Identifying DNA Mutations in Purified Hematopoietic Stem/Progenitor Cells
Summary
Here we describe an in vivo mutagenesis assay for small numbers of purified hematopoietic cells using the LacI transgenic mouse model. The LacI gene can be isolated to determine the frequency, location, and type of DNA mutants spontaneously arisen or after exposure to genotoxins.
Abstract
In recent years, it has become apparent that genomic instability is tightly related to many developmental disorders, cancers, and aging. Given that stem cells are responsible for ensuring tissue homeostasis and repair throughout life, it is reasonable to hypothesize that the stem cell population is critical for preserving genomic integrity of tissues. Therefore, significant interest has arisen in assessing the impact of endogenous and environmental factors on genomic integrity in stem cells and their progeny, aiming to understand the etiology of stem-cell based diseases.
LacI transgenic mice carry a recoverable λ phage vector encoding the LacI reporter system, in which the LacI gene serves as the mutation reporter. The result of a mutated LacI gene is the production of β-galactosidase that cleaves a chromogenic substrate, turning it blue. The LacI reporter system is carried in all cells, including stem/progenitor cells and can easily be recovered and used to subsequently infect E. coli. After incubating infected E. coli on agarose that contains the correct substrate, plaques can be scored; blue plaques indicate a mutant LacI gene, while clear plaques harbor wild-type. The frequency of blue (among clear) plaques indicates the mutant frequency in the original cell population the DNA was extracted from. Sequencing the mutant LacI gene will show the location of the mutations in the gene and the type of mutation.
The LacI transgenic mouse model is well-established as an in vivo mutagenesis assay. Moreover, the mice and the reagents for the assay are commercially available. Here we describe in detail how this model can be adapted to measure the frequency of spontaneously occurring DNA mutants in stem cell-enriched Lin–IL7R–Sca-1+cKit++(LSK) cells and other subpopulations of the hematopoietic system.
Introduction
In most tissues, differentiated cells have a limited life-span. To maintain functional integrity, long-lived, tissue-specific stem cells continuously produce progenitor cells that in turn give rise to the fully differentiated cells required for the function of that particular tissue. Stem cells also replenish their own compartment through a process called self-renewal. Thus, stem cells are responsible for maintaining the functional integrity of the tissue they reside in. Therefore, it is imperative that they are equipped with robust mechanisms to sense and potentially repair damaged DNA. If not, they may acquire multiple genomic (potentially harmful) perturbations, which can be inherited by their progeny. Understanding how stem cells safe-guard their genome during the life span of an organism is an important question and may help us understand why genomic instability is linked with cancer and some other age-related diseases (reviewed in1,2).
Controlling genomic integrity of a tissue at the level of stem cells or early progenitor cell populations can be achieved by either eliminating defective stem (or progenitor) cells via cell death, senescence or differentiation, and/or by efficient repair of damaged DNA. Recent studies have demonstrated that it is possible to measure certain types of DNA repair directly in these rare populations3-6. It was found that, for example in the hematopoietic system, double strand DNA breaks can be repaired by homologous recombination (HR) or non-homologous end joining (NHEJ), the latter being a repair process of lower fidelity and thus increased risk of making errors. Both are being utilized in hematopoietic stem cells (HSCs)4,5, however, in mice it seems it is predominantly NHEJ in HSCs whereas early progenitors cells utilize HR4. A similar observation was made for stem cells in the skin6. Interestingly, in human HSCs HR, not NHEJ, seems to be the repair mechanism of choice for double strand breaks3. Whether this functional difference between the two species is real or merely represents a technological or experimental difference remains to be seen.
A stem cell's repertoire to repair damaged DNA is likely to include other DNA repair mechanisms, such as base excision repair (BER), nucleotide excision repair (NER) and mismatch repair (MMR). BER and NER are responsible for repairing single or multiple base pair lesions in single stranded DNA, while MMR fixes base-base mismatches and insertion/deletion loops; these types of DNA damage cannot be repaired by NHEJ or HR. Supporting this notion are several studies from the hematopoietic system demonstrating a link between alterations in one of these pathways and abnormalities in the HSC compartment7-9, as well as an increased risk of developing myelodysplastic syndrome10-16, a disease that originates in the HSC and that is associated with increasing genomic instability as the disease progresses17. As of yet, measurements of BER, NER, and MMR directly in HSCs have not been reported.
In addition to elucidating the various processes that control tissue integrity at a mechanistic level, it is imperative to be able to measure the extent of mutated DNA, so that the consequences of aberrations in one of these processes can be tested, e.g. in normal versus genetically engineered stem cells or in old versus young. However, the development of a relevant assay is difficult because of the paucity of tissue-specific stem cells and the lack of culture conditions that preserves "stemness". Moreover, such an assay should be amendable to environmental and genetic manipulations. A possible solution to these limitations and requirements is the use of mouse models that are specifically engineered to detect DNA mutations.
Multiple transgenic mouse models for mutation detection have been developed. For example, LacI transgenic mice18 carry a recoverable λ phage vector encoding the LacI reporter system, in which the LacI gene encodes a suppressor of the Lac operator and serves as the mutation reporter. Upon mutation of the LacI gene, the Lac operator is activated and β-galactosidase is produced. β-galactosidase cleaves the chromogenic substrate X-gal (5-bromo-4-chloro-e-indolyl-β-D-galactopyranoside), which turns it blue. The cos sites flanking the LacI vector allows easy recovery by lambda phage proteins and subsequent infection of E. coli. After incubating infected E. coli on agarose that contains the X-gal substrate, plaques can be scored. Blue plaques contain a putative mutant Lac-I carrying phage, while clear plaques harbor non-mutants. The frequency of blue plaques (among the clear ones) indicates the mutant frequency in the original cell population the DNA was extracted from. Moreover, the λ phage hosting the LacI target can be readily sequenced using PCR techniques for relatively high throughput analysis. Sequencing multiple mutant LacI genes will reveal important information about the mutation spectrum, which in turn may point to possible deficiencies in specific DNA repair pathways or to specific genotoxic events. The LacI transgenic system has been standardized across multiple laboratories19 and the reagents are available commercially. One major disadvantage of the LacI system is the limited ability to detect large deletions or rearrangements; therefore, other methods, e.g. multi-color FISH on metaphase spreads need to be used to compliment this deficiency.
Within the λ phage vector of the LacI mouse model, there is a much smaller gene, CII, available for mutation analysis. Its size and the fact that mutants can be selected makes this a less labor-intensive and cheaper assay20 than the LacI gene analysis. However, the LacI gene is more extensively studied for mutagenesis21 and the sensitivity of the gene to mutations has been well characterized so that there is a clear understanding of the amino acid residues that generate a phenotypic response on a chromogenic substrate22-25.
Other mouse models for mutation detection include the use of the ΦX174 or the LacZ transgenes. The ΦX174 transgenic mouse model, with the original A:T→G:C reversion mutation assay26 or the forward mutation assay27 that allows detection of a spectrum of base pair substitutions, represents a less costly system than the LacI model. However, the mutational screen in the forward assay is not trivial and the mutation spectrum of the ΦX174 transgene is not as well-characterized as that of the LacI. In mouse models carrying LacZ transgenes, the LacZ mutational reporter is recovered utilizing E. coli host cells that are sensitive to galactose and medium containing galactose28. A drawback of this system is that recovery of the LacZ target also involves restriction endonuclease digestion followed by ligation and electroporation of E. coli hosts, thereby making it difficult to adapt the system for small numbers of cells. Although it is not an absolute requirement for working with stem/progenitor cell populations (one can always start with more mice), if large numbers of cells are required (e.g. millions or more) it will quickly become impractical and cost-prohibitive. Also, the relatively large size of LacZ, while providing a sensitive mutational reporter, is cumbersome and more costly for DNA sequence analysis and determination of mutation spectra. A major advantage of this model however, is its ability to detect large deletions and insertions, as well as chromosomal rearrangements.
Since all cells in the LacI, ΦX174 and LacZ transgenic mouse models carry the reporter system, any of these mouse models can be used to measure mutagenesis in any cell type of interest, including stem and progenitor cells, as long as they can be reliably harvested and in sufficient numbers. Because we had extensive experience with the LacI mouse model and the LacI mutation assay, we decided to pursue this system further for mutagenesis analysis in hematopoietic stem and progenitor populations.
The hematopoietic tissue is well-characterized in terms of cell surface phenotype of its individual components, including long-term repopulating stem cells, which are identifiable as the extremely rare population of Lin–IL7R–Sca-1+cKit++(LSK)/ Flk2–CD150+CD48– cells29. Mohrin et al.4 demonstrated that the slightly larger population of LSK/Flk2– cells are still good representatives for HSCs and significantly different from the most primitive committed myeloid progenitor (CMP) population when it comes to studying DNA repair. Moreover, when the HSC-enriched LSK (Flk-2+ and Flk-2–) cells were compared to the Lin–IL7R–Sca-1-cKit++ (LS-K progenitor cells), there was still a significant difference in NHEJ ability5 between the less pure, stem cell-enriched LSK population and the progenitor cells. In our study we use HSC-enriched LSK (Flk-2+ and Flk-2–) cells because we found that at least 2 x 105 cells are required for consistent, reliable results in this mutagenesis assay; this cell number is extremely difficult to obtain when one sorts the LSK/Flk2–CD150+CD48 population or even the LSK/Flk2– population (in terms of mice, costs and practicality). This protocol, based on the one originally developed by Kohler et al.18 describes in detail how the spontaneous DNA mutant frequency can be determined in LSK cells and defined populations of differentiated myeloid cells as well as unseparated bone marrow and spleen cells.
Protocol
Leukocytes from LacI transgenic mice on a C57BL/6 background do not express Sca-1 (Figure 1). Therefore, if Sca-1 is a marker used for cell purification, these mice need to be crossed with an appropriate strain to gain Sca-1 expression; in this protocol the F1 of a cross between regular C57BL/6 (B6) mice and LacI (C57BL/6) transgenic mice (LacI) was used (Figure 1). Of the cell populations used in this protocol, LSKs and CMPs represent the smallest populations in the bone marrow. In order to purify at least 2 x 105 of each/sort, combine the marrow from approximately ten mice when harvesting the marrow from only the hindlegs or from at least four mice when also the hip-, front legs-, vertebral column bones, and sternum are used.
Make single cell suspensions from bone marrow and spleen. A small proportion of the bone marrow is used as is, the majority is used to purify LSKs and differentiated myeloid progenitor cells, i.e. CMPs and granulocytic/monocytic progenitors (GMPs) by fluorescence-activated cell sorting (FACS). The isolation of bone marrow cells and FACS-purification of these populations is described elsewhere30-32. Approximately six independent sorts are required to identify significant differences in mutation frequencies between populations.
The following protocol for measuring the spontaneous in vivo mutant frequency in purified hematopoietic subpopulations is adapted from several Stratagene instruction manuals33-35, based on original work from Kohler et al.18 The most critical differences between the existing protocols33-35 and this protocol, required when using relatively small numbers of cells, include differences in volumes of reagents and the times and temperatures used for proteinase K incubation.
1. Sample Storage
After collecting the desired cell populations, aliquot cells in 1-ml centrifuge tubes (for cell number per aliquot, see Table 1). Centrifuge at 266 x g for 7 min, at 4 °C. Aspirate the supernatant carefully. Put the tubes into liquid nitrogen for 5 min and then transfer them to a -80 °C freezer for further use. The samples can be stored for at least 6 months.
2. Isolation of Genomic DNA
- Take the samples from the -80 °C freezer. Add 500 μl of ice-cold DNA lysis buffer (Table 3) to the sample tube. Vortex the tubes for 3-5 sec on medium speed and place the tubes on ice for 10 min.
- Centrifuge the tubes for 12 min, 4,000 x g, 4 °C. Carefully discard ~450 μl of the supernatant. Spin down the tube for 3-5 sec. Use a glass capillary tube to carefully remove the remainder of the supernatant. Air-dry the inside walls of the tube until no droplets are visible anymore (~2 min).
- Add 2 μl of RNace-It ribonuclease cocktail and 10 μl of 1 M DTT to 100 μl of the DNA digestion buffer (Table 3). Adjust the volumes according to the number of samples that will be processed; 20 μl of this DNA digestion solution is required per sample. After adding 20 μl of this to the cell pellet, try to make the pellet detach itself from the bottom, by gently tapping the tube with your finger or a pen.
Note: it may be difficult to see the pellet because of low cell numbers. - Add 20 μl proteinase K solution* (Table 3) to each sample, very gently tap tube again.
*Note: the proteinase K solution should be warmed up in a 50 °C water bath, 2 min prior to use in order to activate the enzyme. - Place the tube immediately in a 50 °C water bath. Digest the sample according to the guidelines in Table 1. Tap the tube very gently every 10 min.
Note: With such small numbers of cells, the temperature of the water bath and the digestion times are absolutely critical for successfully obtaining high quality genomic DNA. - Prepare the DNA dialysis system in the cold room (Figure 2). Pour 600 ml TE buffer (Table 3) into a 600 ml glass beaker, add a small magnetic stir bar so that the buffer can be stirred during dialysis and let the (0.025 mM pore size) membranes float on the surface of the buffer. One membrane per sample; 1-4 membranes can be used in a single dialysis beaker. Make a mark on the margin of the membrane with scissors for identification of each sample.
- After the appropriate digestion time, add the now very viscous genomic DNA* carefully to the center of the floating membrane (Figure 2). Cover the beaker immediately with aluminum foil. Dialyze the genomic DNA at 4 °C for ~16-20 hr, stir the buffer gently.
*Note: When working with viscous DNA solutions, use pipette tips with a wide opening. - The next day pour 600 ml of freshly prepared TE buffer (Table 3) to a clean glass beaker and transfer the membranes to the new beaker with a spoon very carefully, and cover the beaker with foil. Dialyze for another 2 hr.
- Remove the dialysis membrane from the beaker with the TE buffer using a spoon and transfer only the most viscous "clump" of the DNA solution to a new, sterile 1 ml tube. Store the sample at 4 °C. Continue the mutagenesis assay the next day or up to 1.5 months later. For the purified populations, wait at least 1 week.
3. Preparation of Trays for the E. coli/Phage Culture (Day 1)
Each tray will contain two different layers; an agar layer at the bottom and an agarose layer at the top that contains X-gal. The E. coli/phage solution will be added to the latter. The number and type of cells isolated will determine how many trays will be required. Consult Table 1 to calculate the number of trays needed and subsequent amounts of solutions for this part of the protocol. The following will generate ~60 trays, which one experienced person can process readily.
- Prepare the following media:
- for the bottom layer, prepare 6 x 2 L flasks with each containing 1,600 ml ddH2O. Add NZY powder (21 g/L) and agar (15 g/L); mix well.
- for the top layer, prepare 4 x 1 L flasks with each containing 800 ml ddH2O. Add NZY powder (21 g/L) and agarose (7.2 g/L); mix well.
- for culturing E. coli, add 2.5 g of NZY powder to each of 2 x 250 ml flasks and bring the volume to 100 ml with ddH2O;
- for confirmation trays used at step 8, add 8.4 g of NZY powder and 6 g of agar to a 1 L flask and bring the volume to 400 ml with ddH2O.
- Cover the opening of the flasks with aluminum foil. Mix well and autoclave them at 121 °C, 15 psi for 30 min.
- Remove the flasks (VERY HOT!) from the autoclave. Carefully swirl the flasks to mix the agar and agarose, then place them in a 50 °C water bath. When the water bath has reached 50 °C again, pour from the 2 L flasks (step 3.1.1) ~150 ml of NZY agar in each tray (bottom layer).
- Allow the agar to solidify at RT for at least 2 hr, then invert and open the trays. Place the bottom of the trays on the lid, 45° off center (Figure 3) and let dry for 30 min. Close the trays and leave O/N at RT.
- Meanwhile, prepare the SCS-8 E. coli culture for the next day. From one of the two 250 ml flasks* (step 3.1.3), take 5 ml NZY broth (Table 3) and transfer to a sterile 14 ml tube. Supplement with 62.5 μl of maltose/MgSO4 solution and add 10 μl of the SCS-8 E. coli glycerol stock. Incubate at 37 °C for 3-4 hr while shaking at 250-300 rpm. Use 15 μl of this culture to inoculate 95 ml of NZY broth (in a 250 ml sidearm flask), culture O/N at 37 °C in a shaking incubator (250-300 rpm).
*Note: the other 250-ml flask can be used later to adjust the OD of the culture if necessary (step 4.1) - Take the 1 L flask with agar (step 3.1.4), and pour ~6-7 ml of NZY agar in 60 mm dishes (this will be enough for ~50 dishes, needed for step 8). Allow agar to harden (~10 min), then invert and wrap in plastic. These dishes can be kept at 4 °C for up to a month.
4. Preparation of Trays for the E. coli/Phage Culture (Day 2)
- Check the O.D. of the SCS-8 E. coli culture (step 3.5) on the spectrophotometer. Adjust the OD600 to 0.6 with NZY broth (Table 3) (step 3.1.3), and place the flask on ice to stop growth and keep on ice until ready for use. This will be used for steps 6.3 and 8.2. This culture can be kept for 5 days at 4 °C.
- Air dry all assay trays (poured the day before) for ~5 hr (Figure 3).
5. Packaging of Genomic DNA (Day 2; Continued)
- Take the required number of orange Transpack tubes out the -80 °C freezer and place them on dry ice until ready for use; take one orange Transpack tube for each packaging reaction to be performed. Label each tube appropriately. Have the genomic DNA samples (step 2.9) ready on ice.
- This step should be performed one tube at the time. Finish the complete step before moving on to the next DNA sample. Thaw one orange tube*note1 quickly: use your fingers until most of it is thawed, then put on ice. Take the corresponding genomic DNA sample and immediately transfer 8-12 μl sample*note 2,3 to the orange tube. Mix the contents by gently pipetting up and down 3x, as well as by gently tapping the tube with your finger. Try not to introduce bubbles when mixing. Place the tube in a 30 °C water bath for 90 min.
*Note 1: a quick spin in a microcentrifuge may be necessary to collect all contents from the inside walls and the cap.
*Note 2: volume of added sample depends on the number of cells used to generate that sample: 11-12 μl of samples prepared from 2.0-5.0 x 105 cells; 10 μl of 1.0 x 106 cell-samples, and 8 μl of 1.5 x 106 cell-samples.
*Note 3: The DNA is still very viscous; to take the DNA out, push the pipet tip to the bottom of the sample tube and carefully twist the tip around against the inner wall of the tube. - Take 1 or 2 blue Transpack tubes* out the -80 °C freezer and place them on dry ice until further use. Thaw them quickly and transfer 12 μl to each of the orange tubes. Mix the solution by gently pipetting it up and down 3 times. Spin the tube down for 2-3 sec, then tap the tube with your finger for further mixing and immediately return it to the 30 °C water bath for another 90 min.
*Note: Use 1 blue tube for 5-6 reactions and 2 blue tubes for 10-12 reactions. - After 90 min, dilute each reaction with 970 μl SM buffer* (Table 3) to stop the reaction and vortex on medium speed for 5 sec. Put tubes on ice till further use.
*Note: if the number of cells is ≤ 5 x 105, use 500 μl SM buffer (Table 3) to stop the reaction; 2 tubes (of the same sample) can then be combined later (step 6.4).
6. Plating Packaged Genomic DNA (Day 2; Continued)
- Close the inverted agar trays that were opened for drying in the morning. Label the trays for each sample.
- Dissolve 4.8 g of X-gal in 16.8 ml of N, N-dimethylformamide (needed for step 6.5). Stir immediately and put on a shaker platform. Protect against light. Solution should be clear in 20-30 min.
- Aliquot the SCS-8 E. coli cells. For each set of trays*, use one 50 ml conical tube. Label the tube with the name of the packaged DNA sample. Add 2 ml of E. coli suspension for each tray.
* Note: For example 3 x 105 GMPs requires a set of 8 trays (see Table 1). Thus, two aliquots, require 8 x 2 = 16 trays. Keep the aliquots separate and thus prepare two 50 ml tubes, with each 8 x 2 = 16 ml E. coli suspension. - Add the 1 ml* of packaged DNA sample (from step 5.4) to the appropriate 50 ml tube containing the SCS-8 E. coli aliquot and mix well. Incubate this E. coli/phage mixture in a shaking incubator (250-300 rpm), at 37 °C for 23 min.
*Note: If DNA was extracted from ≤ 5 x 105 cells, 500 μl of SM buffer was used to stop the reaction (step 5.4). At this step, the tubes can be combined and added to one 50 ml conical tube. - When the E. coli/phage mixture is incubating, start preparations for the top agarose layer. Add 5 ml of X-gal/N, N-dimethylformamide solution (step 6.2) to each 800 ml flask of top layer agarose solution (from step 3.1.2; kept at 50 °C). Final concentration of X-gal will be 1.5 mg/ml. The X-gal may precipitate a little when added to the agarose. Swirl to dissolve the X-gal and put the bottle back in the 50 °C water bath.
- Take the E. coli/phage mixture out of the incubator (step 6.4). Each sample requires multiple trays; each tray, requires 50 ml X-gal/agarose solution (step 6.5). Pour the required volume of X-gal/agarose for each sample (i.e. 50 ml x number of trays) in a larger sterile plastic bottle and add the appropriate E. coli/phage mixture. Swirl the bottle to mix. Divide the mixture in aliquots of 45-50 ml in 50 ml conical tubes. The number of tubes should be the same as the number of trays required for that sample.
Note: The leftover X-gal/agarose solution will be used in step 8.3. The solution can be stored in a 50 °C water bath until further use. - Pour the 50 ml of top agarose mixture across the bottom half of the assay tray. Quickly spread the agarose by tilting the assay tray slightly in one direction.
Note: the agarose will cool down very quickly and will become impossible to spread out over the tray. Therefore, this step needs to be performed relatively fast and with agarose that has been kept at 50 °C. - Allow the top agarose to harden for at least 15 min. Then, invert and open the assay trays to let them air-dry for 30 min (Figure 3).
- Close the trays, incubate the assay trays inverted (bottom agar layer side on top) at 37 °C for 15-16 hr. Don't stack more than 5 trays.
7. Determination of a Putative Mutant Frequency (Day 3)
- Remove the trays from the 37 °C incubator and let them cool down. Count the translucent plaque forming units (PFU) on each tray; randomly select 2 sites on the trays, draw a square* of 2.5 x 2.5 cm2 or 5 x 5 cm2 and count all the PFUs in each square with a marking cell counter (Figure 4A, B).
*Note: To draw the square use a device as shown in Figure 5. If the number of counted PFUs is ≥40, use the smaller square, otherwise use the larger. - Take the average of the squares counted and multiply by 96 if counting with a smaller square; or multiply this number by 24 if counting with the large one. Add the number of PFUs counted for all trays from the same sample. This is the total number of PFUs generated for that sample.
- Next, count the mutant plaques in each tray. The trays have now cooled down, which will help to locate the mutant plaques. To further facilitate spotting the blue plaques, move the tray over a red and/or white surface; sheets of red and white paper work well. Circle any blue plaque with a marker pen. Record the shape and intensity of the blue color of each mutant (e.g. full, circle, sector; Figure 4C)36,37.
- Determine the putative mutant frequency for each sample by dividing the number of mutant PFUs (step 7.3) by the total number of PFUs (step 7.2).
8. Verification of Putative Mutant Plaques (Day 3-5)
- Using a Pasteur pipette, core each mutant (blue) PFU and transfer the plug into 250 μl sterile SM buffer (Table 3). Add 25 μl of chloroform, vortex for 5 sec and store at 4 °C to continue the next day or leave for 2 hr at RT before continuing with the next step, to verify that the mutant PFU is indeed a mutant. The samples can be stored at 4 °C for at least 1 year.
- Label one 1 ml and one 4 ml sterile tube for each mutant that was plugged. In the new 1 ml tube, dilute the resuspended phage released from the agar plug (step 8.1) 1:50 in sterile SM buffer (2 μl of sample into 100 μl SM buffer (Table 3)), vortex briefly, set aside. To the 4 ml sterile tube add 200 μl SCS-8 E. coli culture (from step 4.1) and 2 μl of the diluted phage from the corresponding 1 ml tube, incubate at 37 °C for 5-10 min.
- Add 2.5 ml of the top agarose containing X-gal (left-over from step 6.6) to each 4 ml tube, swirl tube to mix and pour onto a 60-mm NZY agar plate previously poured (step 3.6). Leave for 10 min (to let the top agarose solidify), invert the dish and incubate O/N at 37 °C.
- The next morning, remove the dish from the incubator and determine the proportion of blue PFUs on each dish (Figure 6). When 70% or more of the PFUs are blue, a mutant is considered confirmed38,39. Core the mutant plaque from the dish and put in a 1.5 ml screw top tube, containing 250 μl SM buffer (Table 3) and 25 μl chloroform; it is now ready for sequencing.
- When 50% or less of the PFU is blue, it is not considered a real mutant38,39. When the frequency of blue PFUs is between 60-70%, check back with in the notes about the shape of the plaque; if the shape of the PFU was "full", proceed with sequencing.
9. Sequencing for Mutations in the LacI Gene
- Set up PCR reactions in a 25 μl reaction volume, including 1.5 μl of plaque supernatant (template, step 8.4), the forward primer SF1 (5'-GGAAACGCCTGGTATCTT-3') and the reverse primer SR2 (5'-GCCAGTGAATCCGTAATCA-3'). Use the PCR extender Taq polymerase kit according to the manufacturer's instructions. Cycling conditions are as follows: 94 °C for 2 min, then 35 cycles of 94 °C for 20 sec, 60 °C for 20 sec, 72 °C for 2 min, followed by a final step of 72 °C for 5 min.
- Take a 5 μl aliquot of each reaction and run on a 0.8% agarose gel, to confirm amplification.
- Clean up the PCR reactions using the Augencourt cleanup kit according to the manufacturer's instructions.
- Quantitate Template DNA.
- Use ~100 ng of PCR product as template DNA for sequencing. Sequence amplicons in both directions using the same PCR primers as sequencing primers (see above).
- Assemble sequences and align with the LacI reference sequence40 to detect mutations.
Representative Results
The in vivo mutagenesis assay measures a rare event (mutant PFUs) among many events (all PFUs). By performing the assay with small number of cells, it is possible that the outcome is considerably influenced by false-positive and false-negative results. To address this issue we performed a serial dilution experiment with unfractionated bone marrow cells, harvested from three different animals. We measured the mutant frequency in the bone marrow of these animals using 1.4 x 106, 7.0 x 105, 3.5 x 105, and 1.75 x 105 cells. The results (Figure 7) show a linear relationship between the input cell number and the number of PFU's generated. Importantly, the mutant frequency is consistent, whether measured with low or high numbers of cells. Although not directly tested (due to the paucity of cells), there is no reason to believe that this is not the case for LSKs and GMPs.
The most important step in this mutagenesis assay is the isolation of genomic DNA (step 2). Although the DNA concentration should ideally be ≥500 ng/μl, this protocol works well with 40-150 ng/μl. More important is the 260/280 ratio (this should be >1.8-2.0) and the molecular weight (should be around 300-500 kb). It is recommended when one is not familiar with the assay to check the quality and the size of the isolated DNA. Figure 8 shows an example of an electrophoresis run of high molecular weight DNA.
The markings of individual PFUs on a tray shown in Figure 4B represent a reasonable and achievable density of plaques when one starts with a small number of purified cells; a tray should hold between 40-150 PFUs/small square. In this particular experiment, the small square in Figure 4B contains 103 plaques. The other square on the tray, shown in the upper corner of Figure 4A contained 111 PFUs. These two counts are close together (less that 10% difference); if this is not the case, count the plaques in 5 squares, drawn at random on the tray, or repeat the experiment because the variation may be due to too few plaques on the tray. On this example tray, the total number of PFUs is ([103+111]/2) x 96 = 10,272 PFUs. This number is lower than the recommended 12,000 PFUs by others41, because we use slightly smaller trays and 12,000 PFUs on these trays did not allow us a good distinction between individual plaques.
Figure 4C shows examples of the different types of blue plaques that can be observed. In the LacI mutagenesis assay, the morphology of the plaques (i.e. full, sector or circle) is an indication of the origin of the mutation; full is a mutation from mouse origin, whereas the other two are likely produced in the E. coli 36,37. Upon replating (see also Figure 6), nearly all "full" plaques reproduce >70% blue plaques again, whereas only a very small portion of the sector plaques do and virtually none of the often smaller, circular plaques.
Table 2 shows the reproducibility of plaque formation with relatively small numbers of purified cells compared to that with high numbers of bone marrow cells (the same pool of cells where the cells were sorted from) and spleen cells.
Mutant PFUs are confirmed, first by replating (Figure 6 shows representative examples of dishes for confirmation of primary mutant (blue) PFUs) and second, by sequencing. The dishes in Figure 6 containing >70% blue PFUs (bottom two) confirm that the mutant originated in the mouse (and not in the E. coli). However, the top left dish shows no blue PFUs and therefore, the primary PFU should not be counted as mutant and sequencing is not necessary. The right dish shows ~65% blue PFUs. Depending on the shape of the primary plaque, this sample will be sent out for sequencing (see step 8.4). Only if a DNA mutation can be confirmed by sequencing will this PFU be counted as mutant. If unsure about the shape of the PFU, sequence!
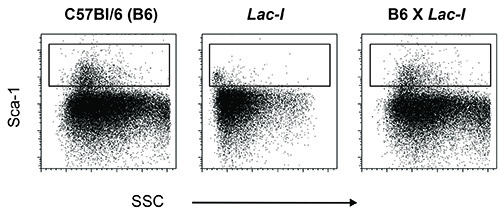
Figure 1. Sca-1 staining on bone marrow cells from wild type C57BL6 mice (B6), transgenic LacI mice (LacI) and the offspring of a cross between these two (B6 x LacI). Bone marrow cells of B6 mice express Sca-1 on their cell surface and this characteristic is used to purify stem- and progenitor cells. Transgenic LacI mice on a B6 background, however, do not express Sca-1; this marker must have been lost while establishing the colony. To reintroduce Sca-1 into transgenic LacI mice, these mice were crossed with regular B6 mice.
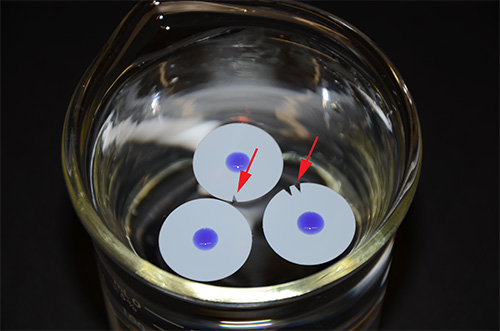
Figure 2. DNA dialysis system. Three membranes holding a viscous DNA solution in the center (colored blue for better visualization) are floating on TE buffer. The red arrows indicate small nicks in the membrane, which are used for sample identification.

Figure 3. Efficient way to dry 60 or more agar/agarose containing trays.
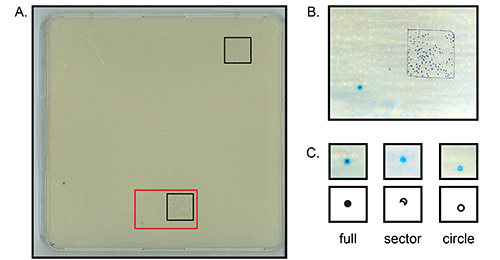
Figure 4. Detection of plaque forming units (PFUs). Depicted are (parts of) large agar trays (depicted in Figure 3) with primary colonies of phage-infected E. coli. (A) Shown is one entire plate with 2 small squares drawn on it for counting plaques. The red insert is shown enlarged in (B). (B) Each individual black marker dot indicates a clear wild type PFU in the agar plate (103 in total). (C) Different shapes of potentially mutant PFUs. Click here to view larger image.
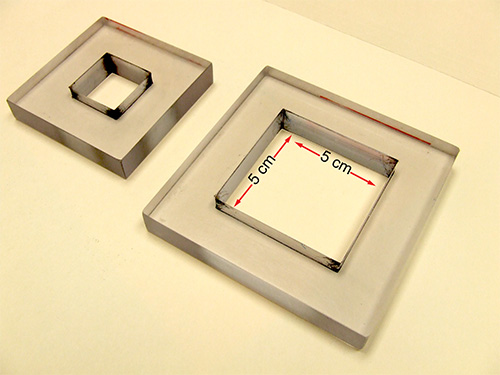
Figure 5. Plexiglas counting squares. Depicted are a small and large counting square. The inner measurements of the large square are 5 cm x 5 cm, that of the small square 2.5 cm x 2.5 cm.
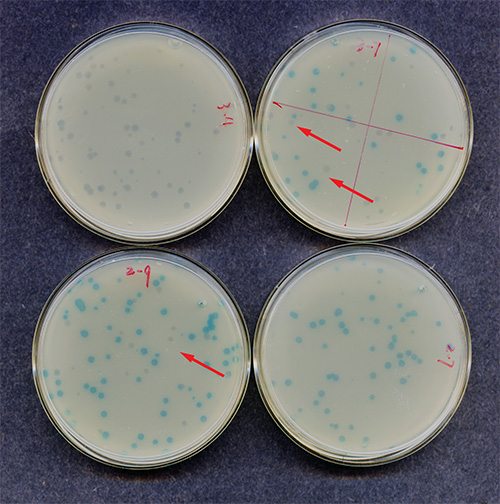
Figure 6. Confirmation of mutant plaque forming units (PFUs). Shown are 4 small dishes inoculated the day before with a diluent of a blue PFU. The upper left dish shows no blue PFUs. The upper right dish shows ~65% blue PFUs. The two lower trays show 80-90% blue PFUs (left) and 100% blue PFUs (right). The red arrows indicate clear (wild type) PFUs.
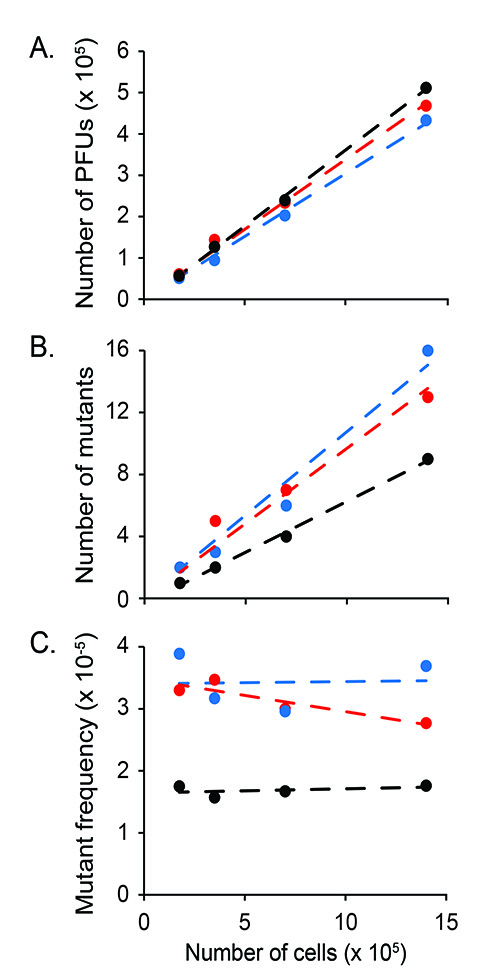
Figure 7. Mutant frequency in unfractionated bone marrow. Depicted are (A) the number of PFUs, (B) the number of mutants and (C) the mutant frequency measured in three different animals (24-26 months old), which are represented by different colors. For each mouse, the measurements were performed on four different sample sizes: 1.4 x 106, 7.0 x 105, 3.5 x 105, and 1.75 x 105 cells. The data show that within this range of cells, sample size does not significantly influence mutant frequency measurements.
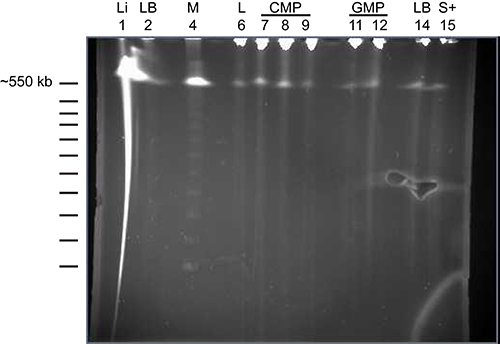
Figure 8. Pulsed-field electrophoresis of high molecular weight DNA samples isolated from purified cells. Genomic DNA was isolated as described in Step 2 of the protocol and run on a Bio-Rad (CHEF-DR III) system. The various samples loaded on the gel are DNA samples isolated from liver (Li; lane 1), bone marrow cells depleted of mature lineage+ cells (LB; lanes 2 and 14), CD34+LSK cells (L; lane 6), CMPs (lanes 7-9), GMPs (lanes 11 and 12) and Sca-1+ cells (S; lane 15). Lane 4 holds the ladder for high molecular weight DNA. The picture shows that the majority of the DNA is high molecular weight (> 600 kb).
Table 1. Miscellaneous sample parameters of purified populations, whole bone marrow and spleen. Use this Table where indicated in the Protocol. For each population indicated on top, the number of cells (x 105) per aliquot, DNA digestion time, volume of DNA sample after dialysis, number of reactions required for each aliquot and number of trays required per aliquot are indicated on the left. The digestion times at 50 °C are critical to the success of the experiment.
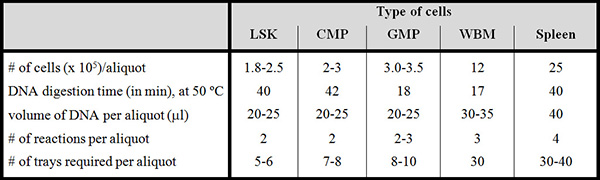
LSK = Lin–Sca-1+Kit++ cells; CMP = committed myeloid progenitor; GMP = granulocytic/ monocytic progenitor; WBM = (unsorted) whole bone marrow. Spleen cells are unsorted.
Table 2. Efficiency of plaque formation is comparable among different cell populations. This table shows the plaque formation among the different populations. Six month-old C57BL/6 mice were used for these experiments (femurs and tibiae of 10-11 mice per experiment, 6 experiments in total). All but one of the populations used in these experiments gave at least 1, up to 24, (confirmed) mutant PFUs (Zhou et al., manuscript in preparation). PFU numbers may vary per mouse strain.
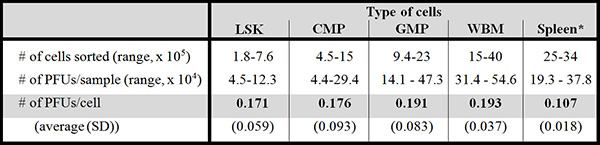
LSK = Lin–Sca-1+Kit++ cells; CMP = committed myeloid progenitor; GMP = granulocytic/ monocytic progenitor; WBM = (unsorted) whole bone marrow. *Spleen cells are unsorted. SD = standard deviation.
Table 3. Instructions for the preparations of additional reagents.
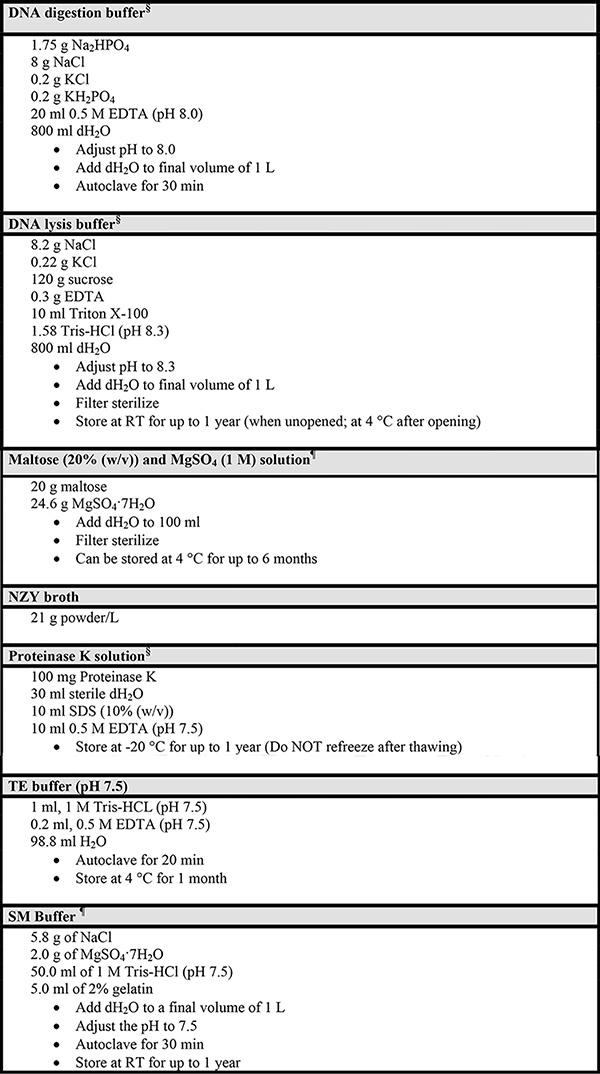
§ From the Stratagene Instruction Manual of the RecoverEase DNA Isolation Kit
¶ From the Stratagene Instruction Manual of the Transpack Packaging Extract
Discussion
The in vivo mutagenesis assay described herein is based on the LacI transgenic mouse model originally generated by Kohler et al.18 This model utilizes a λ phage vector carrying a lacI reporter gene. The two cos sites flanking the vector allow for a relatively simple recovery and subsequent packaging into infectious phage particles, used to infect E. coli. A blue plaque will be generated by phage-infected E. coli that contain a mutated LacI gene. The blue plaque is set against a colorless background, thus greatly simplifying the task of mutant scoring. DNA sequencing techniques can be used to identify the position and type of mutation that has occurred, which may help further investigations into the mechanisms underlying mutagenesis.
The modifications we made to the protocol of this mutagenesis assay allow one to use it with FACS-purified hematopoietic cell populations. However, we found that a reproducible analysis still requires at least 2 x 105 hematopoietic cells to ensure sufficient quantities of high-quality DNA. Since the frequency of long-term repopulating HSCs is extremely low29, performing a mutagenesis analysis on these HSCs is at this point not achievable. The stem cell-enriched population we use in this protocol, LSK cells, contains in addition to Flk-2– long-term repopulating HSCs, also Flk-2– short-term repopulating HSCs and Flk-2+ cells, which represent multipotential progenitor cells (MPPs)42. Despite this limitation we feel that using LSK cells as a read-out for HSCs in this mutagenesis assay is reasonable since it was shown that LSK cells behave more like LSK-Flk-2– HSCs in terms of utilizing NHEJ than progenitor cells5. Moreover, work from Rossi et al.8 suggests that MPPs are better in copying with damaged DNA than HSCs, especially when they age, leading us to hypothesize that the number of mutants found in the LSK population reflects that of the HSCs rather than the MPPs.
Since small numbers of sorted cells are obtained at each experiment, an important issue to consider at the planning stage of this in vivo mutagenesis assay, is the sample size (i.e. the total number of plaques per sample) necessary to achieve statistical significance. In other words, when the goal is to, for example, compare the mutation frequency of manipulated LSK cells to wild-type control LSK cells, how many plaques in total are required for each LSK population to detect a two or three-fold difference in mutation frequency? Of note, the total number of plaques can be combined from all experiments, since we found no significant difference between sorting experiments, at least in wild-type cells, based on a negative binomial regression analysis43. Plaque numbers are important to know because that will determine the number of sorts that need to be performed (~10,000 for WBM and GMPs and ~7,000 for Spleen, LSKs and CMPs). Since the mutation frequencies are small in wild-type tissues44, the normal approximation statistically comparing these is not accurate. For this reason, we use the Poisson distribution, since it approximates the true binomial distribution (of the mutation frequency) when that frequency is small and the sample size is relative large; Huffman45 provides a formula (Equation 4) to calculate sample size based on Poisson distribution. When applied to hypothetical mutation frequencies of 2.5, 4, and 7 mutations per 100,000 cells in the control cells44, we require 456,949 plagues, 285,592 and 163,196, respectively, to detect a significant two-fold difference (two sample test with two tailed significance of 0.05 and power of 0.80). Fewer plaques are required to detect a three-fold difference: 122,148; 76,342; and 43,624, respectively. Thus, the number of plaques required for each population of interest (and thus the number of sorts needed to obtain enough cells) depends on the mutation frequency that can be expected in the control cell population and the predicted fold-change in the comparison population.
The most important step in this mutagenesis assay is the isolation of genomic DNA (step 2). Although it is essential to start this protocol with high quality DNA samples (see "representative results"), when working with sorted cells, cell numbers are often limited and cannot be wasted on determining DNA concentration or size. We found that another good indicator for the quality of the DNA sample is its viscosity; at step 2.9 the sample should be extremely viscous and hard to work with. It requires some practice to get this step right. Therefore, it is recommended to work out the protocol with large cell numbers, e.g. with whole bone marrow or sorted Sca-1+ cells, and get comfortable with isolating DNA from these samples and learn how to judge the quality of the DNA by its viscosity.
The quality and size of the DNA affects the number of PFUs generated. When the DNA isolation step is perfected, the next important element of optimizing the assay is the density of PFUs per tray. Table 1 shows the recommended number of trays to use for each type of sample corresponding to an optimum numbers of cells for that sample. Using this guideline should result in trays where individual plaques can still be distinguished, yet are not spread too thin. A tray should hold between 40-120 PFUs per small square; the example shown in Figure 4B represents a reasonable density. With these optimization aspects worked out, the number of PFUs generated per sample is highly efficient and reproducible (Table 2).
The LacI transgenic mouse model has been used with a variety of other tissues, but not in conjunction with relatively small numbers of highly purified populations. When applied to other (than hematopoietic) tissue-restricted stem cell-enriched populations, it is recommended to pay specific attention to the DNA isolation procedure; they may differ from cell type to cell type and the recommendation for hematopoietic cells may not necessarily work for other tissue cell types. The critical factors, such as the amount of reagents, the temperature at which the proteinase K digestion needs to take place and, most importantly, the proteinase K digestion time will need to be worked out for each cell type; this protocol may serve as a starting point.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We would like to thank David R. Rodriguez, M.A. for the graphic design and photography in this manuscript. This work was supported by funding from the GCCRI, the NIH/NIA (5R21AG033339) and the Cancer Center Support Grant (P30CA054174) to the UTHSCSA Flow Cytometry Core facility and the UTHSCSA Advanced Nucleic Acids Core Facility.
Materials
| LacI transgenic mice | BioReliance Corporation | |
| RecoverEase DNA Isolation Kit, including the RNace-It ribonuclease cocktail | Agilent Technologies (Stratagene) | 720202 |
| Transpack Packaging Extract, including the orange and blue transpack tubes and the SCS-8 E. coli | Agilent Technologies (Stratagene) | 200221 |
| DNA size standards – lambda ladder | Bio Rad | 170-3635 |
| 0.025 mM pore size membranes | Fisher (Millipore) | VSWP 025 00 |
| 245 mm2 bioassay dishes (trays) | Fisher (Corning) | 07-200-600 |
| NZY broth (powder) | Fisher (Teknova) | N1144 |
| Agar | Fisher | BP1423-500 |
| Agarose | Fisher | E-3120-500 |
| N, N-dimethylformamide | Fisher | AC34843-5000 |
| X-gal | Research Products Intl Corp (RPI) | B71800-10.0 |
| Proteinase K | Roche | 3-115-852 |
| PCR Extender Taq Polimerase kit | 5 PRIME | 2200500 |
| Agencourt AMPure XP cleanup kit | Beckman/coulter | A63880 |
References
- Maslov, A. Y., Vijg, J. Genome instability, cancer and aging. Biochim. Biophys. Acta. 1790 (10), 963-969 (2009).
- Aguilera, A., Gomez-Gonzalez, B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9 (3), 204-217 (2008).
- Milyavsky, M., et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell. Stem Cell. 7 (2), 186-197 (2010).
- Mohrin, M., et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell. Stem Cell. 7 (2), 174-185 (2010).
- Shao, L., Feng, W., Lee, K. J., Chen, B. P., Zhou, D. A sensitive and quantitative polymerase chain reaction-based cell free in vitro non-homologous end joining assay for hematopoietic stem cells. PLoS ONE. 7 (3), (2012).
- Sotiropoulou, P. A., et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat. Cell Biol. 12 (6), 572-582 (2010).
- Prasher, J. M., et al. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1-/- mice. EMBO J. 24 (4), 861-871 (2005).
- Rossi, D. J., et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 447 (7145), 725-729 (2007).
- Reese, J. S., Liu, L., Gerson, S. L. Repopulating defect of mismatch repair-deficient hematopoietic stem cells. Blood. 102 (5), 1626-1633 (2003).
- Carney, D. A., et al. Therapy-related myelodysplastic syndrome and acute myeloid leukemia following fludarabine combination chemotherapy. Leukemia. 24 (12), 2056-2062 (2010).
- Jankowska, A. M., et al. Base excision repair dysfunction in a subgroup of patients with myelodysplastic syndrome. Leukemia. 22 (3), 551-558 (2008).
- Tam, C. S., et al. Treatment-related myelodysplasia following fludarabine combination chemotherapy. Haematologica. 91 (11), 1546-1550 (2006).
- Webb, S. Xeroderma pigmentosum. BMJ. 336 (7641), 444-446 (2008).
- Ben-Yehuda, D., et al. Microsatellite instability and p53 mutations in therapy-related leukemia suggest mutator phenotype. Blood. 88 (11), 4296-4303 (1996).
- Casorelli, I., et al. Drug treatment in the development of mismatch repair defective acute leukemia and myelodysplastic syndrome. DNA Repair. 2 (5), 547-559 (2003).
- Olipitz, W., et al. Defective DNA-mismatch repair: a potential mediator of leukemogenic susceptibility in therapy-related myelodysplasia and leukemia. Genes Chrom. Cancer. 34 (2), 243-248 (2002).
- Haase, D., et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 110 (13), 4385-4395 (2007).
- Kohler, S. W., et al. Analysis of spontaneous and induced mutations in transgenic mice using a lambda ZAP/lacI shuttle vector. Environ. Mol. Mutagen. 18 (4), 316-321 (1991).
- Rogers, B. J., Provost, G. S., Young, R. R., Putman, D. L., Short, J. M. Intralaboratory optimization and standardization of mutant screening conditions used for a lambda/lacI transgenic mouse mutagenesis assay. 327 (1-2), 57-66 (1995).
- Jakubczak, J. L., et al. Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc. Natl. Acad. Sci. U.S.A. 93 (17), 9073-9078 (1996).
- Ward, T. L., Prtenjaca, A., Hill, K. A. A novel Escherichia coli-derived mutation detected with the Big Blue cII mutant selectable assay. Environ. Mol. Mutagen. 51 (4), 344-348 (2010).
- Miller, J. H., Coulondre, C., Farabaugh, P. J. Correlation of nonsense sites in the lacI gene with specific codons in the nucleotide sequence. Nature. 274 (5673), 770-775 (1978).
- Provost, G. S., et al. Transgenic systems for in vivo mutation analysis. Mutat. Res. 288 (1), 133-149 (1993).
- Schaaper, R. M., Danforth, B. N., Glickman, B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J. Mol. Biol. 189 (2), 273-284 (1986).
- Schaaper, R. M., Dunn, R. L. Spontaneous mutation in the Escherichia coli lacI gene. Genetics. 129 (2), 317-326 (1991).
- Burkhart, J. G., Burkhart, B. A., Sampson, K. S., Malling, H. V. ENU-induced mutagenesis at a single A: T base pair in transgenic mice containing phi X174. Mutat. Res. 292 (1), 69-81 (1993).
- Valentine, C. R., et al. Characterization of mutant spectra generated by a forward mutational assay for gene A of Phi X174 from ENU-treated transgenic mouse embryonic cell line PX-2. Environ. Mol. Mutagen. 39 (1), 55-68 (2002).
- Gossen, J. A., Molijn, A. C., Douglas, G. R., Vijg, J. Application of galactose-sensitive E. coli strains as selective hosts for LacZ- plasmids. Nucleic Acids Res. 20 (12), 3254 (1992).
- Kiel, M. J., et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121 (7), 1109-1121 (2005).
- Akashi, K., Traver, D., Miyamoto, T., Weissman, I. L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404 (6674), 193-197 (2000).
- Frascoli, M., Proietti, M., Grassi, F. Phenotypic analysis and isolation of murine hematopoietic stem cells and lineage-committed progenitors. J. Vis. Exp. (65), (2012).
- Osawa, M., Hanada, K., Hamada, H., Nakauchi, H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 273 (5272), 242-245 (1996).
- Stratagene, Transpack packaging extract for lambda transgenic shuttle vector recovery. Instruction manual. , (2001).
- Stratagene, RecoverEase DNA isolation kit. Instruction manual. , (2009).
- Stratagene, Big Blue transgenic mouse mutagenesis assay system. Instruction manual. , (1992).
- Nishino, H., Buettner, V. L., Sommer, S. S. Towards validation of the Big Blue transgenic mouse mutagenesis assay: the mutational spectrum of ex vivo pinpoint mutant plaques. Muta. Res. 372 (1), 97-105 (1996).
- Stuart, G. R., Gorelick, N. J., Andrews, J. L., de Boer, J. G., Glickman, B. W. The genetic analysis of lacI mutations in sectored plaques from Big Blue transgenic mice. Environ. Mol. Mutagen. 28, 385-392 (1996).
- Bielas, J. H., Heddle, J. A. Proliferation is necessary for both repair and mutation in transgenic mouse cells. Proc. Natl. Acad. Sci. U.S.A. 97 (21), 11391-11396 (2000).
- Paashuis-Lew, Y., Zhang, X. B., Heddle, J. A. On the origin of spontaneous somatic mutations and sectored plaques detected in transgenic mice. Mutat. Res. 373 (2), 277-284 (1997).
- de Boer, J. G., Glickman, B. W. The lacI gene as a target for mutation in transgenic rodents and Escherichia coli. Genetics. 148 (4), 1441-1451 (1998).
- Piegorsch, W. W., et al. Study design and sample sizes for a lacI transgenic mouse mutation assay. Environ. Mol. Mutagen. 25 (3), 231-245 (1995).
- Yang, L., et al. Identification of Lin(-)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 105 (7), 2717-2723 (2005).
- Hilbe, J. M. . Negative Binomial Regression. second edn. , (2011).
- de Boer, J. G., Provost, S., Gorelick, N., Tindall, K., Glickman, B. W. Spontaneous mutation in lacI transgenic mice: a comparison of tissues. Mutagenesis. 13 (2), 109-114 (1998).
- Huffman, M. D. An improved approximation to-sample poisson test. Appl. Stat. 33 (2), 224-226 (1984).

