Isolation of Lipoprotein Particles from Chicken Egg Yolk for the Study of Bacterial Pathogen Fatty Acid Incorporation into Membrane Phospholipids
Summary
This method provides a framework for studying incorporation of exogenous fatty acids from complex host sources into bacterial membranes, particularly Staphylococcus aureus. To achieve this, protocols for the enrichment of lipoprotein particles from chicken egg yolk and subsequent fatty acid profiling of bacterial phospholipids utilizing mass spectrometry are described.
Abstract
Staphylococcus aureus and other Gram-positive pathogens incorporate fatty acids from the environment into membrane phospholipids. During infection, the majority of exogenous fatty acids are present within host lipoprotein particles. Uncertainty remains as to the reservoirs of host fatty acids and the mechanisms by which bacteria extract fatty acids from the lipoprotein particles. In this work, we describe protocols for enrichment of low-density lipoprotein (LDL) particles from chicken egg yolk and determining whether LDLs serve as fatty acid reservoirs for S. aureus. This method exploits unbiased lipidomic analysis and chicken LDLs, an effective and economical model for the exploration of interactions between LDLs and bacteria. The analysis of S. aureus integration of exogenous fatty acids from LDLs is performed using high-resolution/accurate mass spectrometry and tandem mass spectrometry, enabling the characterization of the fatty acid composition of the bacterial membrane and unbiased identification of novel combinations of fatty acids that arise in bacterial membrane lipids upon exposure to LDLs. These advanced mass spectrometry techniques offer an unparalleled perspective of fatty acid incorporation by revealing the specific exogenous fatty acids incorporated into the phospholipids. The methods outlined here are adaptable to the study of other bacterial pathogens and alternative sources of complex fatty acids.
Introduction
Methicillin-resistant S. aureus (MRSA) is the leading cause of healthcare-associated infection and the associated antibiotic resistance is a considerable clinical challenge1,2,3. Therefore, the development of novel therapeutic strategies is a high priority. A promising treatment strategy for Gram-positive pathogens is inhibiting fatty acid synthesis, a requirement for membrane phospholipid production that, in S. aureus, includes phosphatidylglycerol (PG), lysyl-PG, and cardiolipin4. In bacteria, fatty acid production occurs via the fatty acid synthesis II pathway (FASII)5, which is considerably different from the eukaryotic counterpart, making FASII an attractive target for antibiotic development5,6. FASII inhibitors primarily target FabI, an enzyme required for fatty acid carbon chain elongation7. The FabI inhibitor triclosan is broadly used in consumer and medical goods8,9. Additional FabI inhibitors are being developed by several pharmaceutical companies for the treatment of S. aureus infection10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26. However, many Gram-positive pathogens, including S. aureus, are capable of scavenging exogenous fatty acids for phospholipid synthesis, bypassing FASII inhibition27,28,29. Thus, the clinical potential of FASII inhibitors is debated due to considerable gaps in our knowledge of the sources of host fatty acids and the mechanisms by which pathogens extract fatty acids from the host27,28. To address these gaps, we developed an unbiased lipidomic analysis method to monitor incorporation of exogenous fatty acid from lipoprotein particles into membrane phospholipids of S. aureus.
During sepsis, host lipoprotein particles represent a potential source of host-derived fatty acids within the vasculature, as a majority of host fatty acids are associated with the particles30. Lipoproteins consist of a hydrophilic shell composed of phospholipids and proteins that enclose a hydrophobic core of triglycerides and cholesterol esters31. Four major classes of lipoproteins–chylomicron, very low-density lipoprotein, high-density lipoprotein, and low-density lipoprotein (LDL)—are produced by the host and function as lipid transport vehicles, delivering fatty acids and cholesterol to and from host cells via the vasculature. LDLs are abundant in esterified fatty acid including triglycerides and cholesterol esters31. We have previously demonstrated that highly purified human LDLs are a viable source of exogenous fatty acids for PG synthesis, thus providing a mechanism for FASII inhibitor bypass32. Purifying human LDLs can be technically challenging and time consuming while commercial sources of purified human LDLs are prohibitively expensive to use on a routine basis or to perform large-scale bacterial screens. To address these limitations, we have modified a procedure for the enrichment of LDLs from chicken egg yolk, a rich source of lipoprotein particles33. We have successfully used untargeted, high-resolution/accurate mass spectrometry and tandem mass spectrometry to monitor incorporation of human LDL-derived fatty acids into the membrane of S. aureus32. Unlike previously reported methods, this approach can quantify individual fatty acid isomers for each of the three major staphylococcal phospholipid types. Oleic acid (18:1) is an unsaturated fatty acid present within all host lipoprotein particles that is readily incorporated into S. aureus phospholipids29,30,32. S. aureus is not capable of oleic acid synthesis29; therefore, the quantity of phospholipid-incorporated oleic acid establishes the presence of host lipoprotein-derived fatty acids within the staphylococcal membrane29. These phospholipid species can be identified by the state-of-the-art mass spectrometry method described here, offering unprecedented resolution of the membrane composition of S. aureus cultured in the presence of a fatty acid source it likely encounters during infection.
Protocol
NOTE: The following protocol for enrichment of LDL particles from chicken egg yolk is derived from Moussa et al. 200233.
1. Preparation of chicken egg yolk for enrichment of LDL particles
- Sanitize two large chicken eggs by washing the shells with 70% ethanol solution and allow to air dry.
- Sanitize the egg separator using 70% ethanol solution and allow to air dry. Attach the egg separator onto the lip of a medium sized beaker.
- Crack each egg individually into the egg separator and allow the albumen to flow into the beaker. The intact egg yolk will be retained by the separator.
- Wash the egg yolk twice with 30 mL of sterile phosphate-buffered saline (PBS) to remove residual albumen.
- Gently place the egg yolk onto filter paper.
- Puncture the vitelline membrane with a sterile pipette tip and drain the contents of the membrane into a sterile 50 mL conical centrifuge tube. Discard the membrane and filter paper.
2. Fractionation of LDL-containing plasma from chicken egg yolk
- Add approximately two volumes of 0.17 M NaCl at pH 7.0 to the egg yolk and mix vigorously. Then mix this solution at 4 °C for 60 min.
- Centrifuge the egg yolk dilution at 10 °C at 10,000 x g for 45 min. Remove the plasma fraction (supernatant) from the granular fraction (pellet) into a sterile 50 mL conical tube.
- Repeat step 2.2.
3. Isolation of LDL particles from plasma
- Mix the plasma fraction with 40% ammonium sulfate (w/v) at 4 °C for 60 min.
- Adjust the pH of the plasma fraction with a 420 mM NaOH solution to 8.7.
- Centrifuge the egg yolk dilution at 4 °C at 10,000 x g for a duration of 45 min. Remove the upper semisolid yellow fraction into 7 kDa pore size dialysis tubing. Provide room in the tubing to allow for it to swell.
- Dialyze overnight at 4 °C in 3 L of ultrapure water to remove the ammonium sulfate. Gently agitate the water using a stir bar.
- Transfer dialyzed solution into a sterile 50 mL conical centrifuge tube.
- Centrifuge the solution at 4 °C at 10,000 x g for a duration of 45 min. Carefully remove the upper semisolid yellow fraction to a sterile tube and store at 4 °C.
4. Assessment of chicken LDLs as a source of fatty acids
- Subculture S. aureus cells into 5 mL of fatty acid-free 1% tryptone broth and incubate overnight at 37 °C with shaking (225 rpm). For fatty acid auxotrophs, supplement cultures with a source of fatty acids.
- Dilute overnight cultures to an optical density (OD) at 600 nm (OD600) of 0.1 in 1% tryptone broth. Pipette 50 μL of the cell suspension into each well of a round-bottom 96-well plate.
NOTE: When working with fatty acid auxotrophs, wash the overnight cultures in two volumes of tryptone broth and resuspend in 5 mL of tryptone broth to limit carryover of fatty acids before determining the OD of the culture. - For the wells containing untreated controls, add 50 µL of 1% tryptone broth per well.
- To the wells containing the experimental cell suspensions, add 50 µL of 1% tryptone broth supplemented with 10% egg yolk-derived LDL, 2 μM triclosan, or a mixture of 10% egg yolk-derived LDL and 2 μM triclosan.
NOTE: At this point, each well will contain 100 μL and the final concentration of egg yolk-derived LDL and triclosan per well will be 5% and 1 µM, respectively. - Measure OD600 over time, using a microplate reader set at 37 °C with continuous, linear shaking to monitor growth.
5. Incubation of S. aureus with LDLs for membrane lipid analysis.
- Culture an isolated colony into 5 mL of fatty acid-free 1% tryptone broth and incubate overnight at 37 °C with shaking (225 rpm).
- Dilute overnight cultures 1:100 into a sterile 250 mL baffled flask containing 50 mL of 1% tryptone broth. Incubate to mid-log phase (approximately 4 h) at 37 °C with shaking.
- Transfer 25 mL of culture to a sterile 50 mL centrifuge tube and pellet the cells. Remove the supernatant and resuspend the cell pellet in 750 µL of 1% tryptone broth.
- Combine the resuspended cells and aliquot 300 µL of the cell suspension into a sterile 1.5 mL centrifuge tube.
- Add LDLs to the desired final concentration and incubate at 37 °C with shaking (225 rpm) for 4 h.
- Centrifuge the cultures at 4 °C at 16,000 x g for a duration of 2 min and wash the cell pellets in two volumes of sterile PBS then repeat.
- Record the weight of each wet cell pellet. Snap-freeze the cell pellets on dry ice or in liquid nitrogen and store at -80 °C or proceed directly to section 6.
6. Extraction of S. aureus membrane lipids
- Place frozen S. aureus cell pellets on dry ice. Add 0.5 mm zirconium oxide beads on top of each cell pellet, using a volume of beads approximately equal to the volume of the cell pellet.
NOTE: As an alternative to this method of lipid extraction, researchers can use the well-established Bligh and Dyer or Folch methods for exacting lipids from bacterial cells34. - Add 740 µL of 75% methanol (HPLC grade) chilled to -80 °C directly to the cell pellets.
- Add 2 μL of 50 µM dimyristoyl phosphatidylcholine (prepared in methanol) per 1 mg of cells as an internal standard. Close the test tube and place the 1.5 mL centrifuge tubes containing each sample into an available port in a Bullet Blender tissue homogenizer. Homogenize the samples on low speed, setting 2-3, for 3 min.
- Visually inspect the samples for homogeneity. If clumps of cells are visible, continue homogenization in the Bullet Blender in 2 min increments.
- Remove the samples from Bullet Blender and transfer to a chemical fume hood.
- Add 270 µL of chloroform to each sample tube. Vortex the samples vigorously for 30 min.
CAUTION: Chloroform is a possible carcinogen. - Centrifuge the samples in a benchtop centrifuge for up to 30 min at a minimum 2,000 x g. Faster speeds may be used with compatible centrifuge tubes and the duration of centrifugation may be shortened to 10 min.
- In a chemical fume hood, collect the monophasic supernatant and transfer to a new test tube, while carefully avoiding the protein pellet at the bottom of the extraction tube.
- Add 740 µL of 75% methanol (HPLC grade) and 270 µL of chloroform to the protein pellet, and re-extract each sample as described in steps 6.6-6.8 above. Combine the supernatant from the second extraction with the previously collected supernatant for each sample.
- Evaporate the extraction solvents under a stream of inert gas such as nitrogen or argon, or under vacuum using a centrifuge concentrator (Table of Materials).
- Wash dried lipid extracts three times with 1.0 mL of aqueous 10 mM ammonium bicarbonate solution and re-dry the samples as in step 6.10.
- Resuspend the dried lipid extracts in a suitable nonpolar solvent such as isopropanol. Resuspend samples using 20 µL per 1 mg of fresh cell weight determined in step 5.7 Alternatively, if the weight of the cell pellets is unknown, resuspend the samples in 200 µL of isopropanol and proceed to Section 7.
7. Analysis of S. aureus lipid profiles using high resolution/accurate mass spectrometry
- Prior to conducting a full lipid analysis, select representative test samples from the experimental group(s) and analyze them over a range of sample dilution factors to determine sample dilution ranges in which the total lipid concentrations fall within the linear range of detector response for the mass spectrometer, as previously described35.
- Evaporate aliquots of each sample lipid extract to be subjected to lipid analysis, by drying the aliquots under inert gas or under vacuum in a centrifuge concentrator (Table of Materials).
- Resuspend each dried lipid extract in liquid chromatography–mass spectrometry (LC-MS) grade isopropanol:methanol (2:1, v:v) containing 20 mM ammonium formate, using volumes equivalent to an optimal sample dilution factor as determined in step 7.1.
- For an untargeted lipid analysis, samples may be introduced directly to the high resolution/accurate mass spectrometry platform without the use of chromatography by flow injection or direct infusion of extracts35,36. Transfer the diluted lipid extracts prepared in step 7.3 to an appropriate autosampler vial or 96-well plate.
- For flow injection-based analysis, place the autosampler vials into a temperature-controlled (15 °C) autosampler of an HPLC system capable of capillary/low flow applications, such as a HPLC (Table of Materials) equipped with an electronic flow proportioning and flow monitoring system.
- Fill the HPLC solvent reservoirs with LC-MS grade isopropanol:methanol (2:1, v:v) containing 20 mM ammonium formate.
- Using Agilent Chemstation software, program the HPLC autosampler to perform 5 µL sample injections. From the Instrument menu, select Set Up Injector, and type 5.0 in the Injection Volume field. The units are given as microliters. Ensure that the HPLC is set to isocratic flow at 1 µL per min of 2:1 (v:v) isopropanol:methanol containing 20 mM ammonium formate.
- From the Chemstation Instrument menu, select Set Up Pump… and then select the Micro Flow mode toggle switch.
- In the Timetable fields, enter: Time 0.00, 100% B, Flow 1.0. Hit Enter and create a second row in the Timetable by entering Time 10.0, 100% B, Flow 1.0. Select the OK button at the bottom of the Set Up Pump menu. These settings will enable 10 analytical runs at a flow rate of 1.0 µL per minute.
- Introduce eluate from the HPLC transfer line to the mass spectrometer using an electrospray ionization source fitted with a low-flow (34 G) metal needle.
- Using Thermo Tune Plus instrument control software, select the Setup menu and select Heated ESI Source. Set the ionization voltage to 4000 V and sheath gas to 5 (arbitrary units) by typing these values into the corresponding fields of the dialogue box. Similarly, set the Capillary Temp to 150 °C and the S-lens to 50%.
NOTE: These values need to be optimized for each mass spectrometry platform. - For untargeted lipid analysis, use a high resolution/accurate mass MS platform (Table of Materials) as the detector.
- Using Thermo Tune Plus software, click the Define Scan button, and in the Analyzer menu select FTMS. Set the Mass Range field to Normal and in the Resolution field select 100,000. Ensure that Scan Type is set to Full. Under the Scan Ranges menu, enter 200 in the First Mass (m/z) field, and enter 2000 in the Last Mass (m/z) field.
- Ensure that negative polarity is utilized to detect the most abundant S. aureus lipids.
- Repeat the sample analysis using ion mapping tandem mass spectrometry (MS/MS) fragmentation of all lipid ions within a spectral region of interest in order to confirm lipid structures and fatty acid constituents. Alternatively, selected lipid ions of interest may be subjected to MS/MS analysis after initial lipid identifications have been assigned in Section 8.
8. Database searching to identify endogenous S. aureus and exogenous LDL-derived lipids
- Use Thermo Xcalibur software to further refine observed mass accuracy. In Xcalibur, under the Tools menu, select the Recalibrate Offline. After the Recalibrate Offline window opens, load the mass spectrum file to be recalibrated by selecting the File menu and selecting the Open option.
- Open the file of interest, toggle the Insert Row button at the top of the view window, to view the total ion chromatogram for the MS run. Average the acquired MS signals by left-clicking the computer mouse on one edge of the observed signal peak in the total ion chromatogram and dragging the mouse across the broadest part of the peak.
- Under the Scan Filter menu, select the filter corresponding to full scan MS data. Load a reference file containing the theoretical monoisotopic masses of at least three known S. aureus endogenous lipids by selecting the Load Ref… button and selecting the reference file. Check the Use box next to each lipid monoisotopic mass.
- Click the Search button at the bottom of the viewing window. Re-average the MS signal across the signal peak in the total ion chromatogram as done previously.
- Click the Convert button near the bottom of the viewing window. When the Convert dialogue box opens, click OK. Omit this step if data was collected on mass spectrometry platforms from vendors other than Thermo Scientific.
- Using Xcalibur software, export the recalibrated accurate mass peak lists for each untreated or LDL-treated sample to separate worksheets of an Excel file. Select the Qual Browser icon. Open the recalibrated file of interest by selecting the File menu and selecting the Open… option.
- Average the signal across the broad peak in the total ion chromatogram as described in step 8.2.
- Right click the thumbtack icon in the mass spectrum viewing window and select View | Spectrum List. From the same menu, select Display Options and then All Peaks toggle box in the Display menu. Click the OK button to close the viewing window.
- Right click the thumbtack icon in the mass spectrum viewing window again and select the Export | Clipboard (Exact Mass). Paste the exported data cell A1 of the first worksheet into a new Excel spreadsheet.
- Delete the first 8 rows of text in the exported data file, such that cell A1 of the Excel spreadsheet contains the first mass data point from the mass spectrum. Repeat the exporting of each recalibrated MS file, using a new worksheet in the Excel file for each exported peak list.
- Using the Lipid Mass Spectral Analysis (LIMSA) software37 Add-In for Excel, construct a database containing molecular formulas of known S. aureus lipid species as described by Hewelt-Belka et al. 201438, as well as formulas representing lipid species that could hypothetically be present in LDL.
NOTE: Additionally, care should be taken to include potential molecular formulas in the database for hypothetical bacterial lipids that have incorporated major LDL fatty acids, such as oleic (18:1) and linoleic (18:2) fatty acids32. - To construct the database, open a blank Excel spreadsheet. In cell A1 of the first worksheet, type the theoretical/computed monoisotopic mass of the lipid species to be added to the database, corresponding to the mass of the lipid species in the ionic state observed in the mass spectrometer. In cell B1, enter a name for the lipid species, such as PG(34:0).
- In cell C1, enter the molecular formula for the lipid species, corresponding to the ionic state of the lipid observed in the mass spectrometer. In cell D1, enter the charge of the lipid species as observed in the mass spectrometer. Move to cell A2 to begin a new entry for the next lipid species to be entered in the searchable database.
- Repeat the steps 8.12 and 8.13 until all desired lipid species have been entered into the database. Save the database file and leave it open in Excel.
- In Excel, select the Add-Ins menu in. Select LIMSA to start the LIMSA software. From the main menu, click the Compound Library… button. In the new window that appears, click Import Compounds. This will upload the compound database for use by the LIMSA software.
- Perform accurate-mass based lipid identifications on all MS spectra using the LIMSA software Add-In for Excel according to the vendor’s instructions35. From the LIMSA main menu, select Peak List under the Spectrum type menu. Select Positive mode or Negative mode to correspond with the polarity in which the MS data was acquired.
- In the Peak fwhm (m/z) window, enter the desired mass search window for peak finding. A mass tolerance search window of 0.003-0.005 m/z is recommended for high resolution/accurate mass MS data.
- In the Sensitivity window, enter the desired baseline cutoff (for example, 0.01% relative abundance). In the Isotope correction menu, select Linear fit or Subtract algorithms, either of which may be used with peak list data.
- Highlight lipid compounds to include in the database search by clicking on desired lipid species within the Available Compounds window. Click the Add button to add highlighted lipid species to the search group.
- Define internal standards by clicking on added compounds, then changing the Concentration window to the corresponding concentration of the selected internal standard.
- Ensure that the internal standard and selected lipid species to be quantitated belong to the same class name by selecting each added lipid species and internal standard and typing a class name (such as PG or Lipid) in the Class field.
- To save the searchable group of lipid compounds for future use, click the Save button next to the Groups menu.
- Ensure that the Excel file containing all exported MS peak lists is open to the sheet corresponding to the first S. aureus MS run, and then click the Search button from the LIMSA main menu.
NOTE: The output of the LIMSA database search will include a list of mass spectral features matched to lipids present in the database constructed in steps 8.12-8.13, as well as concentrations for each matched feature following normalization to one or more selected internal standards. - Use Xcalibur software to examine accurate mass MS/MS spectra for m/z corresponding to lipid ions of interest in order to confirm fatty acid constituents present in each identified lipid molecular species. Select the Qual Browser icon. Open the MS/MS file of interest by selecting the File drop-down menu and selecting the Open… option.
- Select the scan filter corresponding to MS/MS analysis of a lipid m/z of interest by right mouse-clicking on the thumbtack icon in the mass spectrum viewing window and select Ranges from the menu. In the new window that appears, select the Filter menu to select the scan.
- Average the signal across the total ion chromatogram as described in step 8.2. Use MetaboAnalyst (www.metaboanalyst.ca) software to perform appropriate statistical tests. Evaluate statistically significant difference in S. aureus lipid composition by comparing normalized lipid abundances across untreated and LDL-treated conditions.
Representative Results
The protocol for the enrichment of LDL from chicken egg yolk is illustrated in Figure 1. This process begins by diluting whole egg yolk with saline and separating the egg yolk solids referred to as granules from the soluble or plasma fraction containing the LDLs (Figure 1)33. The LDL content of the plasma fraction is further enriched by precipitation of the ~ 30-40 kDa β-livetins (Figure 2)33. The presence of protein bands at 140, 80, 65, 60 and 15 kDa correlate with the apoproteins of LDLs (Figure 2)33,39. Treatment with triclosan inhibits growth of S. aureus in fatty acid-free media32. We have previously demonstrated that supplementing cultures with egg yolk plasma or purified human LDLs as exogenous fatty acid sources overcomes triclosan-induced growth inhibition (Figure 3)32. Similarly, supplementation of triclosan-treated cultures with enriched egg yolk LDL restores growth (Figure 3). Further, addition of egg yolk LDLs support the growth of a previously characterized S. aureus fatty acid auxotroph (Figure 4)32. For the most accurate mass spectrometry-based profiling of S. aureus incorporation of exogenous fatty acids, it is important to limit the presence of free fatty acids in the growth medium. The free fatty acid composition of 1% tryptone broth and chicken egg yolk LDLs diluted in tryptone broth was determined by employing flow injection high-resolution/accurate mass spectrometry and found minimal quantities of free fatty acid (Figure 5). The same untargeted mass spectrometry analysis was performed to determine the fatty acid composition of S. aureus phospholipids after exposure to chicken egg yolk LDLs. Orthogonal partial least-squares discriminant analysis (OPLS-DA)40 of abundant S. aureus membrane phospholipids demonstrated clear class separation of untreated and chicken egg yolk LDL-treated conditions, as shown in the OPLS-DA scores plot (Figure 6A). The OPLS-DA loadings plot indicated numerous phosphatidylglycerol species as important variables in the PLS-DA model. Notably, phospholipids containing unsaturated fatty acids, a molecular marker of exogenous fatty acid incorporation, are enriched in the LDL supplemented cultures compared to cells incubated in the absence of LDLs (Figure 6B). Previous studies have found that chicken egg yolks are a rich source of unsaturated fatty acids with oleic acid (18:1) being the most abundant41,42. In agreement with these observations, we found oleic acid to be the most common unsaturated fatty acid utilized for phospholipid synthesis when S. aureus cultures were supplemented with chicken egg yolk LDLs (Figure 6C). Table 1 further illustrates that the fatty acid profiles of membrane phospholipids are altered when S. aureus is grown in the presence of egg yolk LDL.
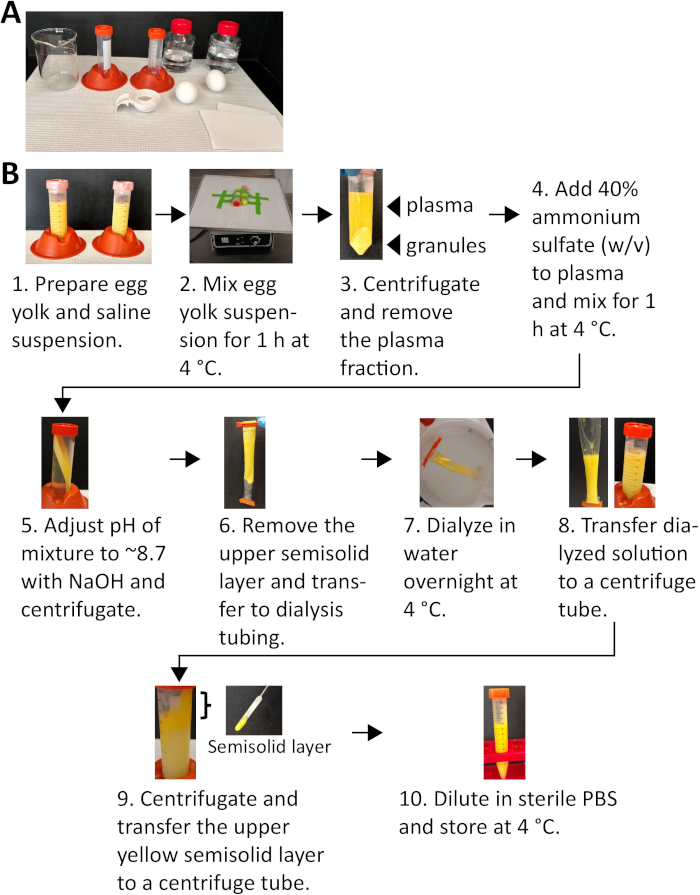
Figure 1: An illustration of LDL enrichment from chicken egg yolk utilizing centrifugation and ammonia sulfate precipitation. (A) The reagents necessary for the enrichment of LDL from chicken egg yolk. (B) The flow chart depicts the significant steps of the LDL enrichment process. Please click here to view a larger version of this figure.

Figure 2: Protein profile of chicken egg yolk prior to and after enrichment for LDL. Protein lysates were prepared using RIPA buffer. Protein lysate (15 µg) was loaded into an 8% acrylamide SDS-PAGE gel. Gels were stained overnight with Bio Rad Protein reagent. The molecular weights in kDa of LDL associated proteins are denoted along the right side of the image. M: protein marker, Y: chicken egg yolk, and LDL: chicken egg yolk LDL enrichment Please click here to view a larger version of this figure.

Figure 3: Egg yolk-derived LDLs protect S. aureus from triclosan-induced FASII inhibition. The growth of S. aureus was monitored over time via measurement of OD600 in 1% tryptone broth in the following conditions: 1% tryptone broth (TB), 1 µM triclosan (TCS), 1 µM triclosan with 1% egg yolk plasma (TCS + EYP), 5% egg yolk LDL (LDL), or 1 µM triclosan with 5% egg yolk LDL (TCS + LDL). The mean from three independent experiments is shown. Error bars represent the standard deviation of the mean. Please click here to view a larger version of this figure.
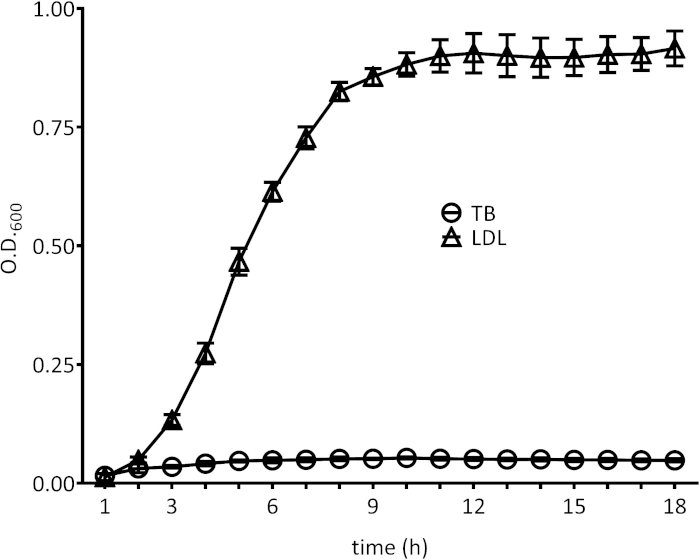
Figure 4: Growth of a S. aureus fatty acid auxotroph is supported by egg yolk-derived LDL. The growth of a fatty acid auxotroph in 1% tryptone broth (TB) with or without 5% egg yolk LDL (LDL) supplementation was monitored over time via measurement of OD600. The mean from three independent experiments is shown. Error bars represent the standard deviation of the mean. Please click here to view a larger version of this figure.

Figure 5: Free fatty acid content measured in 1% tryptone broth or chicken egg yolk LDL. Free fatty acids were detected by flow injection high-resolution/accurate mass spectrometry and tandem mass spectrometry. Normalized numbers of ions per mg of protein was determined for 1% tryptone broth and 1% tryptone broth supplemented with 5% chicken egg yolk LDLs. Please click here to view a larger version of this figure.

Figure 6: Chicken egg yolk low-density lipoproteins are a reservoir of exogenous fatty acids for synthesis of S. aureus phosphatidylglycerol. (A) Scores plot of orthogonal partial least-squares discriminant analysis of chicken egg yolk LDL-treated and untreated S. aureus membrane phospholipids identified using high resolution/accurate mass spectrometry. (B) Percentage of unsaturated phosphatidylglycerol (UPG) compared to total membrane PG of S. aureus grown in the absence (WT) or presence (WT + LDL) of chicken egg yolk LDLs. (C) Unsaturated fatty acid (UFA) profile of membrane PG of S. aureus grown without (WT) or with (WT + LDL) chicken egg yolk LDLs graphed as a percentage of the amount of total PG fatty acids. Please click here to view a larger version of this figure.
| WT cultured in tryptone broth | WT cultured in tryptone broth supplemented with LDLs | |||||
| Phosphatidyl glycerol (TC:TDB)a | Normalized ion abundance/mg of protein | SD | Fatty acidsc | Normalized ion abundance/mg of protein | SD | Fatty Acidsc |
| 24:0 | 0 | 0 | NDb | 0.052031116 | 0.02677 | ND |
| 26:0 | 0 | 0 | ND | 0.009539117 | 0.00362 | ND |
| 28:0 | 0.127937113 | 0.04528 | 15:0_13:0 | 0.167643281 | 0.02392 | 15:0_13:0 |
| 28:1 | 0.006765427 | 0.00157 | ND | 0.002776821 | 0.00372 | 15:0_13:1 |
| 30:0 | 8.680180809 | 2.68375 | 15:0_15:0 | 14.04873592 | 2.4531 | 15:0_15:0 |
| 30:1 | 0 | 0 | ND | 0.010152161 | 0.00449 | 15:1_15:0, 13:1_17:0 |
| 31:0 | 4.150511117 | 1.31658 | 16:0_15:0, 14:0_17:0 | 10.17590926 | 1.88431 | 16:0_15:0, 14:0_17:0, 18:0_13:0 |
| 31:1 | 0.016156004 | 0.01216 | 13:1_15:0, 12:1_19:0 | 0.473478683 | 0.09063 | 13:1_15:0, 18:1_13:0, 12:1_19:0 |
| 32:0 | 29.29259262 | 8.82993 | 15:0_17:0 | 48.24342037 | 8.95664 | 15:0_17:0, 16:0_16:0 |
| 32:1 | 0.02074815 | 0.00941 | ND | 0.307044942 | 0.07305 | 18:1_14:0, 16:1_16:0 |
| 33:0 | 9.000460122 | 2.78194 | 18:0_15:0, 16:0_17:0 | 15.4531776 | 2.98171 | 18:0_15:0, 16:0_17:0 |
| 33:1 | 0.162934812 | 0.04796 | ND | 2.921832928 | 0.30851 | 18:1_15:0 |
| 33:2 | 0 | 0 | ND | 0.167492702 | 0.03211 | 18:1_15:1, 18:2_15:0 |
| 34:0 | 12.3064043 | 3.70242 | 19:0_15:0, 17:0_17:0 | 18.40129157 | 3.21385 | 19:0_15:0, 17:0_17:0 |
| 34:1 | 0 | 0 | ND | 1.423605186 | 0.20066 | 18:1_16:0 |
| 34:2 | 0.000470922 | 0.00082 | ND | 0.156133734 | 0.03929 | 18:2_16:0 |
| 35:0 | 5.727462455 | 1.74583 | 20:0_15:0, 18:0_17:0 | 7.771538992 | 1.28515 | 20:0_15:0, 16:0_19:0, 18:0_17:0 |
| 35:1 | 0.17337586 | 0.05727 | 20:1_15:0 | 0.772202525 | 0.08526 | 20:1_15:0, 18:1_17:0 |
| 35:2 | 0 | 0 | ND | 0.038758757 | 0.01481 | 18:2_17:0, 18:1_17:1 |
| 36:0 | 0.671004303 | 0.2116 | 21:0_15:0, 19:0_17:0 | 0.967295024 | 0.2572 | 21:0_15:0, 20:0_16:0, 19:0_17:0, 22:0_14:0 |
| 36:2 | 0 | 0 | ND | 0.495485065 | 0.04473 | 18:1_18:1, 18:2_18:0 |
| 36:3 | 0 | 0 | ND | 0.059268233 | 0.02291 | 18:2_18:1, 20:3_16:0, 20:2_16:1 |
| 37:0 | 0.060466411 | 0.01961 | 22:0_15:0, 20:0_17:0 | 0.114526894 | 0.01852 | 22:0_15:0, 20:0_17:0, 18:0_19:0 |
| 38:2 | 0 | 0 | ND | 0.079469521 | 0.02872 | 18:2_20:0, 16:1_20:1 |
| aDetected as [M-H]– ions. TC, total chain length; TDB, total number of double bonds. | ||||||
| bND, not determined | ||||||
| cFatty acids are listed in order of isomer abundance. An underscore between fatty acid designations indicates that each fatty acid may be present in either the SN1 or SN2 position, as tandem mass spectrometry alone cannot rule out the possibility that lipid species exist as a mixture of positional isomers. | ||||||
Table 1: Fatty acid profile of S. aureus cultured in the presence of chicken egg yolk LDLs. We used an unbiased lipidomic analysis utilizing high-resolution/accurate MS and MS/MS to determine the fatty acid profile of S. aureus PG. S. aureus was incubated in the presence or absence of chicken egg yolk LDLs, and the PG profile of these cells was compared to that of cells cultured in 1% tryptone broth.
Discussion
S. aureus incorporates exogenous fatty acids into its membrane phospholipids27,32,43. Phospholipid synthesis using exogenous fatty acids bypasses FASII inhibition but also alters the biophysical properties of the membrane27,32,44. While incorporation of exogenous fatty acids into phospholipids of Gram-positive pathogens is well documented, gaps remain in the identity of host fatty acid reservoirs and the structural alterations to each of the three major staphylococcal phospholipid types that result from exogenous fatty acid incorporation. Here, we describe protocols which can be employed to: i) enrich LDL particles from chicken egg yolk, a source of fatty acids, ii) determine the effects of exogenous fatty acids on the growth of S. aureus, and iii) utilize an unbiased lipidomic analysis for monitoring exogenous fatty acid incorporation into membrane phospholipids of S. aureus. The advanced mass spectrometry method provided in this study offers an extraordinary perspective of the membrane composition of S. aureus grown in the presence of exogenous fatty acids.
Several Gram-positive pathogens utilize exogenous fatty acids for membrane synthesis and, as with S. aureus, the possible sources of exogenous fatty acids during infection are poorly understood27,43. The growth analysis described here can be modified to evaluate the proliferation of other Gram-positive pathogens in the presence of lipoproteins if the growth kinetics of each pathogen are considered. Additionally, other complex host sources of exogenous fatty acids could be tested using this protocol if the potential effects of the fatty acid source on background optical density are controlled. Moreover, the described mass spectrometry method for analysis of bacterial lipids is sufficiently flexible to enable lipidome evaluation from virtually any bacterial species. As lipid accurate mass data is collected in ‘full scan’ MS mode, little a priori knowledge of the lipid content of the bacterial species of interest is required, unlike targeted analytical methods based on known fragmentation patterns of specific lipids32,45,46. In the ‘untargeted’ analytical workflow we describe, the downstream data analysis, and particularly the searching of accurate mass peak lists against a lipid database, are key steps that are highly adaptable and may support a broad range of bacterial species and experimental treatments. When constructing or choosing a searchable database to enable identification of lipid species, researchers must consider a wide range of hypothetical endogenous lipid species, while also allowing for detection of novel or unforeseen exogenous lipids derived from the experimental treatment.
In the present study, a high resolution/accurate mass spectrometer (Table of materials) was employed due to its ultra-high resolution/accurate mass capabilities. Alternatively, numerous other high resolution/accurate mass spectrometry platforms could be successfully implemented to perform untargeted lipid analysis. Similarly, a wide range of sample introduction methods including direct infusion, desorption electrospray ionization, or matrix-assisted laser desorption ionization, that enable direct analysis of lipid extracts, could be utilized to rapidly collect untargeted lipidomic data. The inclusion of liquid chromatography prior to sample introduction, when used in combination with high resolution/accurate mass spectrometry, may permit the resolution of some isobaric lipid species during full-scan MS data collection. However, the inclusion of chromatography necessitates ensuring that the chromatographic method of choice is versatile enough to enable separation and detection of unanticipated or novel lipid species that may be present following experimental treatments. Database searching to identify lipids present in the dataset may be performed using any publicly available searchable database. While LIMSA software enables facile development of user-defined databases of tens of thousands of hypothetical lipid species, numerous other options exist for identifying lipids from high resolution/accurate mass MS peak lists. The LIPID MAPS consortium (www.lipidmaps.org) provides tools for searching computational and experimental databases of hypothetical lipids using high resolution/accurate MS-generated peak lists within a user-defined mass tolerance, and many software vendors offer their own solutions for analyzing lipidomics data.
Successful growth curve and exogenous fatty acid analysis is dependent on several factors including LDL purity and limiting background fatty acid levels. Proper identification of the LDL-containing fraction is crucial. The above protocol and Figure 1 illustrate the correct fraction to retain for each step of the enrichment process. We have had success with the use of 40% ammonium sulfate (purity ≥99.5%) for the precipitation and subsequent removal of β-livetins. However, others have reported that the purity and concentration of ammonia sulfate added to the egg yolk plasma can significantly impact this step47. Limiting ammonium sulfate contamination in the LDL enrichment is important for the LDL preparation to be used in bacterial assays, as high concentrations of ammonium sulfate can restrict growth48. During dialysis, ample free space in the dialysis tubing must be provided to allow for the diffusion and removal of the ammonium sulfate. Further optimization of the dialysis process may include additional water changes during the overnight incubation. We have found overnight dialysis to provide the best results, although Moussa et al. report dialysis of 6 h is sufficient. It is critical that the starting concentration of cells in the growth curves are kept consistent between trials. For S. aureus, diluting cells to an initial OD600 of 0.05 has provided the most consistent results. Additionally, high concentrations of FASII inhibitors can result in non-specific effects on bacterial cells. For example, triclosan concentrations above 7 µM induce cytoplasmic membrane damage in S. aureus, therefore it is necessary that the concentration of this compound remain below this level49. We have found a final triclosan concentration of 1 µM results in reproducible growth assays. When evaluating a potential source of exogenous fatty acids for bacterial phospholipid synthesis, it is important to minimize the fatty acid contribution of the culture medium. In the above assays, the culture medium of 1% tryptone supports adequate growth of S. aureus and has minimal fatty acid contamination29,32.
Limiting background fatty acid levels is particularly important for downstream mass spectrometry-based fatty acid profiling. Others have reported the quantity of free fatty acids in chicken egg yolk is naturally low41 and our analysis supports this conclusion (Figure 5). Using tryptone broth and thoroughly washing cells with PBS after incubation are essential. Additionally, it is important to consider the growth phase of the cells. We chose mid-log phase cells to ensure ample bacterial phospholipid synthesis. Other potential contaminating sources of exogenous fatty acids may be introduced after bacterial growth, such as during the lipid extraction steps or subsequent sample preparation prior to mass spectrometry analysis50. Exogenous fatty acids introduced at any step of sample preparation could be detected as free fatty acids during mass spectrometry analysis. Baking laboratory glassware in a high temperature oven (at least 180 °C) overnight can remove exogenous fatty acids from test tubes used for lipid extraction and lipid storage. Additionally, laboratory supplies including plastics may be rinsed with methanol to reduce fatty acid background50. Residual fatty acids from previous analyses may also contaminate internal surfaces of the mass spectrometer itself. Inclusion of analytical blanks during mass spectrometry analysis for determination of background levels of free fatty acids is therefore strongly advised.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank members of the Hammer laboratory for their critical evaluation of the manuscript and support of this work. Dr. Alex Horswill of the University of Colorado School of Medicine kindly provided AH1263. Dr. Chris Waters laboratory at Michigan State University provided reagents. This work was supported by American Heart Association grant 16SDG30170026 and start-up funds provide by Michigan State University.
Materials
| Ammonium sulfate | Fisher | BP212R-1 | ≥99.5% pure |
| Cell culture incubator | Thermo | MaxQ 6000 | |
| Centrafuge | Thermo | 75-217-420 | Sorvall Legen XTR, rotor F14-6×250 LE |
| Costar assay plate | Corning | 3788 | 96 well |
| Filter paper | Schleicher & Schuell | 597 | |
| Large chicken egg | N/A | N/A | Common store bought egg |
| Microplate spectrophotometer | BioTek | Epoch 2 | |
| NaCl | Sigma | S9625 | |
| S. aureus strain AH1263 | N/A | N/A | Provided by Alex Horswill of the University of Colorado |
| Dialysis tubing | Pierce | 68700 | 7,000 MWCO |
| Tryptone | Becton, Dickison and Company | 211705 | |
| 0.5 mm zirconium oxide beads | Next Advance | ZROB05 | |
| Bullet Blender | Next Advance | BBX24B | |
| Methanol (LC-MS grade) | Fisher | A4561 | |
| Chloroform (reagent grade) | Fisher | MCX10559 | |
| Isopropanol (LC-MS grade) | Fisher | A4611 | |
| Dimyristoyl phosphatidylcholine | Avanti Polar Lipids | 850345C-25mg | |
| Ammonium bicarbonate | Sigma | 9830 | ≥99.5% pure |
| Ammonium formate | Sigma | 70221-25G-F | |
| Xcalibur software | Thermo Scientific | OPTON-30801 | |
| LTQ-Orbitrap Velos mass spectrometer | Thermo Scientific | high resolution/accurate mass MS | |
| Agilent 1260 capillary HPLC | Agilent | ||
| SpeedVac Vacuum Concentrators | Thermo Scientific |
References
- Noskin, G. A., et al. National trends in Staphylococcus aureus infection rates: impact on economic burden and mortality over a 6-year period (1998-2003). Clinical Infectious Diseases. 45 (9), 1132-1140 (2007).
- Noskin, G. A., et al. The burden of Staphylococcus aureus infections on hospitals in the United States: an analysis of the 2000 and 2001 Nationwide Inpatient Sample Database. Archives of Internal Medicine. 165 (15), 1756-1761 (2005).
- Laible, B. R. Antimicrobial resistance: CDC releases report prioritizing current threats. South Dakota medicine. 67 (1), 30-31 (2014).
- Zhang, Y. M., Rock, C. O. Membrane lipid homeostasis in bacteria. Nature Reviews Microbiology. 6 (3), 222-233 (2008).
- Zhang, Y. M., White, S. W., Rock, C. O. Inhibiting bacterial fatty acid synthesis. Journal of Biological Chemistry. 281 (26), 17541-17544 (2006).
- Sohlenkamp, C., Geiger, O. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiology Reviews. 40 (1), 133-159 (2016).
- Schiebel, J., et al. Staphylococcus aureus FabI: inhibition, substrate recognition, and potential implications for in vivo essentiality. Structure. 20 (5), 802-813 (2012).
- Heath, R. J., Li, J., Roland, G. E., Rock, C. O. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. Journal of Biological Chemistry. 275 (7), 4654-4659 (2000).
- Heath, R. J., Yu, Y. T., Shapiro, M. A., Olson, E., Rock, C. O. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. Journal of Biological Chemistry. 273 (46), 30316-30320 (1998).
- Park, H. S., et al. Antistaphylococcal activities of CG400549, a new bacterial enoyl-acyl carrier protein reductase (FabI) inhibitor. Journal of Antimicrobial Chemotherapy. 60 (3), 568-574 (2007).
- Schiebel, J., et al. Rational design of broad spectrum antibacterial activity based on a clinically relevant enoyl-acyl carrier protein (ACP) reductase inhibitor. Journal of Biological Chemistry. 289 (23), 15987-16005 (2014).
- Yum, J. H., et al. In vitro activities of CG400549, a novel FabI inhibitor, against recently isolated clinical staphylococcal strains in Korea. Antimicrobial Agents and Chemotherapy. 51 (7), 2591-2593 (2007).
- Kaplan, N., et al. Mode of action, in vitro activity, and in vivo efficacy of AFN-1252, a selective antistaphylococcal FabI inhibitor. Antimicrobial Agents and Chemotherapy. 56 (11), 5865-5874 (2012).
- Karlowsky, J. A., Kaplan, N., Hafkin, B., Hoban, D. J., Zhanel, G. G. AFN-1252, a FabI inhibitor, demonstrates a Staphylococcus-specific spectrum of activity. Antimicrobial Agents and Chemotherapy. 53 (8), 3544-3548 (2009).
- Ross, J. E., Flamm, R. K., Jones, R. N. Initial broth microdilution quality control guidelines for Debio 1452, a FabI inhibitor antimicrobial agent. Antimicrobial Agents and Chemotherapy. 59 (11), 7151-7152 (2015).
- Hunt, T., Kaplan, N., Hafkin, B. Safety, tolerability and pharmacokinetics of multiple oral doses of AFN-1252 administered as immediate release (IR) tablets in healthy subjects. Journal of Chemotherapy. 28 (3), 164-171 (2016).
- Hafkin, B., Kaplan, N., Hunt, T. L. Safety, tolerability and pharmacokinetics of AFN-1252 administered as immediate release tablets in healthy subjects. Future Microbiology. 10 (11), 1805-1813 (2015).
- Flamm, R. K., Rhomberg, P. R., Kaplan, N., Jones, R. N., Farrell, D. J. Activity of Debio1452, a FabI inhibitor with potent activity against Staphylococcus aureus and coagulase-negative Staphylococcus spp., including multidrug-resistant strains. Antimicrobial Agents and Chemotherapy. 59 (5), 2583-2587 (2015).
- Yao, J., Maxwell, J. B., Rock, C. O. Resistance to AFN-1252 arises from missense mutations in Staphylococcus aureus enoyl-acyl carrier protein reductase (FabI). Journal of Biological Chemistry. 288 (51), 36261-36271 (2013).
- Tsuji, B. T., Harigaya, Y., Lesse, A. J., Forrest, A., Ngo, D. Activity of AFN-1252, a novel FabI inhibitor, against Staphylococcus aureus in an in vitro pharmacodynamic model simulating human pharmacokinetics. Journal of Chemotherapy. 25 (1), 32-35 (2013).
- Parsons, J. B., et al. Perturbation of Staphylococcus aureus gene expression by the enoyl-acyl carrier protein reductase inhibitor AFN-1252. Antimicrobial Agents and Chemotherapy. 57 (5), 2182-2190 (2013).
- Kaplan, N., et al. In vitro activity (MICs and rate of kill) of AFN-1252, a novel FabI inhibitor, in the presence of serum and in combination with other antibiotics. Journal of Chemotherapy. 25 (1), 18-25 (2013).
- Kaplan, N., Garner, C., Hafkin, B. AFN-1252 in vitro absorption studies and pharmacokinetics following microdosing in healthy subjects. European Journal of Pharmaceutical Sciences. 50 (3-4), 440-446 (2013).
- Banevicius, M. A., Kaplan, N., Hafkin, B., Nicolau, D. P. Pharmacokinetics, pharmacodynamics and efficacy of novel FabI inhibitor AFN-1252 against MSSA and MRSA in the murine thigh infection model. Journal of Chemotherapy. 25 (1), 26-31 (2013).
- Karlowsky, J. A., et al. In vitro activity of API-1252, a novel FabI inhibitor, against clinical isolates of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrobial Agents and Chemotherapy. 51 (4), 1580-1581 (2007).
- Yao, J., et al. A Pathogen-Selective Antibiotic Minimizes Disturbance to the Microbiome. Antimicrobial Agents and Chemotherapy. 60 (7), 4264-4273 (2016).
- Brinster, S., et al. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature. 458 (7234), 83-86 (2009).
- Balemans, W., et al. Essentiality of FASII pathway for Staphylococcus aureus. Nature. 463 (7279), E3-E4 (2010).
- Parsons, J. B., Frank, M. W., Subramanian, C., Saenkham, P., Rock, C. O. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 108 (37), 15378-15383 (2011).
- Abdelmagid, S. A., et al. Comprehensive profiling of plasma fatty acid concentrations in young healthy Canadian adults. PLoS One. 10 (2), e0116195 (2015).
- Feingold, K. R., Grunfeld, C. Introduction to Lipids and Lipoproteins. Endotext. , (2000).
- Delekta, P. C., Shook, J. C., Lydic, T. A., Mulks, M. H., Hammer, N. D. Staphylococcus aureus utilizes host-derived lipoprotein particles as sources of exogenous fatty acids. Journal of Bacteriology. 200 (11), (2018).
- Moussa, M., Marinet, V., Trimeche, A., Tainturier, D., Anton, M. Low density lipoproteins extracted from hen egg yolk by an easy method: cryoprotective effect on frozen-thawed bull semen. Theriogenology. 57 (6), 1695-1706 (2002).
- Breil, C., Abert Vian, M., Zemb, T., Kunz, W., Chemat, F. Bligh and Dyer and Folch Methods for Solid-Liquid-Liquid Extraction of Lipids from Microorganisms. Comprehension of Solvatation Mechanisms and towards Substitution with Alternative Solvents. International journal of molecular sciences. 18 (4), (2017).
- Lydic, T. A., Busik, J. V., Reid, G. E. A monophasic extraction strategy for the simultaneous lipidome analysis of polar and nonpolar retina lipids. Journal of Lipid Research. 55 (8), 1797-1809 (2014).
- Bowden, J. A., Bangma, J. T., Kucklick, J. R. Development of an automated multi-injection shotgun lipidomics approach using a triple quadrupole mass spectrometer. Lipids. 49 (6), 609-619 (2014).
- Haimi, P., Uphoff, A., Hermansson, M., Somerharju, P. Software tools for analysis of mass spectrometric lipidome data. Analytical Chemistry. 78 (24), 8324-8331 (2006).
- Hewelt-Belka, W., et al. Comprehensive methodology for Staphylococcus aureus lipidomics by liquid chromatography and quadrupole time-of-flight mass spectrometry. Journal of Chromatography A. 1362, 62-74 (2014).
- Jolivet, P., Boulard, C., Beaumal, V., Chardot, T., Anton, M. Protein components of low-density lipoproteins purified from hen egg yolk. Journal of Agricultural and Food Chemistry. 54 (12), 4424-4429 (2006).
- Bylesjö, M., et al. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. Journal of Chemometrics. 20 (8-10), 341-351 (2006).
- Noble, R. C., Cocchi, M. Lipid metabolism and the neonatal chicken. Progress in Lipid Research. 29 (2), 107-140 (1990).
- Cherian, G., Holsonbake, T. B., Goeger, M. P. Fatty acid composition and egg components of specialty eggs. Poultry Science. 81 (1), 30-33 (2002).
- Parsons, J. B., Frank, M. W., Rosch, J. W., Rock, C. O. Staphylococcus aureus Fatty Acid Auxotrophs Do Not Proliferate in Mice. Antimicrobial Agents and Chemotherapy. 57 (11), 5729-5732 (2013).
- Sen, S., et al. Growth-Environment Dependent Modulation of Staphylococcus aureus Branched-Chain to Straight-Chain Fatty Acid Ratio and Incorporation of Unsaturated Fatty Acids. PLoS One. 11 (10), e0165300 (2016).
- Wang, M., Huang, Y., Han, X. Accurate mass searching of individual lipid species candidates from high-resolution mass spectra for shotgun lipidomics. Rapid Communications in Mass Spectrometry. 28 (20), 2201-2210 (2014).
- Peti, A. P. F., Locachevic, G. A., Prado, M. K. B., de Moraes, L. A. B., Faccioli, L. H. High-resolution multiple reaction monitoring method for quantification of steroidal hormones in plasma. Journal of Mass Spectrometry. 53 (5), 423-431 (2018).
- Neves, M. M., Heneine, L. G. D., Henry, M. Cryoprotection effectiveness of low concentrations of natural and lyophilized LDL (low density lipoproteins) on canine spermatozoa. Arquivo Brasileiro de Medicina Veterinaria e Zootecnia. 66 (3), 769-777 (2014).
- Liu, P. V., Hsieh, H. C. Inhibition of Protease Production of Various Bacteria by Ammonium Salts – Its Effect on Toxin Production and Virulence. Journal of Bacteriology. 99 (2), 406 (1969).
- Suller, M. T., Russell, A. D. Triclosan and antibiotic resistance in Staphylococcus aureus. Journal of Antimicrobial Chemotherapy. 46 (1), 11-18 (2000).
- Yao, C. H., Liu, G. Y., Yang, K., Gross, R. W., Patti, G. J. Inaccurate quantitation of palmitate in metabolomics and isotope tracer studies due to plastics. Metabolomics. 12, (2016).

