CRISPR/Cas9 Editing of the C. elegans rbm-3.2 Gene using the dpy-10 Co-CRISPR Screening Marker and Assembled Ribonucleoprotein Complexes.
Summary
The goal of this protocol is to enable seamless CRISPR/Cas9 editing of the C. elegans genome using assembled ribonucleoprotein complexes and the dpy-10 co-CRISPR marker for screening. This protocol can be used to make a variety of genetic modifications in C. elegans including insertions, deletions, gene replacements and codon substitutions.
Abstract
The bacterial Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/Streptococcus pyogenes CRISPR-associated protein (Cas) system has been harnessed by researchers to study important biologically relevant problems. The unparalleled power of the CRISPR/Cas genome editing method allows researchers to precisely edit any locus of their choosing, thereby facilitating an increased understanding of gene function. Several methods for editing the C. elegans genome by CRISPR/Cas9 have been described previously. Here, we discuss and demonstrate a method which utilizes in vitro assembled ribonucleoprotein complexes and the dpy-10 co-CRISPR marker for screening. Specifically, in this article, we go through the step-by-step process of introducing premature stop codons into the C. elegans rbm-3.2 gene by homology-directed repair using this method of CRISPR/Cas9 editing. This relatively simple editing method can be used to study the function of any gene of interest and allows for the generation of homozygous-edited C. elegans by CRISPR/Cas9 editing in less than two weeks.
Introduction
The Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/ Streptococcus pyogenes CRISPR-associated protein (Cas) technology enables efficient targeted genome editing in a wide range of organisms1,2,3,4. The CRISPR system was first discovered as a part of a prokaryotic antiviral immune response5,6,7. The Type II CRISPR system uses an endonuclease such as Cas9, a transactivating RNA (tracrRNA) and a short, target DNA-specific 20-nucleotide long guide CRISPR RNA (crRNA) to recognize an "NGG" Protospacer Adjacent Motif (PAM) and make a double-stranded break in the target DNA5,6,7,8,9,10,11,12. This double-stranded break is recognized as a lesion by the cellular DNA repair machinery. Consequently, the generated double-stranded break can be repaired by one of two pathways- i) Non-homologous end joining (NHEJ) or ii) Homology-directed repair (HDR)13. NHEJ is often error prone and therefore, when this pathway is used to repair the double-stranded break in the target DNA, it often causes inactivating mutations (insertions, deletions) in the gene of interest. On the other hand, by supplying an exogenous repair template with homology to either side of the double-strand break, the cellular DNA repair machinery can be directed to use HDR to repair the break13. The HDR method thus enables precise editing of any locus of interest.
A variety of CRISPR/Cas9 gene editing protocols have been described for C. elegans14,15,16,17,18,19,20. The most commonly employed CRISPR/Cas9 editing methods in C. elegans include both cloning-based and cloning-free protocols to generate repair templates for CRISPR/Cas9 editing14,15,16,17,18,19,20. This protocol discusses in detail a cloning-free CRISPR/Cas9 editing protocol based on using dpy-10 as a co-CRISPR marker for screening. Until now, the only detailed C. elegans-focused CRISPR/Cas9 editing video protocol that exists utilizes a fluorescent marker for screening21. However, using a fluorescent marker for screening requires access to a fluorescence microscope, which many laboratories at small Primarily Undergraduate Institutions (PUIs) may find difficult to access. It is encouraging to note that positive correlative results have been obtained in previous studies between worms carrying fluorescent markers and the presence of the edit21,22. However, additional studies are necessary to determine the overall efficiency of the fluorescence-based screening method for editing a wide variety of loci with different guide RNAs and repair templates. Finally, since the plasmids encoding for these fluorescent markers form extrachromosomal arrays, variable fluorescence is often produced from these arrays that can make positives difficult to identify23. Hence, although the fluorescence-based screening method may be useful to adopt, the above-mentioned issues may limit its applicability.
Using a co-CRISPR marker that produces a visible phenotype greatly reduces the number of progeny that need to be screened to find a positively-edited worm23,24,25,26,27,28. Importantly, the phenotypes that are produced by these markers can be easily detected under a simple dissecting microscope23,24,25,26,27,28,29,30,31,32,33,34. The dpy-10 co-CRISPR marker is one of the best characterized and widely used co-CRISPR markers for performing C. elegans CRISPR/Cas9 genome editing24,27. Therefore, this article will discuss the method of performing CRISPR/Cas9 editing in C. elegans using the direct delivery of ribonucleoprotein complexes with dpy-10 as a co-CRISPR marker27,35. In this method, the prepared editing injection mix consists of a well-characterized dpy-10 crRNA and dpy-10 repair template to mediate the generation of an observable dominant "roller" (Rol) phenotype that is conferred by a known heterozygous dpy-10(cn64) mutation within the dpy-10 gene24,27,29. When present in its homozygous state, the dpy-10(cn64) mutation causes a dumpy (Dpy) phenotype which produces shorter and stouter worms24,29. The Rol phenotype is mediated by the accurate insertion of the co-supplied dpy-10 repair template into a single copy of the dpy-10 gene by HDR-mediated CRISPR/Cas9 editing. Therefore, the appearance of Rol C. elegans indicates a successful injection as well as a successful HDR-mediated editing event within the injected worm's cells. Since the crRNA and the repair template of the target gene of interest are in the same injection mix with the dpy-10 crRNA and dpy-10 repair template, there is a good chance that the identified Rol worms were also simultaneously edited at the target gene of interest. Hence, these Rol worms are then screened for the edit of interest by techniques such as polymerase chain reaction (PCR) (for edits greater than 50 bp) or by PCR followed by restriction digestion (for edits less than 50 bp).
The advantages of using this method for genome editing are: i) CRISPR edits can be generated at a relatively high efficiency (2% to 70%) using this method27; ii) the repair templates and guide RNAs that are used in this method do not involve cloning, thereby reducing the time required for their generation; iii) by assembling ribonucleoprotein complexes in vitro, the concentrations of the assembled editing complexes can be maintained relatively constant, thereby improving reproducibility; iv) some guide RNAs that fail to generate edits when expressed from plasmids have been shown to work for CRISPR/Cas9 genome editing when supplied as in vitro transcribed crRNAs27; v) including the dpy-10 co-CRISPR marker enables easy screening using a dissecting microscope and decreases the number of progeny that must be screened to find positives24,27; and vi) DNA sequencing verified homozygous-edited worm lines can be obtained by this method within a couple of weeks19,27.
Excellent book chapters pertaining to many different C. elegans CRISPR methods have been published14,15,16,17,18,19,20,43. However, the demonstration of the dpy-10 co-CRISPR method in a video format in a laboratory setting is currently lacking. In this article, we describe and demonstrate the process of using the dpy-10 co-CRISPR method to edit a representative target gene named rbm-3.2, a putative RNA-binding protein (WormBase: https://wormbase.org/species/c_elegans/gene/WBGene00011156#0-9fcb6d-10). Specifically, here we describe in detail the method of introducing three premature stop codons within the C. elegans rbm-3.2 gene using in vitro assembled ribonucleoprotein complexes and an exogenously supplied linear single-stranded repair template. These studies have been successful in generating the first C. elegans CRISPR strain with premature stop codons in the rbm-3.2 gene. Since not much is currently known regarding the function of this gene in C. elegans, this strain will serve as a useful tool in dissecting the function of RMB-3.2. This method can also be adopted to make insertions, substitutions, and deletions at any locus within the C. elegans genome.
Protocol
This protocol for CRISPR/Cas9 genome editing was approved by the University of Tulsa Institutional Biosafety Committee following NIH guidelines. Throughout this protocol, sterile technique was practiced. All steps of this protocol were carried out using reagents that were free of nucleases. Special care was taken to prevent RNase contamination such as cleaning gloves as well as workspaces and equipment (e.g. pipettes, exterior of glassware, etc.) with an RNase decontaminating solution.
1. crRNA design
- Find the PAM that enables Cas9 to cut closest to the edit site.
- The PAM site is 5'-NGG-3'.
- Remember that the PAM can be on either strand of the DNA.
- Select 20 bp at the 5' end of the PAM as the crRNA sequence.
NOTE: The actual crRNA sequence that is synthesized by commercial companies is longer than 20 bp as it has an additional generic sequence that is automatically added to the target-specific 20 bp crRNA sequence by the synthesizing company. With regard to off-target effects, recent studies have not found off-target effects of Cas9 in C. elegans36,37,38,39,40. Further, the resultant CRISPR-edited strains are outcrossed. Therefore, we are not very concerned about off-target effects of the designed crRNAs. However, online websites including http://genome.sfu.ca/crispr/ and http://crispor.tefor.net/ can be used to find and select the crRNAs with the least off-target effects38,41. crRNAs that end with a G or GG and those with greater than 50% GC-content are predicted to be more efficient19,23,42. - Order at least 10 nmol of the crRNA from a company (e.g. IDT, Horizon Discovery, etc.).
- Once the lyophilized crRNA arrives, store it at -30°C until the day of the microinjection.
- On the day of performing the microinjection, spin the lyophilized crRNA at maximum speed (17,000 x g) for one minute and resuspend it in 5 mM Tris-Cl (pH 7.4) to make a 8 µg/µL stock of the crRNA.
- Store the unused crRNA stock frozen at -80 °C.
2. Repair template design
- Download a sequence manipulation software (e.g. CLC sequence viewer, APE, Snapgene, Vector NTI, etc.). We use CLC sequence viewer version 8.0 (Qiagen) as it is user-friendly and can be downloaded for free.
- Paste the sequence of the target DNA of interest into the sequence viewer.
- The orientation of the repair template influences the editing efficiency28. For inserting a repair sequence to the 5' end of the PAM, design a single-stranded repair template with DNA sequence from the same DNA strand on which the PAM sequence is located (protospacer strand)28. While for inserting a repair sequence to the 3' end of the PAM, design a single-stranded repair template with DNA sequence from the DNA strand that does not carry the PAM sequence (spacer strand)28.
- Select 35 base pairs of sequence with uninterrupted homology on both sides (on the 5' and 3' end) of the edit.
- Mutate or delete the PAM to prevent cutting of the repair template or the edited genomic DNA by Cas9.
- If mutation of the PAM is not possible, introduce several silent mutations close to the 5' end of the PAM to prevent cutting of the repair template or the edited genomic DNA.
- Make sure to only introduce silent mutations of the PAM if it is present in an exonic region.
- Check the codon usage frequency to ensure that the mutated codon is introduced at a frequency that is comparable to the original non-mutated codon (e.g. https://www.genscript.com/tools/codon-frequency-table). In some cases, it might not be possible to introduce the mutated codon at a similar frequency to the unmutated codon. However, for making mutations of a few amino acids, we do not expect the expression level of the mutant protein to be altered significantly.
- If the edit is small (e.g. mutation of a few bases), introduce a unique restriction site close to the edit by silent mutagenesis. We use http://heimanlab.com/cut2.html to find restriction sites that can be introduced by silent mutagenesis.
- Check the codon usage frequency to ensure that the mutated codon is introduced at a frequency that is comparable to the original non-mutated codon. As stated above, this is not always possible. However, for experiments involving mutations of a few amino acids, we do not expect the expression level of the mutant protein to be altered significantly.
- Synthesize and purchase linear single-stranded repair templates for HDR as 4 nmol ultramer oligos.
- Resuspend the lyophilized oligonucleotide repair template in nuclease-free water to make a 1 µg/µL stock of the repair template and store it at -30 °C until further use.
3. Screening primer design
- Using CLC sequence viewer or other similar applications, design 20 to 23 base pair forward and reverse primers on either side of the edit respectively, such that they produce a band of between 400 to 600 base pairs upon PCR amplification.
- For larger insertions that are several kilobases in size, design the forward primer to be located just outside the left homology arm of the repair template. Design the reverse primer to be located within the repair template itself. In this case, a PCR product will only be obtained in the case of positively-edited worms.
- To identify homozygous-edited worms for longer insertions, amplify the entire region of the insertion by designing forward and reverse primers that are located outside the insertion junctions.
- Test the primers and optimize PCR conditions with wild-type genomic DNA prior to using the primers for genotyping.
- Ensure that a single band of expected size is produced upon PCR amplification of the genomic DNA (where applicable).
4. Preparing young adult worms for injection
- Pick L2-L3 stage C. elegans onto a fresh bacterial lawn on an MYOB plate and incubate at 20 °C overnight.
NOTE: The protocol for making MYOB plates can be found here: http://www.wormbook.org/wli/wbg13.2p12a/ - On the day of microinjection, pick young adult worms with fewer than 10 embryos in the uterus to inject.
5. Preparing the injection mix
- Prepare the injection mix in the same order as shown in Table 1 in sterile nuclease-free tubes.
NOTE: The components of the injection mix can be scaled down to make 5 µL injection mixes (instead of 20 µL) if future injections with this mix are not anticipated. - Mix the injection mix by pipetting.
- Incubate the injection mix at 37 °C for 15 minutes to assemble ribonucleoprotein complexes.
- Spin the injection mix at 4 °C at 17,000 x g for 5 minutes.
6. Microinjection into the C. elegans gonad
- Perform microinjection of the CRISPR injection mix into the C. elegans gonad, as described in Iyer et al. 201943.
- Inject about 30 worms with CRISPR injection mix. The unused injection mix can be re-used by storing at 4°C for a period of about 6 months without loss of efficiency49.
- Inject both gonad arms if possible. Injected worms are considered the P0 generation.
7. Injected worm recovery and transfer
- After microinjection, move the microinjected P0 worms using a worm-pick to a 60 mm MYOB agar plate seeded with OP50 E. coli and let them recover at room temperature for about one hour.
- Note that the recovery temperature will depend upon the genotype of the injected worms. For example, for temperature-sensitive strains, worm recovery at a different temperature may be necessary.
- Using a platinum wire worm pick, transfer each injected worm onto a single seeded 35 mm MYOB agar petri plate and allow them to lay eggs at 20 °C until the next day.
- Note that some worms will die as a result of injury from the microinjection procedure. Only pick those worms that are alive and exhibit movement.
- After 24 hours, transfer the injected worms to new individual plates (1 worm per plate).
8. Picking C. elegans for screening
- 3 to 4 days at 20°C after the injection was performed, monitor all the plates that contain the progeny of the injected worms using a dissecting microscope.
- Identify plates that have F1 progeny exhibiting roller (Rol) and dumpy (Dpy) phenotypes. A Dpy phenotype is where worms appear shorter and stouter than control worms at the same developmental stage (WormBase: https://wormbase.org/species/all/phenotype/WBPhenotype:0000583#0–10). Plates with Rol and Dpy worms represent plates where the F1 progeny have been successfully edited with the dpy-10(cn64) mutation to be present in its heterozygous or homozygous state, respectively.
- Pick the plates with the most roller and dumpy worms for screening.
- Single out 50 to 100 F1 Rol worms from these plates to their own individual plates (1 worm per plate) and allow them to lay eggs.
- Allow L4-staged F1 Rol worms to lay eggs and produce progeny (F2) for 1 to 2 days.
9. Single worm lysis and PCR
- After producing F2 for 1 to 2 days, transfer each singled F1 Rol mother into 2.5 µL of lysis buffer (Table 2) with a 1:100 dilution of 20 mg/mL Proteinase K.
- Freeze the tubes at -30 °C for 20 minutes. The worms can be stored at this stage for an extended period until further analysis.
- Perform single worm lysis on a PCR thermocycler with the following conditions: 60 °C for 1 hour, 95 °C for 15 min and hold at 4 °C.
- Add the following reagents directly to 2.5 µL of the lysed worm containing PCR tube: 12.5 µL of 2x PCR mix containing dNTPs, DNA polymerase, MgCl2 and loading dye, 1 µL of 10 µM forward primer, 1 µL of 10 µM reverse primer, and sterile water to 22.5 µL. The total volume of each PCR reaction is 25 µL.
NOTE: In case of screening multiple F1 worms, it is faster and more convenient to make a PCR master-mix for multiple reactions and add 22.5 µL of the master-mix to each lysis tube. - Set up PCR reactions on a thermocycler with the following conditions: 95 °C for 1 minute, 35 cycles of 95 °C for 15 s, 55 °C for 15 s (optimize for each primer set), 72 °C for 1 minute (optimize for each target DNA)44. Hold reactions at 4 °C.
10. Restriction digestion and agarose gel electrophoresis
NOTE: A restriction digestion is only necessary while screening for small edits (less than 50 bp).
- Transfer 10 µL of the PCR DNA from step 9 to a new tube.
- Add between 2 to 4 units of restriction enzyme and 1x restriction enzyme buffer (1.5 µL of 10x reaction buffer) per 15 µL reaction.
- Incubate at 37 °C (or at other enzyme-specific temperature) for 2 hours (shorter incubation times may be possible with fast acting enzymes).
- Heat inactivate the restriction digest by heating the PCR tubes at 65 °C (or other enzyme-specific temperature) for 10 to 15 minutes.
- Load the entire reaction from each PCR tube into a single well of a 1%-2% agarose gel and run gel at 110 mA until proper band separation is achieved.
NOTE: We use a PCR mix that already has a loading dye included for visualization while running on an agarose gel. This mix does not interfere with the restriction digestion reaction and is therefore convenient to use for screening many worms at one time.
11. Identify positively-edited worms
- Visualize agarose gels under UV light (for ethidium bromide gels) to detect DNA band sizes.
NOTE: Positively-edited worms will display an extra band of DNA at the expected size due to editing using the repair template carrying the restriction site at the desired locus within the worm genome. F1 roller worms are expected to be heterozygous for the edit and are therefore expected to exhibit three bands (one wild-type uncut PCR product and two smaller fragments from the edited cut PCR product). Although rare, it must be noted that homozygotes do arise occasionally. - Save all the original positively-edited plates until the presence of the edit is verified by Sanger sequencing.
12. Homozygose edit of interest
- Pick between 8 and 12 wild-type looking non-rolling F2 worms from the respective positively-edited F1 Rol worm plates and allow them to lay eggs and produce progeny for 1 to 2 days.
NOTE: Since C. elegans are self-fertilizing hermaphrodites, if the edited allele does not affect viability or development, the proportion of expected homozygous mutants should be approximately 25%. Non-rolling F2 worms must be wild-type for the dpy-10 locus. Picking non-rolling worms enables the generation of edited worms that have lost the dpy-10(cn64) mutation and only have the mutation at your gene of interest.- Note that the dpy-10 gene is present on chromosome II. If your gene of interest is linked to dpy-10, you may not be able to easily segregate the dpy-10 mutation away from your gene of interest.
- Perform steps 9 thru 11 to identify homozygous worm lines.
13. Confirm edit by sequencing
- Once homozygous worms are identified, perform lysis and PCR as described in step 9.
- Set up at least 3 PCR reactions for each homozygous line to be sequenced.
- Purify the PCR reactions using a PCR purification kit.
- Measure DNA concentration using a nanodrop spectrophotometer.
- Send sample with the respective forward primer for Sanger sequencing. Ensure that the forward primer is designed to be at least 50 bases away from the edit to be sequenced.
- Analyze the sequencing results using sequence analysis software to confirm the presence of the edit.
Representative Results
rbm-3.2 is a putative RNA-binding protein that has homology to human cleavage stimulation factor subunit 2 tau variant (WormBase: https://wormbase.org/species/c_elegans/gene/WBGene00011156#0-9fcb6d-10). The RBM-3.2 protein was identified as a binding partner of Protein Phosphatase 1 (GSP-1) and its regulators Inhibitor-2 (I-2SZY-2) and SDS-22 in a previous study that identified these proteins as novel regulators of C. elegans centriole duplication (data not shown)45. Presently, very little is known regarding the function of the rbm-3.2 gene in C. elegans. Hence, to further investigate the biological role of the C. elegans rbm-3.2 gene, the rbm-3.2-null allele rbm-3.2(ok688) was obtained from the Caenorhabditis Genetics Center (CGC).
Unfortunately, in addition to possessing a deletion of the entire rbm-3.2 gene, the rbm-3.2(ok688) allele also results in a partial deletion of an overlapping gene, rbm-3.1, thereby complicating genetic analysis. Therefore, in order to accurately investigate the role of the rbm-3.2 gene in C. elegans, we used CRISPR/Cas9 editing to introduce three premature stop codons very close to the start of the C. elegans rbm-3.2 coding region, leaving the overlapping rbm-3.1 gene intact.
To introduce these premature stop codons into the C. elegans rbm-3.2 gene, we designed a crRNA with a PAM motif that was located on the opposite strand (template strand) of DNA (Figure 1A). The Cas9 cut site was located 6 bases away from the rbm-3.2 start codon ATG. To introduce premature stop codons into the rbm-3.2 gene, we designed a repair template with the following five major characteristics: 1) 35 bases of uninterrupted homology to rbm-3.2 upstream of the rbm-3.2 start codon (left homology arm) 2) A short stretch of bases containing the PAM motif was deleted 3) An EcoRI restriction site was introduced immediately after the start codon for screening 4) The second and the third codons of rbm-3.2 were deleted and three stop codons were introduced after the fifth RBM-3.2 codon to stop translation of the rbm-3.2 mRNA 5) 35 bases of uninterrupted homology to the first intron of rbm-3.2 were included downstream of the edit (right homology arm) (Figure 1B).
The injection mix for this CRISPR experiment was prepared as indicated in Table I. The injection mix was incubated at 37 °C for 15 minutes to assemble ribonucleoprotein complexes. The mix was then centrifuged and loaded into the pulled microinjection needle. Microinjection into the C. elegans gonad was performed as described in Iyer et al. 201943.
Figure 2 depicts the experimental timeline for generating edited C. elegans using this protocol. Although we typically inject 30 worms for each CRISPR experiment, some laboratories inject fewer worms (between 10 to 20) for each CRISPR experiment. Since injecting more worms increases the probability of finding positive edits, we prefer to inject a larger number of worms. Many laboratories pick "jackpot" broods (plates with greater than 50% Rol and Dpy progeny) for screening. In our experience after performing several CRISPR experiments, although jackpot broods do arise occasionally, a majority of times, the Rol worms that are picked for screening often come from many different P0 plates, each consisting of a few Rol progeny. No jackpot broods were obtained in this experiment.
In total, we screened the genomic DNA of 73 F1 Rol worms that were obtained from 7 injected P0 worms for the presence of the edit. The sequences of the screening primers and their locations with respect to the start codon of the rbm-3.2 gene are represented in Figure 3A. Through our analysis, 7 out of 73 F1 worms were found to be positive for the edit (9.5%) (Figure 3B). Unedited worms displayed a single DNA fragment of 445 bp upon EcoRI digestion. Whereas, worms carrying a premature stop codon in the rbm-3.2 gene exhibited two fragments of 265 bp and 166 bp respectively upon EcoRI digestion. Heterozygous-edited worms displayed three fragments upon EcoRI digestion: one wild-type unedited DNA fragment of 445 bp and two DNA fragments of 265 bp and 166 bp respectively from the edited copy of the rbm-3.2 gene.
To identify worms carrying homozygous edits, we transferred 12 F2 worms from the identified positive F1 heterozygotes onto new individual plates and allowed them to produce progeny (F3). The F2 worms were then screened for homozygosity as described earlier. 6 out of 12 (50%) screened worms were found to be homozygous for our edit of interest (Figure 4A). The genomic DNA of an identified homozygous-edited worm line was used to set up a DNA sequencing reaction. DNA sequencing analysis confirmed the presence of the edit in its homozygous state (Figure 4B). In general, it is beneficial to sequence multiple homozygous worm lines to ensure that all homozygous-edited worm lines exhibit the same phenotype (if the null-mutation produces a specific phenotype). Further, it might also be necessary to validate gene knockouts using an expression-based assay such as western blotting or RT-PCR or via phenotype analysis. This is because, it is possible that in-spite of introducing premature stop codons at the beginning of the gene, an alternative ATG might be present downstream of the premature stop codons. This ATG could be used to initiate protein expression, resulting in a partially-functional truncated protein.
| Reagent | Concentration | Volume to Add |
| Cas9 protein in nuclease-free water with 20% glycerol | 2 µg/µL | 5 µL |
| tracrRNA in 5 mM Tris-Cl pH 7.5 | 4 µg/µL | 5 µL |
| dpy-10 crRNA in 5 mM Tris-Cl pH 7.5 | 8 µg/µL | 0.4 µL |
| rbm-3.2 crRNA in 5 mM Tris-Cl pH 7.5 | 8 µg/µL | 1 µL |
| dpy-10 repair template in sterile nuclease-free water | 500 ng/µL | 0.55 µL |
| rbm-3.2 repair template in sterile nuclease-free water | 1 µg/µL | 2.2 µL |
| KCl in sterile nuclease-free water | 1 M | 0.5 µL |
| sterile nuclease-free water | – | 5.35 µL |
Table 1: Components of the injection mix for CRISPR/Cas9 editing using ribonucleoprotein complexes and dpy-10 as a co-CRISPR marker. Use sterile techniques and RNase-free reagents while making the injection mix. Please note that sterile, non-DEPC treated nuclease-free water was used to make the injection mix.
| Reagent | Concentration | Volume to add |
| KCl | 1 M | 5 mL |
| Tris-HCl pH 8.3 | 1 M | 1 mL |
| MgCl2 | 1 M | 250 µL |
| NP-40 (or IGEPAL CA-630) | 100% | 450 µL |
| Tween-20 | 100% | 450 µL |
| Gelatin | 2% | 500 µL |
| Water | – | 92.35 mL |
Table 2: Worm lysis buffer recipe. The worm lysis buffer can be made in bulk, autoclaved, filtered and aliquoted for long term storage (NOTE: the lysis buffer can be stored for over a year at room temperature). Add a 1:100 dilution of 20 mg/mL proteinase K to the worm lysis buffer just before each use.
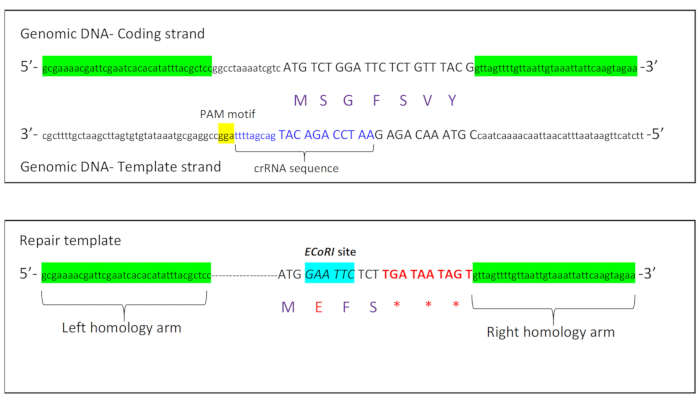
Figure 1: Schematic showing crRNA and repair template design for rbm-3.2 premature stop CRISPR. A. Schematic displaying the coding and template strands of the rbm-3.2 gene with the crRNA sequence and the PAM motif sequence located on the template strand. B. Schematic representing the different characteristics of the repair template that was synthesized to introduce three premature stop codons into the rbm-3.2 gene. Please click here to view a larger version of this figure.

Figure 2: Experimental timeline for generating homozygous-edited C. elegans by CRISPR/Cas9 editing using preassembled ribonucleoprotein complexes and dpy-10 as a co-CRISPR marker. A day-by-day breakdown of the steps that need to be performed to generate homozygous-edited C. elegans using this method of CRISPR/Cas9 editing. Briefly, 30 worms were injected with the CRISPR editing mix using microinjection and segregated onto individual MYOB plates seeded with OP50 E. coli. After 24 hours, the injected P0 worms were transferred onto fresh new MYOB plates and allowed to continue laying eggs. On Days 3 and 4 after microinjection, the plates were examined for the presence of Rol F1 worms. The plates with the maximum number of Rol and Dpy F1 worms were selected and 73 F1 Rol worms (we usually pick between 50 to 100 F1 Rol worms per CRISPR experiment) were singled onto new individual MYOB plates (1 worm per plate) and allowed to lay eggs for about 2 days. On Day 6 after microinjection, worm lysates were prepared from the F1 worms that had produced progeny (F2) and were screened for the presence of the edit by PCR followed by restriction digestion with EcoRI and agarose gel electrophoresis. On Day 7, 12 non-Rol, non-Dpy F2 worms were transferred from the positive plates onto new individual plates and allowed to produce progeny (F3). On Day 9, the F2 worms were screened for homozygosity of the edit as described previously. On Day 10, worm lysis, PCR and PCR cleanup were performed for the homozygous F3 worms and the DNA concentration was measured using a NanoDrop spectrophotometer. On Day 11, Sanger sequencing reactions were set up for positive samples and the reactions were sent for DNA sequencing. On Day 12, the sequencing results were analyzed, and the presence of the edit was verified using a sequence analysis software (e.g. CLC sequence viewer). Please click here to view a larger version of this figure.
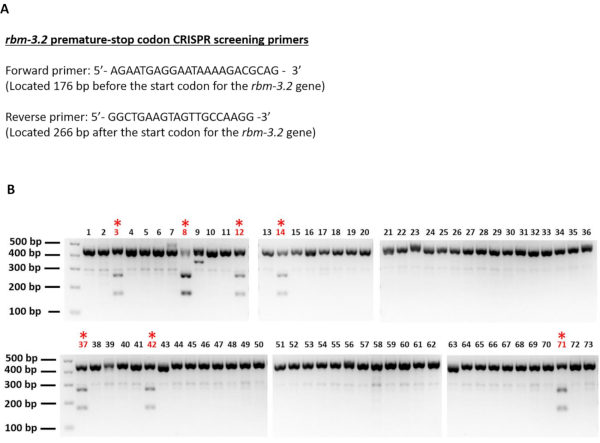
Figure 3: Screening for C. elegans that are heterozygous for the rbm-3.2 premature stop codons. Agarose gel electrophoresis images of C. elegans genomic PCR DNA digested with EcoRI. 73 individual F1 worms were genotyped and screened for the presence of the rbm-3.2 edit. Red numbers and asterisks indicate positively-edited worms. All the 7 identified positively-edited C. elegans were heterozygous for the edit as they exhibited one wild-type unedited DNA fragment of 445 bp and two DNA fragments of 265 bp and 166 bp respectively from the edited copy of the rbm-3.2 gene upon EcoRI digestion. Please click here to view a larger version of this figure.
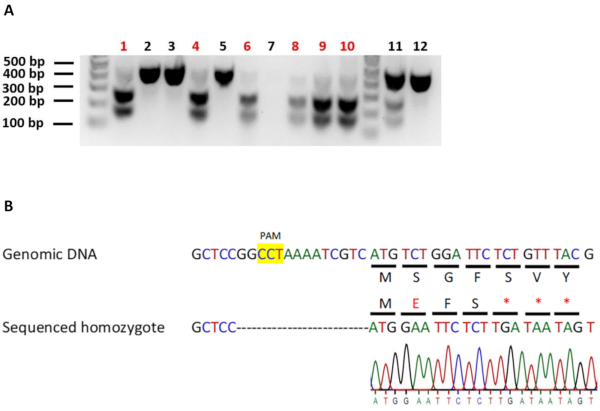
Figure 4: Identifying and verifying homozygous-edited C. elegans carrying the rbm-3.2 premature stop codons. A. Screening for C. elegans that are homozygous for the rbm-3.2 premature stop codons. Agarose gel electrophoresis images of C. elegans genomic DNA digested with EcoRI. 12 individual F2 worms from positive plates were genotyped and screened for homozygosity of the rbm-3.2 edit. Red: homozygous-edited worms. 6 out of the 12 screened F2 worms (50%) were homozygous for the rbm-3.2 premature stop codons. No PCR product was present for worm 7. B. Confirming the insertion of the premature stop codons in rbm-3.2 by DNA sequencing. Schematic showing the comparison of DNA and protein sequences of unedited and edited homozygotes. Analysis of DNA sequencing results of genomic DNA from homozygous-edited worms confirmed the presence of the three premature stop codons in the rbm-3.2 gene upon CRISPR/Cas9 editing. All the resultant amino acid changes after CRISPR/Cas9 editing are indicated in either red letters or red asterisks. Please click here to view a larger version of this figure.
Discussion
We have used the above protocol to edit several genes besides rbm-3.2. Our editing efficiencies for different loci, guide RNAs and repair templates (single-stranded and double-stranded) have varied between 2% and 58% (data not shown). The observed editing efficiencies are comparable to the previously reported editing efficiencies of 2% to 70% for this protocol27. We have also been successful in using this technique in making gene deletions. Using two crRNAs we replaced a gene that is close to 6 kb in length with the coding sequence for green fluorescent protein (GFP) (data not shown). For this experiment, about 14% of the Rol F1 worms that were analyzed were found to be positive for the gene deletion and replacement with GFP (data not shown). However, additional experiments are required to determine the maximum length of gene deletions and replacements that can be performed using this technique.
This technique can be used to make insertions that are about 1.6 kb in length19. A recent study has shown that for making insertions that are over 1.6 kb in length using this protocol, generating two double-strand breaks and using repair templates with longer homology arms can enable the insertion of much larger fragments of DNA (~10 Kb)28. Alternatively, multiple rounds of gene editing with this protocol may be performed to generate larger edits. Other plasmid-based C. elegans CRISPR/Cas9 gene editing protocols may also be adopted for CRISPR experiments involving the insertion of DNA fragments larger than 1.6 kb46,47,48.
Prior to using this protocol for gene editing, it is necessary to ensure that the gene of interest is not linked to the dpy-10 locus on chromosome II. In the case of a target gene being linked to dpy-10, it may be problematic to segregate the dpy-10 mutation away from your edit of interest. Hence, in the event that the gene of interest is linked to the dpy-10 locus, other co-CRISPR markers such as unc-58 (X-chromosome), unc-22 or zen-4 (chromosome IV), and ben-1 or pha-1 (chromosome III) that are located on different chromosomes may be used24,25,26,28. Data from the Meyer lab demonstrate that ben-1 and zen-4 mutations can be used as successful co-CRISPR markers for screening with this method28. However, it is important to note that using zen-4 and pha-1 as co-CRISPR markers necessitates performing the CRISPR experiment in non-wild-type zen-4(cle10ts) or pha-1(e2123ts) backgrounds respectively26,28. Further, CRISPR experiments involving some co-CRISPR markers such as ben-1 may require the preparation of special plates (e.g. plates containing benzimidazole)28. We have successfully used the unc-58 co-CRISPR marker to screen for positive edits for a gene that is linked to the dpy-10 using this method (data not shown). The unc-58(e665) mutation confers a visible phenotype (paralysis) that can be effectively used to screen for positively-edited worms24. Alternatively, if access to a fluorescent microscope is available, a fluorescently-tagged gtbp-1 gene can also be used as a co-CRISPR marker for this protocol19.
For a high editing efficiency while using this method of genome editing, the edit site must be within 10 to 30 bases from the Cas9 cutting site. If the edit site is over 30 bases away from the Cas9 cutting site, the editing efficiency drops drastically19,35,37. However, a recent study from the Meyer lab has demonstrated that creating two double-strand breaks at a distance from one another can enable the insertion of edits far away from the Cas9 cut site using this protocol28.
In the current protocol, the repair template was designed so that all the three inserted stop codons appear in the same reading frame. However, an alternative strategy to create null-mutants would be to use a universal 43-bases long knock-in STOP-IN cassette that has been described previously50. Importantly, this cassette has stop codons in all the three possible reading frames and causes frameshift mutations. This is an especially useful strategy for generating null-mutants when improper or incomplete repair occurs at the edit site.
In conclusion, due to its short duration and the recent advances in this method, this is an excellent method for routine laboratory experiments involving the addition of short immunogenic epitope tags, fluorescent tags, making gene deletions, gene replacements and codon substitutions.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by University of Tulsa (TU) start-up funds and the TU Faculty Development Summer Fellowship awarded to Jyoti Iyer. We would like to thank the Chemistry Summer Undergraduate Research Program (CSURP) and the Tulsa Undergraduate Research Challenge (TURC) for awarding stipends to the students involved in this study. We would like to acknowledge Ms. Caroline Dunn for her technical assistance. Finally, we would like to thank Drs. Jordan Ward, Aimee Jaramillo-Lambert, Anna Allen, Robert Sheaff, William Potter and Saili Moghe for their critical comments on this manuscript.
Materials
| Barcoded DNA sequencing tubes | Eurofins Genomics | SimpleSeq Kit Premixed | N/A |
| Cas9 protein | PNA Bio | CP01 | Lyophilized Cas9 protein was resuspended in 20%glycerol containing nuclease-free water, aliquoted and stored at -80 °C |
| crRNAs | Horizon Discovery/ GE Dharmacon | N/A | Lyophilized crRNAs were resuspended in 5 mM Tris-Cl pH 7.5, aliquoted and stored at -80 °C |
| ECoRI | New England Biolabs | R3101S | Stored at -30 °C |
| Minelute PCR purification kit | Qiagen | 28004 | Spin columns stored at 4 °C |
| MyTaq Redmix | Meridian Bioscience | BIO-25043 | Stored at -30 °C |
| Primers | IDT | N/A | Standard 20 nmol oligos were synthesized, resuspended in sterile nuclease-free water and stored at -30 °C |
| Proteinase K | Gold Bio | P-480-SL4 | Stored at -30 °C |
| Repair template oligos | IDT | N/A | 4 nmol Ultramer oligos were synthesized, resuspended in sterile nuclease-free water and stored at -30 °C |
| tracrRNA | Horizon Discovery/ GE Dharmacon | U-002005-50 | Lyophilized tracrRNA was resuspended in 5 mM Tris-Cl pH 7.5, aliquoted and stored at -80 °C |
References
- Sander, J. D., Joung, J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology. 32 (4), 347-355 (2014).
- Carroll, D. Genome engineering with targetable nucleases. Annual Review of Biochemistry. 83, 409-439 (2014).
- Chandrasegaran, S., Carroll, D. Origins of Programmable Nucleases for Genome Engineering. Journal of Molecular Biology. 428 (5), 963-989 (2016).
- Knott, G. J., Doudna, J. A. CRISPR-Cas guides the future of genetic engineering. Science. 361 (6405), 866-869 (2018).
- Barrangou, R., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 315 (5819), 1709-1712 (2007).
- Marraffini, L. A., Sontheimer, E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 322 (5909), 1843-1845 (2008).
- Brouns, S. J., et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 321 (5891), 960-964 (2008).
- Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 155 (3), 733-740 (2009).
- Garneau, J. E., et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 468 (7320), 67-71 (2010).
- Gasiunas, G., Barrangou, R., Horvath, P., Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences. 109 (39), 2579-2586 (2012).
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816-821 (2012).
- Makarova, K. S., Koonin, E. V. Annotation and classification of CRISPR-Cas systems. CRISPR, Methods in Molecular Biology. 1311, 7-75 (2015).
- Pawelczak, K. S., Gavande, N. S., VanderVere-Carozza, P. S., Turchi, J. J. Modulating DNA Repair Pathways to Improve Precision Genome Engineering. ACS Chemical Biology. 13 (2), 389-396 (2018).
- Frøkjær-Jensen, C. Exciting prospects for precise engineering of Caenorhabditis elegans genomes with CRISPR/Cas9. Génétique. 195 (3), 635-642 (2013).
- Waaijers, S., Boxem, M. Engineering the Caenorhabditis elegans genome with CRISPR/Cas9. Methods. 68 (3), 381-388 (2014).
- Xu, S. The application of CRISPR-Cas9 genome editing in Caenorhabditis elegans. Journal of Genetics and Genomics. 42 (8), 413-421 (2015).
- Dickinson, D. J., Goldstein, B. CRISPR-based methods for Caenorhabditis elegans genome engineering. Génétique. 202 (3), 885-901 (2016).
- Nance, J., Frøkjær-Jensen, C. The Caenorhabditis elegans transgenic toolbox. Génétique. 212 (4), 959-990 (2019).
- Paix, A., Folkmann, A., Seydoux, G. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods. 121, 86-93 (2017).
- Kim, H. M., Colaiácovo, M. P. CRISPR-Cas9-Guided Genome Engineering in Caenorhabditis elegans. Current Protocols in Molecular Biology. 129 (1), 106 (2019).
- Prior, H., MacConnachie, L., Martinez, J. L., Nicholl, G. C., Beg, A. A. A Rapid and Facile Pipeline for Generating Genomic Point Mutants in C. elegans Using CRISPR/Cas9 Ribonucleoproteins. Journal of Visualized Experiments. (134), e57518 (2018).
- Prior, H., Jawad, A. K., MacConnachie, L., Beg, A. A. Highly efficient, rapid and Co-CRISPR-independent genome editing in Caenorhabditis elegans. G3: Genes, Genomes, Genetics. 7 (11), 3693-3698 (2017).
- Farboud, B., Meyer, B. J. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Génétique. 199 (4), 959-971 (2015).
- Arribere, J. A., Bell, R. T., Fu, B. X., Artiles, K. L., Hartman, P. S., Fire, A. Z. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Génétique. 198 (3), 837-846 (2014).
- Kim, H., et al. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Génétique. 197 (4), 1069-1080 (2014).
- Ward, J. D. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Génétique. 199 (2), 363-377 (2015).
- Paix, A., Folkmann, A., Rasoloson, D., Seydoux, G. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Génétique. 201 (1), 47-54 (2015).
- Farboud, B., Severson, A. F., Meyer, B. J. Strategies for efficient genome editing using CRISPR-Cas9. Génétique. 211 (2), 431-457 (2019).
- Levy, A. D., Yang, J., Kramer, J. M. Molecular and genetic analyses of the Caenorhabditis elegans dpy-2 and dpy-10 collagen genes: a variety of molecular alterations affect organismal morphology. Molecular Biology of the Cell. 4 (8), 803-817 (1993).
- Kramer, J. M., Johnson, J. J., Edgar, R. S., Basch, C., Roberts, S. The sqt-1 gene of C. elegans encodes a collagen critical for organismal morphogenesis. Cell. 55 (4), 555-565 (1988).
- Schnabel, H., Schnabel, R. An organ-specific differentiation gene, pha-1, from Caenorhabditis elegans. Science. 250 (4981), 686-688 (1990).
- Granato, M., Schnabel, H., Schnabel, R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Research. 22 (9), 1762-1763 (1994).
- Chalfie, M., Thomson, J. N. Structural and functional diversity in the neuronal microtubules of Caenorhabditis elegans. Journal of Cell Biology. 93 (1), 15-23 (1982).
- Severson, A. F., Hamill, D. R., Carter, J. C., Schumacher, J., Bowerman, B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Current Biology. 10 (19), 1162-1171 (2000).
- Paix, A., Schmidt, H., Seydoux, G. Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Research. 44 (15), 128 (2016).
- Chiu, H., Schwartz, H. T., Antoshechkin, I., Sternberg, P. W. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Génétique. 195 (3), 1167-1171 (2013).
- Paix, A., et al. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Génétique. 198 (4), 1347-1356 (2014).
- Au, V., et al. CRISPR/Cas9 methodology for the generation of knockout deletions in Caenorhabditis elegans. G3: Genes, Genomes, Genetics. 9 (1), 135-144 (2019).
- Dickinson, D. J., Ward, J. D., Reiner, D. J., Goldstein, B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature Methods. 10 (10), 1028-1034 (2013).
- Friedland, A. E., Tzur, Y. B., Esvelt, K. M., Colaiácovo, M. P., Church, G. M., Calarco, J. A. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature Methods. 10 (8), 741-743 (2013).
- Haeussler, M., et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biology. 17 (1), 148 (2016).
- Gagnon, J. A., et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PloS One. 9 (5), e98186 (2014).
- Iyer, J., DeVaul, N., Hansen, T., Nebenfuehr, B. Using microinjection to generate genetically modified Caenorhabditis elegans by CRISPR/Cas9 editing. Microinjection, Methods in Molecular Biology. 1874, 431-457 (2019).
- JoVE Science Education Database. Basic Methods in Cellular and Molecular Biology. PCR: The Polymerase Chain Reaction. Journal of Visualized Experiments. , (2017).
- Peel, N., Iyer, J., Naik, A., Dougherty, M. P., Decker, M., O’Connell, K. F. Protein phosphatase 1 down regulates ZYG-1 levels to limit centriole duplication. PLoS Genetics. 13 (1), 1006543 (2017).
- Dickinson, D. J., Pani, A. M., Heppert, J. K., Higgins, C. D., Goldstein, B. Streamlined genome engineering with a self-excising drug selection cassette. Génétique. 200 (4), 1035-1049 (2015).
- Norris, A. D., Kim, H. M., Colaiácovo, M. P., Calarco, J. A. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Génétique. 201 (2), 449-458 (2015).
- Schwartz, M. L., Jorgensen, E. M. SapTrap, a toolkit for high-throughput CRISPR/Cas9 gene modification in Caenorhabditis elegans. Génétique. 202 (4), 1277-1288 (2016).
- Ghanta, K. S., Mello, C. C. Melting dsDNA Donor Molecules Greatly Improves Precision Genome Editing in Caenorhabditis elegans. Génétique. 216 (2), (2020).
- Wang, H., Park, H., Liu, J., Sternberg, P. W. An efficient genome editing strategy to generate putative null mutants in Caenorhabditis elegans using CRISPR/Cas9. G3: Genes, Genomes, Genetics. 8 (11), 3607-3616 (2018).

