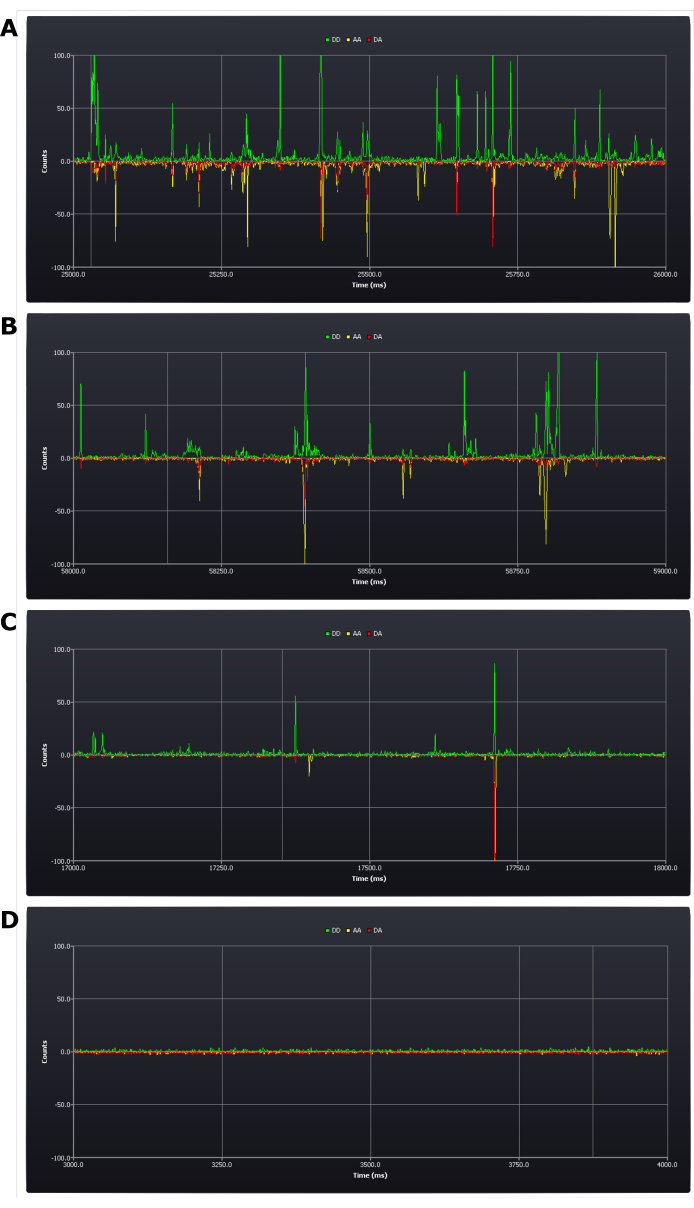
Figure 2: Screen shots from live trace during experimental setup showing different concentrations of doubly labeled duplex DNA samples. (A) too high, (B) upper acceptable limit, (C) target concentration, (D) too low. Photon counts (1 ms bins) are shown in the three detection channels; donor emission after donor excitation (DD), acceptor emission after donor excitation (DA), and acceptor emission after acceptor excitation (AA). Please click here to view a larger version of this figure.
A static single-species system would typically require 30 to 60 min of measurement to obtain the necessary ~1,000 bursts needed for robust data analysis. The length of time and number of bursts required will increase with multiple species or dynamic systems. Following data collection and analysis using the protocol figures are exported from the Jupyter notebooks. The alternation plot (Figure 3A) should match the ALEX period of the experimental setup. The time trace (Figure 3B) is used to qualitatively assess that the sample concentration is reasonable. The background plot (Figure 3C) shows the distribution of inter-photon delay periods with a linear fit to the longer times to estimate the background rate26. The background trace (Figure 3D) can identify if there were changes in the sample over the duration of the experiment; primarily this would be due to evaporation during longer acquisition times. ES histograms are generated for all photons (Figure 3E) and doubly labeled species (Figure 3F). Finally, a 1D E histogram (Figure 3G) is generated with gaussian fitting of the burst data.
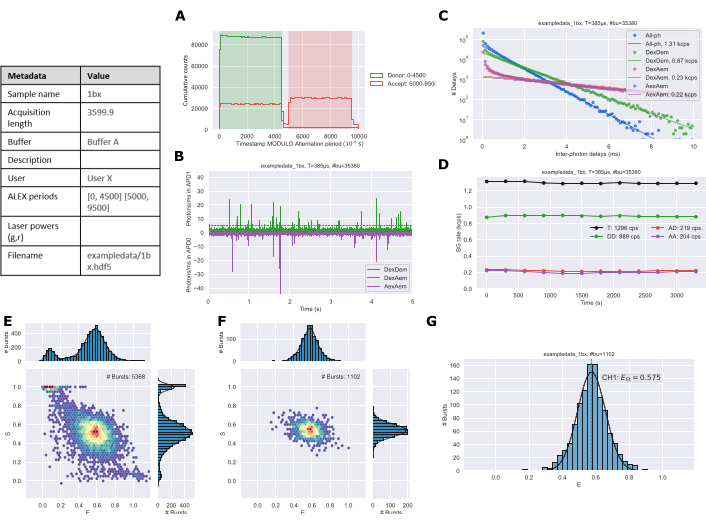
Figure 3: Example figure output of analyzed data generated by the Jupyter Notebooks. (A) Alternation plot, (B) Time trace, (C) Background determination, (D) Background rates, (E) All photon ES histogram, (F) Dual channel ES histogram, and (G) 1D E histogram. Please click here to view a larger version of this figure.
| Name | Sequence |
| 1a | 5’- GAG CTG AAA GTG TCG AGT TTG TTT GAG TGT TTG TCT GG – 3’ |
| 3’- CTC GAC TTT CAC AGC TCA AAC AAA CTC ACA AAC AGA CC – 5’ | |
| 1b | 5’- GAG CTG AAA GTG TCG AGT TTG TTT GAG TGT TTG TCT GG – 3’ |
| 3’- CTC GAC TTT CAC AGC TCA AAC AAA CTC ACA AAC AGA CC – 5’ | |
| 1c | 5’- GAG CTG AAA GTG TCG AGT TTG TTT GAG TGT TTG TCT GG – 3’ |
| 3’- CTC GAC TTT CAC AGC TCA AAC AAA CTC ACA AAC AGA CC – 5’ |
Table 1: DNA sequences used in the protocol. Nucleotides are highlighted in blue and red representing C2 amino modified thymine residues labeled with Atto-550 and Atto-647N, respectively.
Correction Factor Finder Alpha-Delta: Please click here to download this File.
Correction Factor Finder Gamma-Beta: Please click here to download this File.
FRET Analysis 1.4 Corrected: Please click here to download this File.
FRET Analysis 1.4 Uncorrected: Please click here to download this File.
Discussion
The most critical steps in the protocol are the alignment of the microscope and adjusting the sample concentration to the correct dilution. If alignment is off, then there might be insufficient signal to identify bursts and plot histograms, and if misalignment occurs between samples then accurate FRET correction may fail due to changes in leakage and detection / excitation efficiencies. The use of an appropriate concentration is also important, too high a concentration will give coincident bursts, containing multiple molecules with potentially different FRET efficiencies or labeling stoichiometries. Too low a concentration will give too few bursts for robust data analysis.
The protocol described here is for measuring distances in static single FRET species. If the sample has more than one peak in the FRET efficiency histogram, or peaks appear broad (which can happen with dynamic species), then more bursts may be needed to fit histograms to the same degree of precision. For two well-separated peaks then approximately twice as much data will be needed, but if the populations overlap slightly then even more data is required.
If the two populations interconvert on the time scale of the experiment, the dynamics and kinetics of the system can potentially be determined. Tests such as BVA27 and 2CDE28 can confirm that the intermediate bursts are dynamic in nature, whereas analyses including dPDA29,30 or H2MM31 can determine the rates of interconversion. Jupyter notebooks for BVA and 2CDE are available on the FRETBursts26 website, and the MATLab based software PAM32 can run BVA, 2CDE, and PDA analyses.
Confocal single-molecule FRET can easily observe states much more shortly lived (~1 ms) than TIRF; however, the short observation times, limited by diffusion, give no molecular history, and so cannot determine longer dwell times, or complex transition networks in the way that surface immobilized experiments can.
As the protocol measures freely diffusing molecules at a very low concentration, it works best when measuring intramolecular distances on the same molecule. Intermolecular distances between transiently bound molecules can be measured provided that the Kd of the two molecules is low enough that the complex exists at a significant quantity at the low working concentration required by the experiment (~100 pM). If the Kd is much higher than this, then only singly labeled molecules will be seen. This problem can be overcome by using microfluidics to mix the two labeled components together at a high concentration and then rapidly diluting and flowing over the objective before the complex dissociates33,34.
Measuring FRET efficiencies at the single-molecule level has a significant advantage over ensemble techniques, as it informs on heterogeneous subpopulations, which in an ensemble experiment would be averaged. Furthermore, single-molecule FRET with ALEX gives access to accurate FRET efficiencies, which can be converted to accurate distances. This enables determination of more detailed structural information rather than simply probing relative distance changes. The smfBox carries all these benefits and capabilities but can be constructed on a much lower budget than comparable commercially available microscopes capable of confocal smFRET23.
The smfBox represents a much lower barrier to entry for smFRET techniques, allowing researchers to measure conformational changes, and accurate distances within and between proteins and nucleic acids7,8,9,10,11,35.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The authors gratefully acknowledge the following funding sources: BBSRC (BB/T008032/1); EPSRC (Studentship to B.A.) and MRC (Studentship to A. R.-T.).
Materials
| Amino modified oligonucleotide | Eurogentec | N/A | May be ordered from various suppliers or synthesised; amino modification enables labeling with NHS-ester modified dyes |
| Avalanche photodiode (APD) | Excelitas | SPCM-AQRH-14 | Two APDs are required for the smfBox setup |
| Bovine Serum Albumin (BSA) | Merck | A2153 | System dependant; imaging buffer component (0.1 mg/mL in buffer) |
| Compact Laser Combiner | OMICRON | LightHUB-2 | 515 nm (80 mW) and 638 nm (100 mW) lasers |
| Coverglass | VWR | 630-2742 | Thickness: 0.17 ± 0.01 mm, LxW: 22×22 mm |
| Cy3B | Cytiva | PA63101 | 1 mg, PA63100 (5 mg), PA96106 (25 mg) |
| FRETBursts Python Package | N/A | N/A | Open-source python package for burst analysis of freely-diffusing single-molecule FRET data: https://fretbursts.readthedocs.io |
| Imaging Buffer | N/A | System dependant; 5 mM NaCl, 20 mM MgCl2, 5 mM Tris pH 7.5 and 0.1 mg/mL BSA | |
| Immersion Oil | Olympus | IMMOIL-F30CC | |
| Jupyter notebooks | Project Jupyter | N/A | Open-source web application to create and share documents that contain live code, equations, visualizations and text; data analysis notebooks for smfBox can be found in the SI |
| Lens Tissue | ThorLabs | MC-5 | MC-50E is same item in bulk |
| Magnesium Chloride | Merck | M2670 | System dependant; imaging buffer component (20 mM in buffer) |
| MilliQ/Ultrapure water | N/A | ||
| Nanopoistioner | Piezoconcept | FOC300 | Nanopositioner for accurate positioning of microscope objective |
| NHS-ester modified ATTO-550 | ATTO-TEC | AD 550-31 | 1 mg, AD 550-35 (5 mg) |
| NHS-ester modified ATTO-647N | ATTO-TEC | AD 647N-31 | 1 mg, AD 647N-35 (5 mg) |
| Objective lens | Olympus | N1480700 | Olympus objective series from orignal smfBox discontinued; replaced by N5702300 |
| OMICRON Control Center (OCC)- laser control center | OMICRON | N/A | v3.5.34 – OMICRON laser driver software |
| Press-To-Seal silicone isolator | Grace Bio-Labs | 664201 | 8-9 mm Diameter x 1.7 mm Depth |
| smOTTER | N/A | N/A | Open-source acquisition software for the Craggs Lab smfBox: https://github.com/craggslab/smOTTER |
| Sodium Chloride | Merck | S7653 | System dependant; imaging buffer component (5 mM in buffer) |
| Tris base | Merck | 93362 | System dependant; imaging buffer component (5 mM, pH 7.5 in buffer) |
| Type I ultrapure water | Merck | ZIQ7000T0 | Milli-Q® IQ 7000 Ultrapure Water System |
Referencias
- Forster, T. Intermolecular energy migration and fluorescence. Annals of Physics. 437 (1-2), 55-75 (1948).
- Stryer, L., Haugland, R. P. Energy transfer: a spectroscopic ruler. Proceedings of the National Academy of Sciences. 58 (2), 719-726 (1967).
- Hohlbein, J., et al. Conformational landscapes of DNA polymerase I and mutator derivatives establish fidelity checkpoints for nucleotide insertion. Nature Communications. 4 (1), 2131 (2013).
- Lerner, E., et al. Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science. 359 (6373), (2018).
- Hohlbein, J., Craggs, T. D., Cordes, T. Alternating-laser excitation: single-molecule FRET and beyond. Chemical Society Reviews. 43 (4), 1156-1171 (2014).
- Lerner, E., et al. The FRET-based structural dynamics challenge — community contributions to consistent and open science practices. arXiv. , (2020).
- Hellenkamp, B., Wortmann, P., Kandzia, F., Zacharias, M., Hugel, T. Multidomain structure and correlated dynamics determined by self-consistent FRET networks. Nature Methods. 14 (2), 174-180 (2017).
- Craggs, T. D., et al. Substrate conformational dynamics facilitate structure-specific recognition of gapped DNA by DNA polymerase. Nucleic Acids Research. 47 (20), 10788-10800 (2019).
- Tsytlonok, M., et al. Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation. Nature Communications. 10 (1), 1676 (2019).
- Nagy, J., et al. Complete architecture of the archaeal RNA polymerase open complex from single-molecule FRET and NPS. Nature Communications. 6 (1), 6161 (2015).
- LeBlanc, S. J., et al. Coordinated protein and DNA conformational changes govern mismatch repair initiation by MutS. Nucleic Acids Research. 46 (20), 10782-10795 (2018).
- Segal, M., et al. High-throughput smFRET analysis of freely diffusing nucleic acid molecules and associated proteins. Methods. 169, 21-45 (2019).
- Kapanidis, A. N., et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proceedings of the National Academy of Sciences. 101 (24), 8936-8941 (2004).
- Kapanidis, A. N., et al. Alternating-laser excitation of single molecules. Accounts of Chemical Research. 38 (7), 523-533 (2005).
- Müller, B. K., Zaychikov, E., Brauchle, C., Lamb, D. C. Pulsed interleaved excitation. Biophysical Journal. 89 (5), 3508-3522 (2005).
- Laurence, T. A., Kong, X., Jager, M., Weiss, S. Probing structural heterogeneities and fluctuations of nucleic acids and denatured proteins. Proceedings of the National Academy of Sciences of the United States of America. 102 (48), 17348-17353 (2005).
- Pollina, T., et al. PlanktonScope: Affordable modular imaging platform for citizen oceanography. bioRxiv. , 056978 (2020).
- Collins, J. T., et al. Robotic microscopy for everyone: the OpenFlexure microscope. Biomedical Optics Express. 11 (5), 2447-2460 (2020).
- Courtney, A., Alvey, L. M., Merces, G. O. T., Burke, N., Pickering, M. The Flexiscope: a low cost, flexible, convertible and modular microscope with automated scanning and micromanipulation. Royal Society Open Science. 7 (3), 191949 (2020).
- Martens, K. J. A., et al. Visualisation of dCas9 target search in vivo using an open-microscopy framework. Nature Communications. 10 (1), 3552 (2019).
- Auer, A., et al. Nanometer-scale multiplexed super-resolution imaging with an economic 3D-DNA-PAINT microscope. ChemPhysChem. 19 (22), 3024-3034 (2018).
- Li, H., et al. Squid: Simplifying quantitative imaging platform development and deployment. bioRxiv. , 424613 (2020).
- Ambrose, B., et al. The smfBox is an open-source platform for single-molecule FRET. Nature Communications. 11 (1), 5641 (2020).
- Ingargiola, A., Laurence, T., Boutelle, R., Weiss, S., Michalet, X. Photon-HDF5: An open file format for timestamp-based single-molecule fluorescence experiments. Biophysical Journal. 110 (1), 26-33 (2016).
- Hellenkamp, B., et al. Precision and accuracy of single-molecule FRET measurements-a multi-laboratory benchmark study. Nature Methods. 15 (9), 669-676 (2018).
- Ingargiola, A., Lerner, E., Chung, S., Weiss, S., Michalet, X. FRETBursts: An open source toolkit for analysis of freely-diffusing single-molecule FRET. PLOS One. 11 (8), 0160716 (2016).
- Torella, J. P., Holden, S. J., Santoso, Y., Hohlbein, J., Kapanidis, A. N. Identifying molecular dynamics in single-molecule FRET experiments with burst variance analysis. Biophysical Journal. 100 (6), 1568-1577 (2011).
- Tomov, T. E., et al. Disentangling subpopulations in single-molecule FRET and ALEX experiments with photon distribution analysis. Biophysical Journal. 102 (5), 1163-1173 (2012).
- Santoso, Y., Torella, J. P., Kapanidis, A. N. Characterizing single-molecule FRET dynamics with probability distribution analysis. ChemPhysChem. 11 (10), 2209-2219 (2010).
- Kalinin, S., Valeri, A., Antonik, M., Felekyan, S., Seidel, C. A. M. Detection of structural dynamics by FRET: A photon distribution and fluorescence lifetime analysis of systems with multiple states. The Journal of Physical Chemistry B. 114 (23), 7983-7995 (2010).
- Pirchi, M., et al. Photon-by-photon hidden Markov model analysis for microsecond single-molecule FRET kinetics. The Journal of Physical Chemistry B. 120 (51), 13065-13075 (2016).
- Schrimpf, W., Barth, A., Hendrix, J., Lamb, D. C. PAM: A framework for integrated analysis of imaging, single-molecule, and ensemble fluorescence data. Biophysical Journal. 114 (7), 1518-1528 (2018).
- Zijlstra, N., et al. Rapid microfluidic dilution for single-molecule spectroscopy of low-affinity biomolecular complexes. Angewandte Chemie International Edition. 56 (25), 7126-7129 (2017).
- Hellenkamp, B., Thurn, J., Stadlmeier, M., Hugel, T. Kinetics of transient protein complexes determined via diffusion-independent microfluidic mixing and fluorescence stoichiometry. The Journal of Physical Chemistry B. 122 (49), 11554-11560 (2018).
- Bennet, I. A., et al. Regional conformational flexibility couples substrate specificity and scissile phosphate diester selectivity in human flap endonuclease 1. Nucleic Acids Research. 46 (11), 5618-5633 (2018).

