Motility of Single Molecules and Clusters of Bi-Directional Kinesin-5 Cin8 Purified from S. cerevisiae Cells
Summary
The bi-directional mitotic kinesin-5 Cin8 accumulates in clusters that split and merge during their motility. Accumulation in clusters also changes the velocity and directionality of Cin8. Here, a protocol for motility assays with purified Cin8-GFP and analysis of motile properties of single molecules and clusters of Cin8 is described.
Abstract
The mitotic bipolar kinesin-5 motors perform essential functions in spindle dynamics. These motors exhibit a homo-tetrameric structure with two pairs of catalytic motor domains, located at opposite ends of the active complex. This unique architecture enables kinesin-5 motors to crosslink and slide apart antiparallel spindle microtubules (MTs), thus providing the outwardly-directed force that separates the spindle poles apart. Previously, kinesin-5 motors were believed to be exclusively plus-end directed. However, recent studies revealed that several fungal kinesin-5 motors are minus-end directed at the single-molecule level and can switch directionality under various experimental conditions. The Saccharomyces cerevisiae kinesin-5 Cin8 is an example of such bi-directional motor protein: in high ionic strength conditions single molecules of Cin8 move in the minus-end direction of the MTs. It was also shown that Cin8 forms motile clusters, predominantly at the minus-end of the MTs, and such clustering allows Cin8 to switch directionality and undergo slow, plus-end directed motility. This article provides a detailed protocol for all steps of working with GFP-tagged kinesin-5 Cin8, from protein overexpression in S. cerevisiae cells and its purification to in vitro single-molecule motility assay. A newly developed method described here helps to differentiate between single molecules and clusters of Cin8, based on their fluorescence intensity. This method enables separate analysis of motility of single molecules and clusters of Cin8, thus providing the characterization of the dependence of Cin8 motility on its cluster size.
Introduction
A large number of motility events within eukaryotic cells are mediated by the function of molecular motor proteins. These motors move along the cytoskeletal filaments, actin filaments, and microtubules (MTs), and convert the chemical energy of ATP hydrolysis into kinetic and mechanical forces required to drive biological motility within cells. The MT-based S. cerevisiae Cin8 is a bipolar, homotetrameric kinesin-5 motor protein that crosslinks and slides spindle MTs apart1. Cin8 performs essential functions during mitosis, in spindle assembly2,3,4 and spindle elongation during anaphase5,6,7. Previously, it had been demonstrated that Cin8 is a bi-directional motor, which switches directionality under different experimental conditions. For instance, under high ionic strength conditions, single Cin8 motors move toward the minus-end of the MTs, while in clusters, in multi-motor MT gliding assays, and between antiparallel MTs, Cin8 motors move mainly toward the plus-ends of the MTs8,9,10,11,12. These findings were highly unexpected because of several reasons. First, Cin8 carries its catalytic motor domain at the amino-terminus and such motors were previously believed to be exclusively plus-end directed, whereas Cin8 was shown to be minus-end directed at the single-molecule level. Second, kinesin motors were believed to be unidirectional, either minus-end or plus-end directed, whereas Cin8 was shown to be bi-directional, depending on the experimental conditions. Finally, because of the MT orientation at the mitotic spindle, the classical role of kinesin-5 motors in the separation of spindle poles during spindle assembly and anaphase B could only be explained by their plus-end directed motility on the MTs they crosslink1,13. Following the first reports on the bi-directionality of Cin8, a few other kinesin motors were demonstrated to be bi-directional14,15,16, indicating that the bi-directional motility of kinesin motors may be more common than earlier believed.
It has been previously reported that in cells, Cin8 also moves in a bi-directional manner8, supporting the notion that the bi-directional motility of some kinesin-5 motors is important for their intracellular functions. In addition, since the three kinesin-5 motors that were reported to be bi-directional are from fungal cells, a possible role for the bi-directionality of kinesin-5 motors has been recently proposed in such cells10. According to this model, in closed mitosis of fungal cells, where the nuclear envelope doesn't break down during mitosis, kinesin-5 motors provide the initial force that separates the spindle poles apart prior to spindle assembly. To perform this task, prior to spindle pole separation, kinesin-5 motors localize near the spindle poles, by their minus-end directed motility on single nuclear MTs. Once at this position, kinesin-5 motors cluster, switch directionality, capture, and cross-link MTs from neighboring spindle poles. Subsequently, kinesin-5 motors provide the initial separation of the poles by plus-end directed motility on the MTs they crosslink. By this model, both minus-end directed motility on single MTs and plus-end directed motility on cross-linked MTs during antiparallel sliding are required for fungal kinesin-5 motors to perform their roles in spindle assembly1,13.
The overall goal of the described method is to obtain high-purity fungal GFP-tagged kinesin-5 Cin8 and to perform single-molecule motility assays (Figure 1) while separately analyzing the motility of single molecules and clusters of Cin8. The separation between single molecules and clusters is important since one of the factors that had been demonstrated to affect the directionality of Cin8 is its accumulation in clusters on the MTs10,12. Alternative motility assays, such as the MT surface gliding and MT sliding assays do not provide information regarding the activity of single motor proteins17,18. The robust single-molecule motility assay and analysis methods described here have been successfully applied to characterize different aspects of kinesin-5 motors, Cin8 and Kip110,11,12,14,19,20.
Here, a detailed protocol is presented for Cin8 overexpression and purification, polymerization of MTs, and the single-molecule motility assay. Furthermore, the analyses to differentiate between single molecules and clusters of Cin8, and to determine single motor and cluster velocities by mean displacement (MD) and mean square displacement (MSD) analysis are also described. This protocol aims to help researchers to visualize all the steps of the procedures and assist with troubleshooting this type of assays.
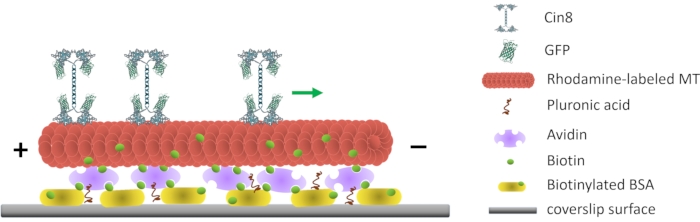
Figure 1: Schematic representation of the single-molecule motility assay. Biotinylated fluorescent MTs are attached to the glass surface, coated with Avidin that interacts with the surface-attached biotinylated-BSA. The green arrow represents the movement direction of single Cin8 molecules under high ionic strength conditions. +/- represent the polarity of the MT. Please click here to view a larger version of this figure.
Protocol
Representative Results
Discussion
In this work, a protocol for single-molecule motility assay with the bi-directional kinesin-5 Cin8 and the motility analysis are presented. The full-length Cin818 including the native nuclear localization signal (NLS) at the C-terminal has been purified from the native host S. cerevisiae. As the Cin8 is a nuclear motor protein, grinding the S. cerevisiae cells under liquid nitrogen is found to be the most efficient method for cell lysis. After lysis, by combining metal affinity a…
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This research was supported in part by the Israel Science Foundation grant (ISF-386/18) and the Israel Binational Science Foundation grant (BSF-2019008), awarded to L.G.
Materials
| Adenine | FORMEDIUM | DOC0230 | |
| ATP | Sigma | A7699 | |
| Biotinylated-BSA | Sigma | A8549 | |
| Casein | Sigma | C7078 | |
| Catalase (C40) | Sigma | C40 | |
| Creatine-Kinase | Sigma | C3755 | |
| Dithiothreitol (DTT) | Sigma | D0632 | |
| EDTA | Sigma | E5134 | |
| EGTA | Sigma | E4378 | |
| Fluorescence filter set for GFP | Chroma | 49002: ET-EGFP (FITC/Cy2) | |
| Fluorescence filter set for Rhodamine | Chroma | 49004: ET-CY3/TRITC | |
| Fluorescence inverted microscope | Zeiss | Axiovert 200M | |
| Galactose | Tivan Biotech | GAL02 | |
| Glucose | Sigma | G8270 | |
| Glucose Oxidase | Sigma | G7141 | |
| Glycerol | Sigma | G5516 | |
| GlycylGlycine | Merck | G0674 | |
| GMPCPP | Jana Bioscience | Nu-405L | |
| GTB | Cytoskeleton | BST01-010 | |
| GTP | Sigma | G8877 | |
| Histidine | Duchefa Biochemie | H0710.0100 | |
| ImageJ-FIJI software | https://imagej.net/plugins/trackmate/ | version 2.1.0/1.53c; Java 1.8.0_172 [64-bit] for Windows 10 | |
| Imidazole | Sigma | I0125 | |
| InstantBlue Coomassie Protein Stain | Abcam | ab119211 | |
| Lens | Zeiss | 100x/1.4 oil DIC objective | |
| Lysine | FORMEDIUM | DOC0161 | |
| Magnesium Chloride | Sigma | M8266 | |
| Methionine | Duchefa Biochemie | M0715.0100 | |
| Neo | Andor Technologies | sCMOS camera | |
| NeutraAvidin | Life | A2666 | |
| Ni-NTA Agarose | Invitrogen | R901-15 | |
| Phospho-Creatine | Sigma | P1937 | |
| Pipes | Sigma | P1851 | |
| Pluronic acid F-127 (poloxamer) | Sigma | P2443 | |
| Potassium Chloride | Sigma | P9541 | |
| Raffinose | Tivan Biotech | RAF01 | |
| Size Exclusion chromatography instument | GE Healthcare | AKTA Pure | |
| Spectrophotometer | ThermoFisher Scientific | NanoDrop | |
| Superose-6 10/300 GL | GE Healthcare | 17-5172-01 | |
| Tris | Roshe | 10708976001 | |
| Triton X-100 | Sigma | T8787 | |
| Tryptophan | Duchefa Biochemie | T0720.0100 | |
| Tubulin protein | Cytoskeleton | T240 | |
| Tubulin, biotinylated | Cytoskeleton | T333P | |
| Tubulin, TRITC Rhodamine | Cytoskeleton | TL530M | |
| Uracil | Sigma | U0750-100G | |
| Yeast nitrogen base | FORMEDIUM | CYN0401S | |
| α-GFP antibody | Santa Cruz Biotechnology | SC8036 | |
| β-mercaptoethanol | Sigma | M3148 |
Referencias
- Singh, S. K., Pandey, H., Al-Bassam, J., Gheber, L. Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles. Cellular and Molecular Life Sciences. 75 (10), 1757-1771 (2018).
- Hoyt, M. A., He, L., Totis, L., Saunders, W. S. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genética. 135 (1), 35-44 (1993).
- Saunders, W. S., Hoyt, M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 70 (3), 451-458 (1992).
- Hoyt, M. A., He, L., Loo, K. K., Saunders, W. S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. Journal of Cell Biology. 118 (1), 109-120 (1992).
- Gerson-Gurwitz, A., et al. Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cellular and Molecular Life Sciences. 66 (2), 301-313 (2009).
- Fridman, V., Gerson-Gurwitz, A., Movshovich, N., Kupiec, M., Gheber, L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Reports. 10 (4), 387-393 (2009).
- Movshovich, N., et al. Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. Journal of Cell Science. 121 (15), 2529-2539 (2008).
- Gerson-Gurwitz, A., et al. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. Embo Journal. 30 (24), 4942-4954 (2011).
- Roostalu, J., et al. Directional switching of the kinesin Cin8 through motor coupling. Science. 332 (6025), 94-99 (2011).
- Shapira, O., Goldstein, A., Al-Bassam, J., Gheber, L. A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. Journal of Cell Science. 130 (4), 725-734 (2017).
- Goldstein-Levitin, A., Pandey, H., Allhuzaeel, K., Kass, I., Gheber, L. Intracellular functions and motile properties of bi-directional kinesin-5 Cin8 are regulated by neck linker docking. eLife. 10, 71036 (2021).
- Pandey, H., et al. Drag-induced directionality switching of kinesin-5 Cin8 revealed by cluster-motility analysis. Science Advances. 7 (6), 1687 (2021).
- Pandey, H., Popov, M., Goldstein-Levitin, A., Gheber, L. Mechanisms by which kinesin-5 motors perform their multiple intracellular functions. International Journal of Molecular Sciences. 22 (12), 6420 (2021).
- Fridman, V., et al. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. Journal of Cell Science. 126, 4147-4159 (2013).
- Edamatsu, M. Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochemical and Biophysical Research Communications. 446 (1), 231-234 (2014).
- Popchock, A. R., et al. The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nature Communications. 8, 13999 (2017).
- Bodrug, T., et al. The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding. eLife. 9 (9), 51131 (2020).
- Gheber, L., Kuo, S. C., Hoyt, M. A. Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. Journal of Biological Chemistry. 274 (14), 9564-9572 (1999).
- Pandey, H., et al. Flexible microtubule anchoring modulates the bi-directional motility of the kinesin-5 Cin8. Cellular and Molecular Life Sciences. 78 (16), 6051-6068 (2021).
- Shapira, O., Gheber, L. Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Scientific Reports. 6, 25597 (2016).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Britto, M., et al. Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proceedings of the National Academy of Sciences of the United States of America. 113 (47), 7483-7489 (2016).
- Kapitein, L. C., et al. Microtubule cross-linking triggers the directional motility of kinesin-5. Journal of Cell Biology. 182 (3), 421-428 (2008).
- Furuta, K., Edamatsu, M., Maeda, Y., Toyoshima, Y. Y. Diffusion and directed movement in vitro motile properties of fission yeast kinesin-14 Pkl1. Journal of Biological Chemistry. 283 (52), 36465-36473 (2008).
- Katrukha, E. A., et al. Probing cytoskeletal modulation of passive and active intracellular dynamics using nanobody-functionalized quantum dots. Nature Communications. 8, 14772 (2017).
- Tinevez, J. Y., et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115, 80-90 (2017).
- Jakobs, M. A. H., Dimitracopoulos, A., Franze, K. KymoBulter, a deep learning software for automated kymograph analysis. eLife. 8, 42288 (2019).

