Reconstitution of Septin Assembly at Membranes to Study Biophysical Properties and Functions
Summary
Cell-free reconstitution has been a key tool to understand the cytoskeleton assembly, and work in the last decade has established approaches to study septin dynamics in minimal systems. Presented here are three complementary methods to observe septin assembly in different membrane contexts: planar bilayers, spherical supports, and rod supports.
Abstract
Most cells can sense and change their shape to carry out fundamental cell processes. In many eukaryotes, the septin cytoskeleton is an integral component in coordinating shape changes like cytokinesis, polarized growth, and migration. Septins are filament-forming proteins that assemble to form diverse higher-order structures and, in many cases, are found in different areas of the plasma membrane, most notably in regions of micron-scale positive curvature. Monitoring the process of septin assembly in vivo is hindered by the limitations of light microscopy in cells, as well as the complexity of interactions with both membranes and cytoskeletal elements, making it difficult to quantify septin dynamics in living systems. Fortunately, there has been substantial progress in the past decade in reconstituting the septin cytoskeleton in a cell-free system to dissect the mechanisms controlling septin assembly at high spatial and temporal resolutions. The core steps of septin assembly include septin heterooligomer association and dissociation with the membrane, polymerization into filaments, and the formation of higher-order structures through interactions between filaments. Here, we present three methods to observe septin assembly in different contexts: planar bilayers, spherical supports, and rod supports. These methods can be used to determine the biophysical parameters of septins at different stages of assembly: as single octamers binding the membrane, as filaments, and as assemblies of filaments. We use these parameters paired with measurements of curvature sampling and preferential adsorption to understand how curvature sensing operates at a variety of length and time scales.
Introduction
The shapes of cells and many of their internal compartments are dependent on the lipid membranes that surround them. Membranes are viscoelastic structures that can be deformed through interactions with proteins, lipid sorting, and acting internal and external forces to generate a variety of shapes1,2,3,4. These shapes are often described in terms of membrane curvature. Cells use a diverse suite of proteins capable of preferentially assembling onto, or "sensing", particular membrane curvatures to ensure defined spatio-temporal control over processes including cell trafficking, cytokinesis, and migration5,6. The dynamics of cell machinery at the membrane are notably difficult to observe due to the difficulty of balancing time and spatial resolution with cell health. While super-resolution techniques can offer a detailed view of such structures, they require lengthy acquisitions that are not amenable to the timescales of assembly/disassembly for most machinery. Additionally, the molecular complexity of these assemblies in their native environment and the multitude of roles a single component can play make minimal reconstitution systems a valuable tool for studying the functional capacity of molecules.
Minimal membrane mimetics have been developed to study membrane properties and protein-membrane interactions outside of the cell. Membrane mimetics vary from free-standing lipid bilayers, such as liposomes or giant unilamellar vesicles, to supported lipid bilayers (SLBs)7,8,9,10. SLBs are biomimetic membranes anchored to underlying support, typically composed of glass, mica, or silica11,12. A variety of geometries can be used, including planar surfaces, spheres, rods, and even undulating or micropatterned substrates to probe protein-membrane interactions on both concave and convex curvatures simultaneously13,14,15,16,17,18. Bilayer formation begins with vesicle adsorption onto a hydrophilic surface, followed by fusion and rupture to form a continuous bilayer (Figure 1)19. Supported bilayers are particularly amenable to light and electron microscopy, providing both better time and spatial resolution than is often achievable in cells. Curved SLBs especially provide an attractive means to probe protein curvature sensitivity in the absence of significant membrane deformation, allowing one to distinguish between curvature sensing and curvature induction, which are often impossible to separate in free-standing systems.
Septins are a class of filament-forming cytoskeletal proteins well known for their ability to assemble on positively curved membranes6,18,20. Over the course of the cell cycle in yeast, septins assemble into a ring and must rearrange to form the hourglass and double ring structures associated with bud emergence and cytokinesis, respectively21. While beautiful work has been done using platinum replica electron microscopy to observe septin architecture at varying cell cycle stages22, watching septin assembly over time using light microscopy in yeast has met with limited spatial resolution. Previous work on septins using lipid monolayers visualized by transmission electron microscopy (TEM) was able to reconstitute several interesting septin structures such as rings, bundles, and gauzes23. However, EM techniques are likewise limited in their temporal resolution, unlike fluorescence microscopy. In order to better resolve the kinetic parameters of the multi-scale process of septin assembly, we turned to supported membrane mimetics, where one can carefully control membrane geometry, sample conditions, and imaging modality.
The protocols described here use planar or curved SLBs, purified protein, and a combination of microscopy techniques. Quantitative fluorescence confocal microscopy and total internal reflection fluorescence microscopy (TIRFM) were used to measure both bulk protein binding onto various membrane curvatures, as well as to measure the binding kinetics of single molecules. Furthermore, this protocol has been adapted to be used with scanning electron microscopy (SEM) to examine protein ultrastructure on different membrane curvatures. While the focus of these protocols is on the septin cytoskeleton, the protocols can be easily modified to investigate the curvature sensitivity of any protein the reader finds interesting. Additionally, those working in fields such as endocytosis or vesicular trafficking may find these techniques useful for probing the curvature-dependent assemblies of multi-protein complexes.
Protocol
NOTE: Forming supported lipid bilayers requires the preparation of monodispersed small unilamellar vesicles (SUVs). Please refer to a previously published protocol24 on SUV formation. Briefly, all SUVs are formed by probe sonication for 12 min in total at 70% amplitude via 4 min sonication periods followed by 2 min rest periods in ice-water. SUV solutions must be well clarified and monodispersed in size. Size distributions of SUVs can be measured, for example, by dynamic light scattering25.
1. Planar lipid bilayers
- Plasma cleaning of the slides
NOTE: Plasma cleaning removes organic contaminants and increases the hydrophilicity of the glass, ensuring efficient lipid adsorption. The following steps may change or need to be optimized depending on the plasma cleaner and glass coverslips used.- Purge the plasma cleaner for 5 min with oxygen to remove air from the lines and chamber. This is to ensure that, when the plasma cleaner is run, there is primarily oxygen in the lines and chamber, providing more consistent bilayer preparations.
- While purging, stream an inert gas, such as N2 or Ar, over the micro cover glass slides to remove dust and particulates.
OPTIONAL: Cover glass slides can be washed by spraying each side with 100% isopropanol followed by water. Repeat the isopropanol-water wash 3x and then dry with an N2 stream. - Arrange dry cover glass slides into a ceramic cradle. Place the cradle at the back of the plasma chamber so that the coverslips are parallel with the long edge of the chamber (this keeps them in place during plasma cleaning). Run the plasma cleaner for 15 min with oxygen at maximum power.
- Chamber preparation
NOTE: The method described here uses homemade chambers from plastic PCR tubes to support the reaction volumes, but other low adhesion materials, such as silicone wells, may also be suitable.- Using a razor blade or scissors, cut off the cap just below the frosted part of a 0.2 mL PCR tube (Figure 2).
- Paint the rim of the PCR tube with UV-activated adhesive, avoiding the inside of the tube. Gently place the PCR tube glue-down in the center of a plasma-cleaned coverslip.
NOTE: To avoid glue inside the chamber, use the minimum amount of glue to get a seal, and promptly treat with UV without bumping or disturbing the chamber. - Place the chamber under long-wavelength UV light for 5-7 min to cure the adhesive.
- Bilayer formation
NOTE: Monodispersed SUVs should be used in this step for efficient bilayer formation. Table 1 provides recipes for all buffers used in the bilayer formation, including stock and final buffer concentrations.- Add 40 µL of supported lipid bilayer buffer (SLBB: 300 mM KCl, 20 mM HEPES, pH 7.4, 1 mM MgCl2; Table 1), 10 µL of 5 mM SUVs, and 1 µL of 100 mM CaCl2 to the well. Gently shake the chambers from side to side to disrupt the SUVs, and then incubate at 37 °C for 20 min.
NOTE: To limit evaporation, incubate in a covered Petri dish with a wet wipe.
- Add 40 µL of supported lipid bilayer buffer (SLBB: 300 mM KCl, 20 mM HEPES, pH 7.4, 1 mM MgCl2; Table 1), 10 µL of 5 mM SUVs, and 1 µL of 100 mM CaCl2 to the well. Gently shake the chambers from side to side to disrupt the SUVs, and then incubate at 37 °C for 20 min.
- Washing away excess lipids
NOTE: This step removes excess SUVs and acts as a buffer exchange step. It is very important not to scrape the bilayer with the pipette tip while washing; if the glass is exposed, septins and many other proteins will bind irreversibly, reducing the effective protein concentration.- After incubation, rinse the bilayer 6x with 150 µL of SLBB, pipetting up and down to mix each time while avoiding bubbles.
NOTE: Add 150 µL directly to the 50 µL of buffer and SUVs already present for a total of 200 µL in the reaction well. - When ready to start incubating with the septins or a different protein of choice, wash the bilayer 6x with 150 µL of reaction buffer (RXN: 33.3 mM KCl, 50 mM HEPES, pH 7.4, 1.4 mg/mL BSA, 0.14% methylcellulose, 1 mM BME).
NOTE: For best results, make the reaction buffer fresh and be very careful while mixing in the reaction well to avoid bubbles. - On the last wash step, remove 125 µL of reaction buffer, leaving 75 µL in the well. Add 25 µL of septins diluted in septin storage buffer (SSB: 300 mM KCl, 50 mM HEPES, pH 7.4, 1 mM BME) at the desired concentration and image by TIRF microscopy26.
NOTE: When setting up this assay for the first time or incorporating a new lipid composition, it is good practice to ensure lipids are freely diffusing on the surface using fluorescence recovery after photobleaching (FRAP). While the rate of recovery will be different for different lipid compositions and inherently lower than for free-standing systems, the lipids should not be immobile.
- After incubation, rinse the bilayer 6x with 150 µL of SLBB, pipetting up and down to mix each time while avoiding bubbles.
| Supported Lipid Bilayer Buffer (SLBB) | ||
| Stock | Volume | Final concentration |
| 2 M KCl | 1.5 mL | 300 mM |
| 1 M HEPES | 200 µL | 20 mM |
| 500 mM MgCl2 | 20 µL | 1 mM |
| Water | 8 mL | |
| Pre-Reaction Buffer (PRB) | ||
| Stock | Volume | Final concentration |
| 2 M KCl | 166 µL | 33.3 mM |
| 1 M HEPES | 500 µL | 50 mM |
| Water | 9.33 mL | |
| Reaction Buffer | ||
| Stock | Volume | Final concentration |
| 2 M KCl | 166 µL | 33.3 mM |
| 1 M HEPES | 300 µL | 50 mM |
| 10 mg/mL BSA | 1.39 mL | 1.39 mg/mL |
| 1% Methylcellulose | 1.39 mL | 0.0014 |
| Water | Up to 10 mL | |
| BME | 0.7 µL | 1 mM |
| Septin Storage Buffer (SSB) | ||
| Stock | Volume | Final concentration |
| 2 M KCl | 1.5 mL | 300 mM |
| 1 M HEPES | 500 µL | 50 mM |
| Water | Up to 10 mL | |
| BME | 0.7 µL | 1 mM |
Table 1: Buffer components for preparation of supported lipid bilayer and reactions. Volumes of stock solutions that are incorporated into buffers and the final concentrations of each component are shown. SLB and PRB can be stored at room temperature and reused between experiments. Reaction buffer and SSB are made fresh for each experiment.
2. Spherical supported lipid bilayers
NOTE: This protocol uses silica microspheres suspended in ultrapure water at 10% density. For any work on the kinetic parameters of protein assembly, it is important to strictly control the total membrane surface area between experiments and curvatures. Table 2 shows the corrected volumes of beads and buffer to maintain 5 mm2 of total membrane surface area. This protocol expands on a previously published method8,18.
- Bilayer formation
NOTE: As for planar bilayers, it is important to have high-quality SUVs for robust bilayer formation. Table 2 lists the volume of beads from a 10% density solution and their respective SLBB volumes that are used to obtain a final surface area of 5 mm2 for a range of bead diameters.- Silica microspheres (also referred to as beads) tend to settle and clump together; to mix the beads and break up any clusters, vortex the bottle for 15 s, then bath-sonicate for 1 min, and vortex again for 15 s.
- Mix the appropriate volume of beads (Table 2) with the corresponding volume of SLBB and 10 µL of 5 mM SUVs in a 0.5 mL low-adhesion microcentrifuge tube.
- Rock the bead-lipid mixture end-over-end for 1 h at room temperature. Ensure that this incubation is done on a rotator to prevent sedimentation.
- Prepare chambers on PEGylated coverslips
NOTE: As for planar bilayers, homemade chambers from plastic PCR tubes are used to support the reaction volumes, but other low adhesion materials, such as silicone wells, may work just as well.- While the beads are rocking, thaw a PEGylated coverslip at room temperature. Cut a 0.2 mL PCR tube and glue it to the coverslip as described in step 1.2. For 24 mm x 50 mm slides, up to 10 chambers can feasibly fit on one slide.
NOTE: This protocol uses 24 mm x 50 mm PEGylated glass coverslips that are prepared in advance. Briefly, glass coverslips are thoroughly cleaned through sonication in acetone, methanol, and 3 M KOH. Coverslips are then functionalized with n-2-aminoethyl-3-aminopropyltrimethoxysilane and covalently linked to mPEG succinimidyl valerate. Finished PEGylated coverslips are stored vacuum-sealed at −80 °C. For a similar passivation protocol, please see the work of Gidi et al.27. While PEGylated glass coverslips are suggested here, other means of passivation, such as BSA or poly-L-lysine methods, may be effective.
- While the beads are rocking, thaw a PEGylated coverslip at room temperature. Cut a 0.2 mL PCR tube and glue it to the coverslip as described in step 1.2. For 24 mm x 50 mm slides, up to 10 chambers can feasibly fit on one slide.
- Washing away excess lipids
NOTE: This step functions to remove excess SUVs and perform a buffer exchange. Beads are washed 4x in total with pre-reaction buffer (PRB: 33.3 mM KCl, 50 mM HEPES, pH 7.4). Table 3 lists the spin speeds in RCF for a range of bead diameters.- Spin beads at the designated RCF for 30 s (Table 3). If using multiple bead sizes, keep beads rocking until it is their time to be washed.
NOTE: It is not worth sedimenting larger beads in the same spin as smaller beads with the smaller bead sedimentation velocity to save time. This can cause beads to stick to each other and compromise the integrity of the bilayer. - After the first spin, remove 50 µL of supernatant and add 200 µL of PRB. After the second and third spins, remove 200 µL and add 200 µL of PRB. After the fourth spin, remove 200 µL and add 220 µL of PRB. Pipette vigorously for each wash.
NOTE: The final resuspension volume is different than the washes. Do not vortex the beads after incubation with the lipids as this can leave gaps in the bilayers. To avoid sedimentation while working with the other bead sizes or setting up other parts of the assay, place the bead mixtures back on the end-over-end rotator.
- Spin beads at the designated RCF for 30 s (Table 3). If using multiple bead sizes, keep beads rocking until it is their time to be washed.
- Preparing the reaction
NOTE: This reaction is designed to preserve the total membrane surface area of the reaction at 5 mm2 and to produce a final reaction condition of 100 mM KCl, 50 mM HEPES, 1 mg/mL BSA, 0.1% methylcellulose, and 1 mM BME.- If doing a competition assay with multiple bead sizes, make a 1:1 mixture by mixing equal volumes of each bead size together. Then, mix 29 µL of the desired bead mixture (or of a single bead) with 721 µL of RXN buffer.
NOTE: It is important to thoroughly mix the beads at each step with a pipette to avoid dense clusters of beads; this will result in a more even bead distribution and accurate total surface area. - Mix 75 µL of diluted beads and 25 µL of protein diluted in SSB to the wells.
NOTE: The authors typically image yeast septin complexes at 1-50 nM. - If measuring septins at a steady-state, incubate at room temperature for 1 h, and then image by either near-TIRF or confocal microscopy.
- If doing a competition assay with multiple bead sizes, make a 1:1 mixture by mixing equal volumes of each bead size together. Then, mix 29 µL of the desired bead mixture (or of a single bead) with 721 µL of RXN buffer.
| Bead Diameter (µm) | Volume of well-mixed beads (µL) | Volume of SLB buffer (µL) | Volume of SUVs (µL) |
| 6.46 | 8.94 | 61.1 | 10 |
| 5.06 | 7 | 63 | 10 |
| 3.17 | 4.39 | 65.6 | 10 |
| 0.96 | 1.33 | 68.7 | 10 |
| 0.54 | 0.75 | 69.3 | 10 |
| 0.31 | 0.43 | 69.6 | 10 |
Table 2: Normalized volumes of microspheres. In order to maintain an equal surface area of each bead size and to keep the total membrane surface area consistent between experiments, volumes for each bead size and buffer that normalized the total surface area were calculated.
| Bead diameter (µm) | Sedimentation velocity (RCF) |
| 0.31 | 4.5 |
| 0.54 | 4.5 |
| 0.96 | 2.3 |
| 3.17 | 0.8 |
| 5.06 | 0.3 |
Table 3: Sedimentation velocities for microspheres of varying diameters. For each bead diameter, the shown minimum sedimentation velocities were used to pellet the beads for washing away unbound liposomes.
3. Rod supported lipid bilayers
NOTE: In contrast to the other assays presented here, the rod assay does not allow for careful control of the total membrane surface area. One can be consistent in amounts and volumes between experiments, but because this results in rods of different lengths and diameters, it is difficult to extrapolate the total membrane surface area in the reaction. Thus, while this is an excellent assay for exploring curvature sensing with multiple curvatures on a single surface and has been useful for exploring septin ultrastructure, it is not recommended for kinetic measurements. This method was previously reported18 and is being expanded upon here.
- Obtaining borosilicate rods from glass microfiber filters
- Tear a single 42.5 mm grade GF/C glass microfiber filterinto small pieces and place them into a 100 mL beaker with 60 mL of 100% ethanol. Bath-sonicate until the solution becomes opaque; this can take as little as 20-30 min with a finely shredded filter.
NOTE: Sonicate thoroughly to break up big chunks; the more dissociated/opaque, the better. - Cover the solution with film and store the solution at room temperature overnight.
- Tear a single 42.5 mm grade GF/C glass microfiber filterinto small pieces and place them into a 100 mL beaker with 60 mL of 100% ethanol. Bath-sonicate until the solution becomes opaque; this can take as little as 20-30 min with a finely shredded filter.
- Forming bilayers on the rods
NOTE: This step is performed with excess lipids to achieve complete rod coverage as it is impossible to quantify the total surface area in this method.- The next day the ethanol solution will be settled (step 3.1.2.), so mix it well before using. Combine 10 µL of rod solution and 70 µL of SLBB.
- Spin at top speed in a mini centrifuge for 30 s and remove 50 µL of the supernatant. Resuspend in another 50 µL of SLBB and repeat the spin for a total of four washes to dilute out the ethanol.
NOTE: Rod supports will not form a solid pellet at the bottom of the tube. They are instead smeared along the side of the tube; this makes them difficult to completely preserve when washing. Since the total surface area in this assay is not controlled, it is fine if they are disturbed. - Combine 10 µL of well-mixed rod solution with 50 µL of 5 mM SUVs and 20 µL of SLBB. Incubate for 1 h at room temperature, rotating end-over-end as when forming bilayers on the microspheres.
- Washing away excess lipids
- Similar to step 3.2.2., spin the lipid-rod solution at top speed in a mini centrifuge for 30 s to pellet the membrane-coated rods out of the solution.
- After the first spin, remove 50 µL of the supernatant and add 200 µL of PRB. After the second and third spins, remove 200 µL and add 200 µL of PRB. After the fourth spin, remove 200 µL, add 220 µL of PRB, and pipette vigorously to mix.
- Preparing the reaction
- In a 1.5 mL microcentrifuge tube, mix 721 µL of reaction buffer with 29 µL of rod solution. Mix well.
- In a 0.2 mL PCR tube, combine 75 µL of rods in reaction buffer and 25 µL of septins at the desired concentration, diluted in SSB. Allow it to incubate at room temperature for at least 1 h.
- Preparing for scanning electron microscopy
- After incubation, add the 100 µL septin-rod mixture onto a round, PEG-coated 12 mm coverslip and incubate at room temperature for 1 h in a closed Petri dish.
- Fix the sample by filling the bottom of the Petri dish (10 mL should be enough) with 2.5% glutaraldehyde in 0.05 M sodium cacodylate (NaCo), pH 7.4, for 30 min. Aspirate the liquid with a glass pipette. Wash the sample by incubating it with 10 mL of 0.05 M NaCo for 5 min and repeat for a total of two washes.
- Postfix in 0.5% OsO4 cacodylate buffer for 30 min, and then wash 3x in 0.05 M NaCo (5 min/wash).
- Incubate the sample with 1% tannic acid for 15 min, and then wash in NaCo 3x. Incubate for 15 min in 0.5% OsO4 and wash 3x with NaCo.
- Dehydrate the samples using increasing ethanol concentrations: 30% EtOH for 5 min, 2x; 50% EtOH for 5 min; 75% EtOH for 5 min; and 100% EtOH for 5 min, 2x. Follow with another 10 min incubation.
- Incubate in transition fluid (hexamethyldisilazane) 3x (5 min, 10 min, then 5 min).
- Allow to air dry, then place the samples in a desiccator until sputter-coating. Sputter-coat the 4 nm layer with a gold/palladium alloy and then image on a scanning electron microscope.
Representative Results
Following the preparation of each SLB, septins or the protein of interest may be incubated with the desired support and imaged via TIRFM, confocal microscopy, or SEM. The results shown here use septins recombinantly expressed and purified from E. coli17. Using TIRFM on planar SLBs, it is possible to determine the length of filaments and their flexibility, measure the diffusion coefficients and observe assembly over time28,29. In order to collect the highest quality measurements, it is first necessary to ascertain the quality of bilayers, especially when preparing them for the first time or when changing lipid compositions. A visual inspection of the protein distribution on the bilayers by TIRFM can help identify regions of the bilayer that have been scratched or are malformed. Protein distribution should be homogeneous (Figure 3A), and there should not be holes or gaps in the bilayer (Figure 3B). It is best to avoid membranes with holes, which can form from dust contamination or smudges from slide handling, as this can change the protein distribution in other areas of the bilayer. To visualize the membrane itself, trace rhodamine-PE can be incorporated into SUVs for bilayer formation. In high-quality bilayers, the field will appear even (images not shown), but if liposomes are old or if washes are not stringent enough, unburst and tubulated liposomes may accumulate on the surface (Figure 3C). Additionally, while solid supports will hamper the free diffusion of lipids8, the lipids should not be immobile. FRAP experiments can be used to assess the mobility of lipids on planar bilayers, which may vary by composition and should show recovery rates in the order of seconds30.
Spherical supports can be used to examine protein binding on membranes of defined curvatures either in isolation (one curvature) or with several membrane curvatures in the same well to observe competition between curvatures6,18. Near-TIRFM on beads >1 µm can also be used to measure the number of association events for a given area27. As with planar bilayers, we use rhodamine-PE to look for smooth bilayer deposition (Figure 4A), i.e., no lipid clumps, which indicate multi-lamellarity or unburst liposomes (Figure 4B), and no gaps in the bilayer (Figure 4C). For measuring the total protein adsorption or the protein adsorption over time on curved surfaces, it is necessary to isolate individual beads from each other in order to create a discrete volume for which sum lipid intensity and sum septin intensity can be measured. Thus, the beads should be well-separated rather than clumped together as in Figure 4D. We address troubleshooting options for all of these potential issues in the discussion section.
This SLB assay can also be applied to rod supports, which offer an environment for proteins to sample multiple curvatures on a single surface. Pairing this assay with scanning electron microscopy allows the user to examine curvature preference, alignment, and length distribution of septins18 or other proteins of interest. While similar to the other assays presented here, the rod assay does not allow for careful control of total membrane surface area because of the heterogeneous nature of the substrate that is derived from filter paper. The material properties of the filter paper used here result in rods of different lengths and diameters (Figure 5A); this is useful for exploring protein curvature sensing and organization on curved surfaces (Figure 5B), but because the ratio of protein to the membrane cannot be controlled, the rods are of limited utility for generating saturation-binding curves or measuring parameters such as binding constants.
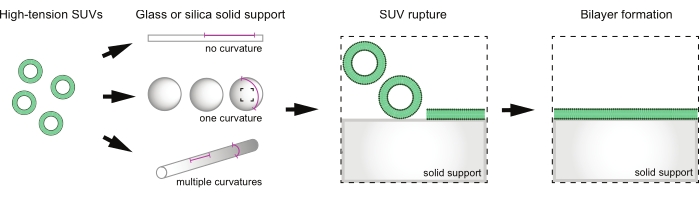
Figure 1: Overview of supported lipid bilayer formation on supports with various curvatures. SUVs are incubated with solid supports of different geometries in order to change the curvatures available for sampling on a given membrane. SUVs adsorb onto the solid support surface and rupture to create lipid bilayers. Please click here to view a larger version of this figure.
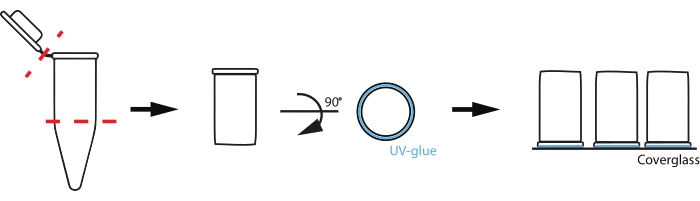
Figure 2: Schematic of reaction chamber set up. To prepare custom chambers, a 0.2 mL PCR tube is cut where the tube begins to taper and at the cap (red dashed lines). The uncut rim of the cut tube is then coated with a thin layer of UV-activated glue (blue) and placed glue-down on a coverslip. Please click here to view a larger version of this figure.
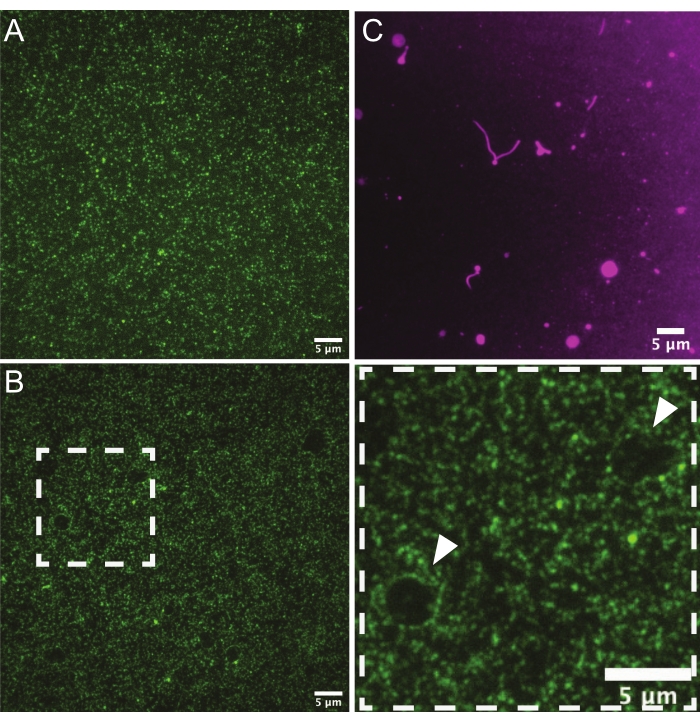
Figure 3: Representative TIRF micrographs of planar bilayers. (A) A representative image of a high-quality lipid bilayer (75% DOPC, 25% Soy PI)with non-polymerizable septins bound (green). Protein is not clustering in any specific regions, and there are no liposomes attached to the membrane and no holes. (B) This bilayer was made with poorly cleaned glass coverslips and exhibits what appear to be "holes" (white arrows) in the membrane where there are fewer septins. Septins can be seen crowded at the edges of these defects in the bilayer (denoted by the white arrows in the zoomed region on the right). (C) A representative image of a low-quality bilayer using trace rhodamine-PE. Unburst liposomes and tubulated lipids are visible on the surface. Please click here to view a larger version of this figure.
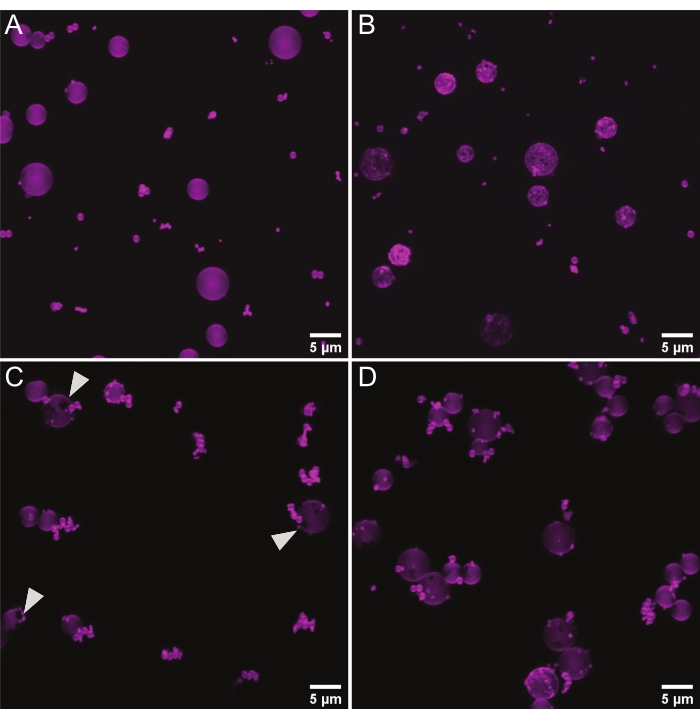
Figure 4: Representative micrographs of lipid-coated microspheres. Representative images from competition assays where lipid-coated beads (75% DOPC, 25% Soy PI, 0.1% Rhodamine PE)with diameters of 0.3 µm, 0.5 µm, 1 µm, 3 µm, and 5 µm were mixed. All images are maximum Z projections. (A) Representative image of a high-quality mixture of lipid-coated microspheres. The spherical supports are evenly coated by the membrane, and there are few bead clusters. (B) Representative image of uneven membrane coating, likely caused by insufficient washing of excess lipids. (C) Representative image of beads with gaps in membrane coverage (white arrows) due to improper handling. (D) Representative image of densely clustered beads, likely caused by insufficient mixing throughout the procedure, especially when combining the different beads together. Please click here to view a larger version of this figure.
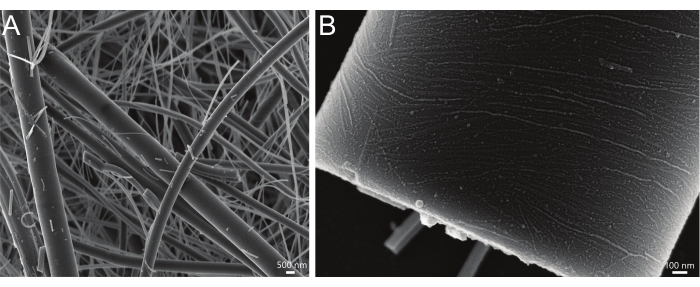
Figure 5: Representative electron micrographs of lipid and septin-coated rods. (A) SEM image showing the distribution of lipid-coated (75% DOPC, 25% Soy PI, 0.1% Rhodamine PE) rod lengths and diameters. (B) The edge of an isolated membrane-coated rod with septin filaments aligned along the axis of positive curvature. Please click here to view a larger version of this figure.
Discussion
Cell membranes take on many different shapes, curvatures, and physicochemical properties. In order to study the nanometer-scale machinery through which cells build micrometer-scale assemblies, it is necessary to design minimal reconstitution systems of membrane mimetics. This protocol presents techniques that precisely control both membrane curvature and composition while allowing the user to easily take quantitative fluorescence measurements using widely available microscopy techniques.
The most critical components of this protocol are assembling wells on the appropriate surface and handling the lipids. The wells described here are handmade using PCR tubes and UV-activated adhesives; it is important not to get glue inside of the reaction area during assembly. This can be checked for during the experiment by imaging along the edges of the well and looking for high protein adsorption onto the cover glass. The user may choose to use alternative materials for wells, such as silicone, which can be ordered commercially, but the presented method provides sturdy wells that will not shift or leak and can support larger reaction volumes. The surface of the cover glass, in our hands, has been very important for forming planar bilayers. Optimization of plasma cleaning times or even the brand of cover glass may be needed to see even formation of the bilayers, as appropriate surface treatment and charge are critical to vesicle adsorption and fusion31,32,33. When setting up these experiments for the first time or working with new lipid compositions, fluorescence recovery after photobleaching (FRAP) experiments should be used to assess the fluidity of the membrane compared to free-standing systems30. It is expected that support systems will be less fluid than their free-standing counterparts but not immobile12.
When assessing the bilayer quality by fluorescence, it is important to look for unburst liposomes or defects in the bilayer (Figure 3C), as these can alter local protein adsorption. The following troubleshooting options may be considered. a) One can adjust the SUV preparation to improve bilayer quality. Sonication, freeze-thaw, and extrusion are all commonly used methods that may vary in ease and success based on lipid compositions. In our hands, a fully clarified SUV solution with a monodispersed size distribution results in the most homogeneous bilayers. Dynamic light scattering (DLS) can be used to assess size distribution25. b) One can increase the difference in salt concentration between the inside of the SUVs and the external buffer. By reducing the internal ion concentration (or increasing the external one), osmotic stress on the SUVs increases the likelihood of rupture31. Alternatively, altering the monovalent cation present may change the kinetics of SLB formation and aid in optimizing SLBs of new lipid compositions34. c) One can increase the number or force of washes. In the authors' experience, vigorous mixing during the washes leads to cleaner bilayers and does not appear to be disruptive; instead, the biggest issue comes from accidentally scraping the bilayer with pipette tips, which will appear as a gash in the membrane. For curved bilayers, it is best to minimize handling with narrow pipette tips and mix gently when performing the wash steps, especially for larger (>1 µm) beads, as the membrane may be sheared from the glass beads. If persistent gaps in the spherical membrane bilayers are observed (Figure 4C), increasing the width of the pipette tips (by cutting off the end of the tip or buying wide tips) and reducing handling as much as possible, while still completely mixing the bead solutions, can help. One common technical issue with this technique is the tendency of the beads to clump together (Figure 4D). This can be partially resolved by increasing the sonication time before bilayer formation; however, some clustering is unavoidable as membrane-coated beads may also tend to clump together over time in the well. Clumped beads should be avoided for quantitative analyses.
Limitations to quantitative measurements of single-molecule dynamics on planar bilayers include the need to stay at low concentrations in order to accurately track and count the particles. However, this can be rectified by under-labeling the protein of interest during acquisition to reduce the number of particles visible during tracking35. Additionally, spherical and rod SLBs were developed specifically to quantify the curvature sensitivity of proteins on micrometer-scale membranes, and the silica microspheres used here are only available down to 100 nm in diameter, at which point they are diffraction-limited puncta when viewed by light microscopy. Thus, those looking to assess precise curvature specificity on smaller diameters will be unable to do so using this method. However, this technique will still provide valuable information about the trend of curvature preference and allow for the dissection of curvature-dependent assemblies.
Compared to free-standing lipid bilayer systems, this technique is amenable to a wide range of buffer conditions and does not require careful osmotic balancing during preparation. Additionally, because it uses glass supports, the bilayers readily sink to the bottom of the well, removing the need for tethering or the use of sucrose or other thickening agents36. While free-standing lipid bilayer systems provide a useful means of investigating membrane deformation by membrane-binding proteins, it is difficult to study the relationship between membrane curvature and complex assembly. Unlike easily deformable membrane systems, this approach allows the user to use a variety of lipid compositions without the intrinsic shape of individual lipid species dictating the global geometry of the membrane. Lastly, rod-shaped SLBs allow sampling of multiple curvatures on the same continuous membrane for proteins of interest (Figure 5B), which is uniquely informative for evaluating curvature-dependent assemblies or the colocalization of multiple components.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) Grant no. R01 GM-130934 and National Science Foundation (NSF) Grant MCB- 2016022. B.N.C, E.J.D.V., and K.S.C. were supported in part by a grant from the National Institute of General Medical Sciences under award T32 GM119999.
Materials
| 0.2 mL PCR Tubes with flat cap, Natural | Watson | 137-211C(EX) | |
| 0.5 mL low adhesion tubes | USA Scientific | 1405-2600 | |
| Beta mercaptoethanol (BME) | Sigma-Aldrich | M6250-100ML | |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A4612-25G | |
| Coverglass for making PEGylated coverslips | Thermo Scientific | 152450 | Richard-Allan Scientific SLIP-RITE Cover Glass 24×50 #1.5 |
| DOPC | Avanti Polar Lipids | 850375 | |
| Egg Liss Rhodamine PE | Avanti Polar Lipids | 810146 | |
| EMS Glutaraldehyde Aqueous 25%, EM Grade | VWR | 16220 | |
| EMS Sodium Cacodylate Buffer | VWR | 11652 | |
| Ethanol, 200 proof | Fisher Scientific | 04-355-223EA | |
| HEPES | Sigma Aldrich | H3375-1KG | |
| Hexamethyldisilazane | Sigma-Aldrich | 440191 | |
| Magnesium chloride | VWR | 7791-18-6 | |
| Methyl cellulose 4000cp | Sigma-Aldrich | M052-100G | |
| Microglass coverslips for planar bilayers | Matsunami | Discontinued | 22×22 |
| Mini centrifuge | |||
| Non-Functionalized Silica Microspheres | Bangs Laboratories, Inc. | Depends on size: SS0200*-SS0500* | Silica in aqueous suspension |
| Optical Adhesive | Norland Thorlabs | NOA 68 | Flexible adhesive for glass or plastics |
| Osmium tetroxide | Millipore Sigma | 20816-12-0 | |
| Parafilm | VWR | 52858-000 | |
| Plasma Cleaner | Plasma Etch | PE-25 | Voltage: 120V, 60Hz. Current: 15 AMPS |
| Potassium chloride | VWR | 0395-1kg | |
| Round coverglass, #1.5 12mm | VWR | 64-0712 | |
| Sonicator bath | Branson | 1510R-MT | Bransonic Ultrasonic cleaner. 50-60 Hz. Output: 70W |
| Soy PI | Avanti Polar Lipids | 840044 | |
| Tabletop centrifuge | Eppendorf | 22331 | |
| UV Lamp | Spectroline | ENF-260C | 115 Volts, 60 Hz, 0.20 AMPS |
| WhatmanGlass Microfiber Filter Paper | VWR | 28455-030 | 42.5 mm diameter, Grade GF/C |
Riferimenti
- Zimmerberg, J., Kozlov, M. M. How proteins produce cellular membrane curvature. Nature Reviews Molecular Cell Biology. 7 (1), 9-19 (2006).
- Parthasarathy, R., Groves, J. T. Curvature and spatial organization in biological membranes. Soft Matter. 3 (1), 24-33 (2007).
- Mao, Y., Baum, B. Tug of war-The influence of opposing physical forces on epithelial cell morphology. Biologia dello sviluppo. 401 (1), 92-102 (2015).
- Ranganathan, R., Alshammri, I., Peric, M. Lipid organization in mixed lipid membranes driven by intrinsic curvature difference. Biophysical Journal. 118 (8), 1830-1837 (2020).
- Bigay, J., Casella, J. -. F., Drin, G., Mesmin, B., Antonny, B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. The EMBO Journal. 24 (13), 2244-2253 (2005).
- Bridges, A. A., Jentzsch, M. S., Oakes, P. W., Occhipinti, P., Gladfelter, A. S. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. Journal of Cell Biology. 213 (1), 5-6 (2016).
- Picard, F., Paquet, M. -. J., Dufourc, &. #. 2. 0. 1. ;. J., Auger, M. Measurement of the lateral diffusion of dipalmitoylphosphatidylcholine adsorbed on silica beads in the absence and presence of melittin: A 31P two-dimensional exchange solid-state NMR study. Biophysical Journal. 74 (2), 857-868 (1998).
- Fu, R., et al. Spherical nanoparticle supported lipid bilayers for the structural study of membrane geometry-sensitive molecules. Journal of the American Chemical Society. 137 (44), 14031-14034 (2015).
- Vanni, S., Hirose, H., Barelli, H., Antonny, B., Gautier, R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nature Communications. 5 (1), 4916 (2014).
- Gill, R. L., et al. Structural basis for the geometry-driven localization of a small protein. Proceedings of the National Academy of Sciences. 112 (15), 1908-1915 (2015).
- Pan, J., Dalzini, A., Song, L. Cholesterol and phosphatidylethanolamine lipids exert opposite effects on membrane modulations caused by the M2 amphipathic helix. Biochimica et Biophysica Acta (BBA) – Biomembranes. 1861 (1), 201-209 (2019).
- Beckers, D., Urbancic, D., Sezgin, E. Impact of nanoscale hindrances on the relationship between lipid packing and diffusion in model membranes. The Journal of Physical Chemistry B. 124 (8), 1487-1494 (2020).
- Lee, A. A., et al. Stochasticity and positive feedback enable enzyme kinetics at the membrane to sense reaction size. Proceedings of the National Academy of Sciences. 118 (47), 2103626118 (2021).
- Ferhan, A. R., et al. Nanoplasmonic sensing architectures for decoding membrane curvature-dependent biomacromolecular interactions. Analytical Chemistry. 90 (12), 7458-7466 (2018).
- Beber, A., et al. Membrane reshaping by micrometric curvature sensitive septin filaments. Nature Communications. 10 (1), 420 (2019).
- Lou, H. -. Y., et al. Membrane curvature underlies actin reorganization in response to nanoscale surface topography. Proceedings of the National Academy of Sciences. 116 (46), 23143-23151 (2019).
- Bridges, A. A., et al. Septin assemblies form by diffusion-driven annealing on membranes. Proceedings of the National Academy of Sciences. 111 (6), 2146-2151 (2014).
- Cannon, K. S., Woods, B. L., Crutchley, J. M., Gladfelter, A. S. An amphipathic helix enables septins to sense micrometer-scale membrane curvature. Journal of Cell Biology. 218 (4), 1128-1137 (2019).
- Johnson, J. M., Ha, T., Chu, S., Boxer, S. G. Early steps of supported bilayer formation probed by single vesicle fluorescence assays. Biophysical Journal. 83 (6), 3371-3379 (2002).
- Lobato-Márquez, D., et al. Mechanistic insight into bacterial entrapment by septin cage reconstitution. Nature Communications. 12 (1), 4511 (2021).
- Gladfelter, A. S., Pringle, J. R., Lew, D. J. The septin cortex at the yeast mother-bud neck. Current Opinion in Microbiology. 4 (6), 681-689 (2001).
- Ong, K., Wloka, C., Okada, S., Svitkina, T., Bi, E. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nature Communications. 5, 1-10 (2014).
- Bertin, A., et al. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. Journal of Molecular Biology. 404 (4), 711-731 (2010).
- Bridges, A. A., Gladfelter, A. S. In vitro reconstitution of septin assemblies on supported lipid bilayers. Methods in Cell Biology. 136, 57-71 (2016).
- Hupfeld, S., Holsæter, A. M., Skar, M., Frantzen, C. B., Brandl, M. Liposome size analysis by dynamic/static light scattering upon size exclusion-/field flow-fractionation. Journal of Nanoscience and Nanotechnology. 6 (9), 3025-3031 (2006).
- Johnson, D. S., Jaiswal, J. K., Simon, S. Total internal reflection fluorescence (TIRF) microscopy illuminator for improved imaging of cell surface events. Current Protocols in Cytometry. (1), Chapter 12, Unit 12.29 (2012).
- Gidi, Y., Bayram, S., Ablenas, C. J., Blum, A. S., Cosa, G. Efficient one-step PEG-silane passivation of glass surfaces for single-molecule fluorescence studies. ACS Applied Materials & Interfaces. 10 (46), 39505-39511 (2018).
- Woods, B. L., et al. Biophysical properties governing septin assembly. bioRxiv. , (2021).
- Cannon, K. S., et al. A gene duplication of a septin provides a developmentally-regulated filament length control mechanism. bioRxiv. , (2021).
- Pincet, F., et al. FRAP to characterize molecular diffusion and interaction in various membrane environments. PLOS One. 11 (7), 0158457 (2016).
- Reimhult, E., Höök, F., Kasemo, B. Intact vesicle adsorption and supported biomembrane formation from vesicles in solution: Influence of surface chemistry, vesicle size, temperature, and osmotic pressure. Langmuir. 19 (5), 1681-1691 (2003).
- Cha, T., Guo, A., Zhu, X. -. Y. Formation of supported phospholipid bilayers on molecular surfaces: Role of surface charge density and electrostatic interaction. Biophysical Journal. 90 (4), 1270-1274 (2006).
- Andrews, J. T., et al. Formation of supported lipid bilayers (SLBs) from buffers containing low concentrations of group I chloride salts. Langmuir. 37 (44), 12819-12833 (2021).
- Gurtovenko, A. A., Vattulainen, I. Effect of NaCl and KCl on phosphatidylcholine and phosphatidylethanolamine lipid membranes: Insight from atomic-scale simulations for understanding salt-induced effects in the plasma membrane. The Journal of Physical Chemistry B. 112 (7), 1953-1962 (2008).
- Danuser, G., Waterman-Storer, C. M. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annual Review of Biophysics and Biomolecular Structure. 35 (1), 361-387 (2006).
- Chan, Y. -. H. M., Boxer, S. G. Model membrane systems and their applications. Current Opinion in Chemical Biology. 11 (6), 581-587 (2007).

