Isolation and Flow Cytometric Characterization of Murine Small Intestinal Lymphocytes
Summary
There is growing interest in the quantitative characterization of intestinal lymphocytes owing to increasing recognition that these cells play a critical role in a variety of intestinal and systemic diseases. In this protocol, we describe how to isolate single cell populations from different small-intestinal compartments for subsequent flow cytometric characterization.
Abstract
The intestines — which contain the largest number of immune cells of any organ in the body — are constantly exposed to foreign antigens, both microbial and dietary. Given an increasing understanding that these luminal antigens help shape the immune response and that education of immune cells within the intestine is critical for a number of systemic diseases, there has been increased interest in characterizing the intestinal immune system. However, many published protocols are arduous and time-consuming. We present here a simplified protocol for the isolation of lymphocytes from the small-intestinal lamina propria, intraepithelial layer, and Peyer's patches that is rapid, reproducible, and does not require laborious Percoll gradients. Although the protocol focuses on the small intestine, it is also suitable for analysis of the colon. Moreover, we highlight some aspects that may need additional optimization depending on the specific scientific question. This approach results in the isolation of large numbers of viable lymphocytes that can subsequently be used for flow cytometric analysis or alternate means of characterization.
Introduction
The principal task of the small intestine is often considered to be the digestion and absorption of nutrients1. While this metabolic function is clearly essential, the small intestine has an equally significant role in protecting the host from the continual barrage of environmental antigens found within the lumen2. The intestinal tract separates the outside world (e.g., luminal antigens) from the internal environment of the host with an epithelial layer that is only a single cell layer thick. As such, the small-intestinal immune system has the formidable task of balancing its threshold for reactivity, allowing foreign antigens from the diet and commensal microbes to enter the mucosa with minimal, if any, immune response while mounting a robust response against invading pathogens and other "harmful" antigens. Excessive or inappropriate immune responses to these antigens can lead to pathologic disease (e.g., inflammatory bowel disease, type I diabetes, multiple sclerosis) and must be avoided3-6.
Overall, the gastrointestinal tract is thought to represent the largest immune organ in the body, containing over 70% of all antibody-secreting cells7. The small-intestinal immune system is comprised of 3 main compartments — the lamina propria (LP), the intraepithelial layer, and Peyer's patches (PPs) — that each contains a distinctive group of lymphocytes2. The LP lymphocytes (LPLs) are primarily TCRαβ+ T cells with ~20% B cells; intraepithelial lymphocytes (IELs) contain very few B cells with more TCRγδ+ T cells than TCRαβ+ T cells; and PPs, which are secondary lymphoid organs embedded in the small-intestinal wall, contain ~80% B cells. Although each of these anatomical regions has slightly distinct functions and ontological bases, they function in a harmonized fashion to protect the host from pathogenic insults.
Furthermore, there is growing appreciation that the microbiota is a critical determinant for the development of the intestinal immune system, with increasing recognition of the cognate relationship between specific microbes and the ontogeny of particular cell lineages8,9. Moreover, given that education of the intestinal immune system affects immune responses in anatomically distant sites (e.g., arthritis, multiple sclerosis, pneumonia), it has become clear that development of the intestinal immune system is relevant to more disease processes than previously recognized10-12. As such, interest in quantitatively assessing the intestinal immune system has extended beyond host-pathogen interactions to now include host-commensal interactions and the pathogenesis of many systemic diseases as well.
Given the variability of current methods in the isolation of intestinal lymphocytes, a method that is optimized for yield, viability, and consistency while balancing the time required is increasingly critical. Protocols that involve Percoll gradients are time and labor intensive and potentially more prone to human error, leading to variable yield and viability13. Herein, we provide an optimized protocol for the isolation and characterization of lymphocytes from all 3 small-intestinal immune compartments. Additionally, given increasing interest in microbe-induced alterations in the mucosal immune system, we include steps that can be used to allow for the horizontal transmission of microorganisms between mice to assess how these changes quantitatively affect the intestinal immune system.
Protocol
All studies were conducted under strict review and guidelines according to the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School, which meets the veterinary standards set by the American Association for Laboratory Animal Science (AALAS).
1. Horizontal Transmission of Bacteria via Co-housing (Optional)
- To minimize exogenous contamination (particularly if using gnotobiotic mice), practice aseptic technique while assembling sterile disposable cages, using food and water that have both been autoclaved.
- Use ear punches or tags to individually mark 6 week old C57BL/6 mice that harbor different microbiotas and house them in the same cage. Given that mice are coprophagic and eat fecal pellets from the cage floor, microbes will naturally be transmitted horizontally between the mice.
- Co-house mice for ≥1 week to allow time for microbial transfer and immunologic change prior to analysis.
2. Preparation of a Single Cell Suspension from the Small-intestinal Intraepithelial Layer and Lamina Propria
- Preparation of Solutions
- Prepare extraction media (per small intestine): 30 ml RPMI + 93 µl 5% (w/v) dithiothreitol (DTT) + 60 µl 0.5 M EDTA + 500 µl fetal bovine serum (FBS). Add the DTT immediately before use.
- Prepare digestion media (per small intestine): 25 ml RPMI + 12.5 mg dispase + 37.5 mg collagenase II + 300 µl FBS. Add the dispase and collagenase immediately before use.
- For all incubations performed at 37 °C, prewarm solutions to 37 °C.
- Euthanize the mouse by CO2 asphyxiation followed by cervical dislocation.
- Place the mouse dorsal side down and spray the abdomen with 70% ethanol. Use scissors to perform a laparotomy by sequentially cutting the skin and then peritoneal fascia along the ventral midline from the pubic symphysis to the xiphoid process, thus exposing the peritoneal cavity.
- Use scissors to separate the small intestine from the stomach by transecting the pyloric sphincter. Gently remove the small intestine from the peritoneum, teasing away the mesenteric fat.
- To fully remove the small intestine, make a second cut at the ileo-cecal junction. Place the isolated small intestine into cold (i.e., 4 °C) RPMI containing 10% FBS to maximize cell viability.
- Gently remove large pieces of fat using curved forceps. Use caution to avoid tearing the intestinal tissue itself while trying to remove fat.
- To remove intestinal contents, gently flush intestines with 15 – 20 ml of cold PBS using an 18 G feeding needle affixed to a syringe.
- Use scissors to excise PPs, and place them into cold RPMI containing 5% FBS. PPs are located on the antimesenteric side of the small intestine and appear as a multi-lobulated white mass. Depending on the specific strain, a mouse typically has 8 – 12 PPs.
- Cut the small intestine into 3 – 4 inch segments.
- Remove residual fat by rolling each small-intestinal segment on a paper towel moistened with RPMI, using a dull scalpel to tease the fat away from the tissue.
- Turn the tissue inside out by cannulating the intestinal segments with curved forceps and grasping the distal end of the tissue. Then use a pair of straight forceps to gently remove the tissue segment from the curved forceps (beginning at the proximal end), resulting in the tissue being inverted.
- Place tissue segments in a cup containing 30 ml of extraction media and a stir bar. Secure the lid on the cup, and stir at 500 rpm for 15 min at 37 °C; stirring should be vigorous but not turbulent.
- After incubation, use a steel strainer to separate tissue pieces from the epithelium-containing supernatant, which should appear cloudy. This supernatant contains intraepithelial lymphocytes that can be further analyzed, if desired, by proceeding to filtering the sample in steps 2.18 – 2.23.
- Manually agitate the tissue pieces in RPMI to wash away residual extraction media.
- Place tissue on a dry paper towel and flip it end over end several times to facilitate removal of residual mucus that was not liberated by the extraction medium.
- Place tissue fragments in a 1.5 ml tube with 600 µl of digestion medium.
- Use scissors to mince the tissue until pieces no longer stick to the scissors and the solution appears homogenous. This step is critical to ensure complete enzymatic digestion of the tissue.
- Add the minced small intestine to a cup containing 25 ml of digestion media. Stir at 500 rpm for 30 min at 37 °C. Halfway through the digestion (i.e., at 15 min), pipet up and down with a serological pipet to help break up any large chunks of tissue.
- Filter digested tissue (or epithelial layer from step 2.12 if processing IELs) through a 100 µm cell strainer into a 50 ml tube. Rinse the strainer with 20 ml of RPMI containing 10% FBS.
- Centrifuge the filtered solution at 500 x g for 10 min at 4 °C.
- Carefully decant supernatant and resuspend pellet in 1 ml of RPMI containing 10% FBS.
- Filter resuspended cells through a 40 µm cell strainer into a 50 ml tube. Rinse strainer with 20 ml of RPMI containing 10% FBS.
- Centrifuge filtered solution at 500 x g for 10 min at 4 °C.
- Carefully decant supernatant, and resuspend pellet in 1 ml of RPMI containing 2% FBS.
3. Preparation of a Single Cell Suspension from Peyer's Patches
- Transfer excised PPs to a cup with 25 ml of digestion media and a stir bar. Secure lid, and spin at 500 rpm for 10 min at 37 °C.
- Filter digested PPs through a 40 µm cell strainer into a 50 ml tube. If any clumps remain, press them through the strainer using the flat end of the plunger from a 1 ml syringe.
- Rinse strainer with 10 ml of RPMI containing 10% FBS.
- Centrifuge filtered solution at 500 x g for 10 min at 4 °C.
- Carefully decant supernatant, and resuspend pellet in 1 ml of RPMI containing 2% FBS.
4. Surface Staining for Flow Cytometry
- Aliquot appropriate volume of cells (e.g., 200 µl of LPLs) into a 96-well round-bottom plate.
- Centrifuge at 500 x g for 5 min at 4 °C. Decant supernatant by inverting the plate.
- Resuspend cells in 90 µl of RPMI containing 2% FBS and a 1:100 dilution of anti-CD16/32 (Fc block). Incubate for 10 min at 4 °C.
- Add 10 µl PBS. Centrifuge at 500 x g for 5 min at 4 °C. Decant supernatant by inverting the plate.
- Resuspend cells in 250 µl PBS. Centrifuge at 500 x g for 5 min at 4 °C. Decant supernatant by inverting the plate.
- (Optional) Stain cells with viability dye, if desired, by following the manufacturer's protocol.
- Resuspend cells in 100 µl 1% formalin to fix the cells. Incubate for 1 hr in the dark at 4 °C.
- Add 200 µl PBS. Centrifuge at 500 x g for 5 min at 4 °C. Decant supernatant by inverting the plate.
- Resuspend cells in 250 µl PBS. Centrifuge at 500 x g for 5 min at 4 °C. Decant supernatant by inverting the plate.
- Resuspend cells in 200 µl PBS. Analyze cells using a flow cytometer.
Representative Results
Flow cytometric analysis of single cell suspensions of small-intestinal lymphocytes should yield a discrete population of cells that have similar forward and side scatter characteristics as splenocytes (Figures 1A and 1B). The lymphocytes may begin to die if the tissue is not maintained at 4 °C during the initial stages of the isolation, resulting in the lymphocyte population having a lower forward scatter and being more difficult to separate from other epithelial and dead cells (Figure 1C). Moreover, if the small-intestinal tissue is not completely cleaned of its mesenteric fat, there is virtually a complete loss of lymphocytes (Figure 1D). These features illustrate how working quickly (i.e., not allowing the tissue to warm up) and paying attention to detail (i.e., removing even small amounts of mesenteric fat) is necessary to ensure sample quality. Typically, we obtain ~80% viability for small-intestinal LPLs (Figure 1E).
Figure 2A demonstrates how the composition of the microbiota can greatly affect the numbers of specific cell populations. As we have previously demonstrated, germ-free (GF) mice, which are completely devoid of any microorganisms, have a small-intestinal immune system that is markedly immature as compared to specific pathogen-free (SPF) mice, and colonization of GF mice with a normal mouse microbiota (MMb) results in restoration of the small-intestinal immune system14. In contrast, gnotobiotic mice harboring a normal human microbiota (HMb) — and that have a similar number and diversity of fecal bacteria as MMb mice — have a small-intestinal immune system that is indistinguishable from that of GF mice14.
Taking advantage of the coprophagic nature of mice, co-housing mice together represents a simple, yet powerful method for allowing horizontal transmission of organisms between mice. This approach allows one to test whether the resulting change in the microbiota affects the small-intestinal immune system. As an example, we demonstrated that co-housing HMb mice with MMb mice for 4 weeks resulted in the normalization of the number of CD3+ CD8+ T cells in the SI LP (Figure 2B)14. This result confirms that the differences between MMb and HMb mice observed in Figure 2A is due to variations in the microbiota between these mice — variations that are overcome by co-housing the mice together.
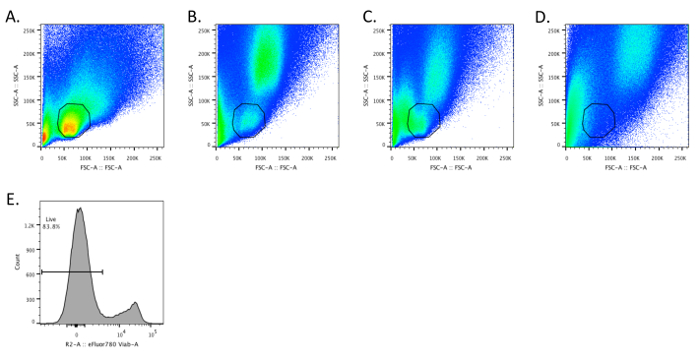
Figure 1. Intestinal Lymphocytes have FSC and SSC Characteristics Similar to Splenocytes. A – D. Dot plots depicting SSC and FSC for optimally prepared splenocytes (A) and small-intestinal lamina propria (SI LP) cells (B – D). B. The SI LP sample was prepared according to the described protocol. C. The intestinal tissue was intentionally allowed to warm to demonstrate that SI LP cells begin to have a lower FSC. D. The mesenteric fat was not cleaned off the small intestine prior to preparing single cells, resulting in the loss of a discernible SI LP lymphocyte population. E. The sample depicted in panel B was stained in step 4.7 with fixable viability dye (e.g., eFluor 780) according to the manufacturer's instructions. Please click here to view a larger version of this figure.
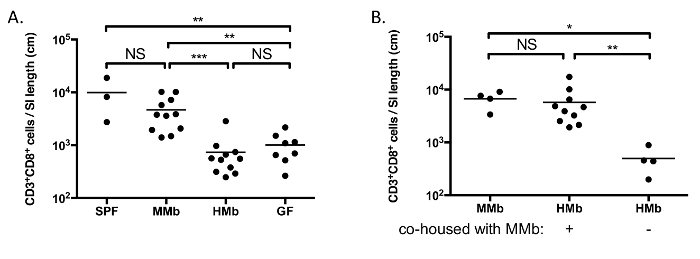
Figure 2. The Microbiota Impacts Maturation of the Small-intestinal Immune System. A. CD3+ CD8+ T cells were enumerated in the SI LP of SPF, MMb, HMb, and GF mice. B. HMb mice were co-housed with MMb mice for 4 weeks prior to enumerating CD3+ CD8+ T cells in the SI LP. MMb and HMb mice that were not co-housed were analyzed for comparison. Each data point represents an individual mouse, and the horizontal bars reflect the mean. NS, not significant; *, p <0.05; **, p <0.01; ***, p <0.001. Figures are reproduced-with permission-from reference 14, with slight modification of B. Please click here to view a larger version of this figure.
Discussion
We present a protocol for the isolation and flow cytometric characterization of small-intestinal lymphocytes, including the LPLs, IELs, and lymphocytes in the PPs. For those interested in evaluating how changes in the microbiota affect the small-intestinal immune system, we detail the straightforward steps involved in the horizontal transmission of organisms between mice harboring different microbiotas. Although this protocol focuses on the small intestine, the procedure is the same for analysis of the large intestine, with the only difference being that there are no PPs to remove (although colonic patches are present, these are typically not grossly visible).
There are several key steps in achieving optimal isolation of lymphocytes from small-intestinal tissue. Meticulous removal of adipose tissue is absolutely critical to ensuring lymphocyte viability and optimal yield. Additionally, DTT is a crucial mucolytic used in the extraction of epithelial cells. Given its unstable nature, we suggest storing single-use aliquots of a 5% solution at ≤-20 °C and not re-freezing any unused volume. Although EDTA is used in the extraction medium to aid in the removal of epithelial cells15, its presence — as well as excess FBS or mucus — during the digestion process can inhibit the collagenase. The concentration of collagenase used has been demonstrated to differentially affect surface marker expression and cell viability13,16,17. Moreover, depending on the surface marker of interest, it may be that a different formulation of collagenase (e.g., collagenase type I, II, VI, or VIII) is optimal13,17. Some empiric optimization may be required depending on the specific experimental question and, owing to lot-to-lot variation, the specific lot and relative activity level of collagenase. Furthermore, additional optimization may be required depending on the specific strain and age of mouse used. For example, the times listed here have been optimized for a ~6 week old C57BL/6 mouse; for a ~8 week old Swiss Webster mouse, the digestion time may be increased to 40 minutes.
In summary, we have provided a detailed protocol for the isolation of small-intestinal lymphocytes from the lamina propria, intraepithelial layer, and PPs. Once mastered, one can reasonably process 4 mice and have single cell suspensions ready for staining in ~4 hr. Although we describe the immediate staining of these cells for flow cytometric characterization, one can alternatively stimulate these cells to assess cytokine production, sort cells for transcriptional and/or proteomic analyses, or culture the cells to look at in vitro interactions with other cell types. Moreover, while we focused on the lymphocyte population, this protocol can also be used to examine myeloid cells. Taken together, this streamlined approach to isolating single cell populations from various intestinal compartments allows one to interrogate both how various factors (e.g., microbiota, dietary antigens) influence the ontogeny of the intestinal immune system and also how these cells may be relevant in extra-intestinal diseases.
Divulgations
The authors have nothing to disclose.
Acknowledgements
NKS is supported by NIH award K08 AI108690.
Materials
| Sterile Gloves | Kimberly-Clark | 555092 | |
| sterile mouse cage | Innovive | MS2-AD | contains lid, cage bottom, and alpha-dri bedding |
| metal feeder | Innovive | M-FEED | |
| water bottle | Innovive | M-WB-300 | |
| card holder | Innovive | CRD-HLD-H | |
| autoclavable rodent chow (NIH-31M) | Zeigler | 4131207530 | |
| RPMI medium 1640 | Gibco | 11875-119 | |
| dithiothreitol (DTT) | Sigma | D5545-5G | |
| 0.5 M EDTA (pH 8.0) | Ambion | AM9262 | |
| fetal bovine serum (FBS) | GemBio | 100-510 | |
| dispase II | Invitrogen | 17105-041 | the concentration in the protocol is based on an activity level of 1.878 U/mg |
| collagenase, type II | Invitrogen | 17101-015 | the concentration in the protocol is based on an activity level of 245 U/mg |
| dissecting scissors | Roboz | RS-5882 | |
| feeding needle (18 G, 2" length) | Roboz | FN-7905 | |
| 10 ml syringe | BD | 305482 | |
| PBS | Gibco | 14190-250 | |
| Disposable Scalpel (15 blade) | Miltex | 4-415 | |
| curved forceps | Roboz | RS-5211 | |
| straight forceps | Roboz | RS-5132 | |
| multi-purpose cups, 120 ml | VWR | 89009-662 | |
| stir bar | VWR | 58949-062 | |
| multi-position stir plate, 9-position | VWR | 12621-048 | |
| stainless steel conical strainer, 3 inch | RSVP | ||
| 1.5 ml tube | Eppendorf | 0030 125.150 | |
| 100 μm cell strainer | Falcon | 08-771-19 | |
| 40 μm cell strainer | Falcon | 08-771-1 | |
| 50 mL conical tube | Falcon | 352098 | |
| 1 ml syringe | BD | 309659 | |
| 96-well plate, round-bottom | Corning | 3799 | |
| anti-mouse CD16/32 (Fc block) | Biolegend | 101320 | |
| (optional) fixable viability dye eFluor 780 | eBiosciences | 65-0865-18 | |
| 10% formalin, neutral buffered | Thermo Scientific | 5725 |
References
- Cummings, D. E., Overduin, J. Gastrointestinal regulation of food intake. J Clin Invest. 117 (1), 13-23 (2007).
- Mowat, A. M., Agace, W. W. Regional specialization within the intestinal immune system. Nat Rev Immunol. 14 (10), 667-685 (2014).
- Round, J. L., Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 9 (5), 313-323 (2009).
- Sartor, R. B. Microbial influences in inflammatory bowel diseases. Gastroenterology. 134 (2), 577-594 (2008).
- Tlaskalova-Hogenova, H., et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 93 (2-3), 97-108 (2004).
- Wen, L., et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 455 (7216), 1109-1113 (2008).
- Pabst, R., Russell, M. W., Brandtzaeg, P. Tissue distribution of lymphocytes and plasma cells and the role of the gut. Trends Immunol. 29 (5), 206-208 (2008).
- Surana, N. K., Kasper, D. L. The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunol Rev. 245 (1), 13-26 (2012).
- Surana, N. K., Kasper, D. L. Deciphering the tete-a-tete between the microbiota and the immune system. J Clin Invest. 124 (10), 4197-4203 (2014).
- Gauguet, S., et al. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect Immun. , (2015).
- Ochoa-Reparaz, J., et al. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 3 (5), 487-495 (2010).
- Wu, H. J., et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 32 (6), 815-827 (2010).
- Goodyear, A. W., Kumar, A., Dow, S., Ryan, E. P. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J Immunol Methods. 405, 97-108 (2014).
- Chung, H., et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 149 (7), 1578-1593 (2012).
- Resendiz-Albor, A. A., Esquivel, R., Lopez-Revilla, R., Verdin, L., Moreno-Fierros, L. Striking phenotypic and functional differences in lamina propria lymphocytes from the large and small intestine of mice. Life Sci. 76 (24), 2783-2803 (2005).
- Carrasco, A., et al. Comparison of lymphocyte isolation methods for endoscopic biopsy specimens from the colonic mucosa. J Immunol Methods. 389 (1-2), 29-37 (2013).
- Van Damme, N., et al. Chemical agents and enzymes used for the extraction of gut lymphocytes influence flow cytometric detection of T cell surface markers. J Immunol Methods. 236 (1-2), 27-35 (2000).

