Paired Whole Cell Recordings in Organotypic Hippocampal Slices
Summary
Pair recordings are simultaneous whole cell patch clamp recordings from two synaptically connected neurons, enabling precise electrophysiological and pharmacological characterization of the synapses between individual neurons. Here we describe the detailed methodology and requirements for establishing this technique in organotypic hippocampal slice cultures in any laboratory equipped for electrophysiology.
Abstract
Pair recordings involve simultaneous whole cell patch clamp recordings from two synaptically connected neurons, enabling not only direct electrophysiological characterization of the synaptic connections between individual neurons, but also pharmacological manipulation of either the presynaptic or the postsynaptic neuron. When carried out in organotypic hippocampal slice cultures, the probability that two neurons are synaptically connected is significantly increased. This preparation readily enables identification of cell types, and the neurons maintain their morphology and properties of synaptic function similar to that in native brain tissue. A major advantage of paired whole cell recordings is the highly precise information it can provide on the properties of synaptic transmission and plasticity that are not possible with other more crude techniques utilizing extracellular axonal stimulation. Paired whole cell recordings are often perceived as too challenging to perform. While there are challenging aspects to this technique, paired recordings can be performed by anyone trained in whole cell patch clamping provided specific hardware and methodological criteria are followed. The probability of attaining synaptically connected paired recordings significantly increases with healthy organotypic slices and stable micromanipulation allowing independent attainment of pre- and postsynaptic whole cell recordings. While CA3-CA3 pyramidal cell pairs are most widely used in the organotypic slice hippocampal preparation, this technique has also been successful in CA3-CA1 pairs and can be adapted to any neurons that are synaptically connected in the same slice preparation. In this manuscript we provide the detailed methodology and requirements for establishing this technique in any laboratory equipped for electrophysiology.
Introduction
Glutamate receptors mediate the majority of excitatory synaptic transmission at central nervous system synapses. The two major subtypes of ionotropic glutamate receptors localized at the spine head of the postsynaptic membrane are N-methyl-D-aspartate (NMDA) and α-amino-3-hydroxy-5-methylisoxazole-4-proprionic acid (AMPA) receptors. At resting membrane potentials, AMPA receptors carry most of the postsynaptic current during synaptic transmission. In the hippocampus, the NMDA receptor plays a key role in triggering changes in the number of AMPA receptors in the postsynaptic membrane: by acting as a ”coincidence detector”1 to initiate changes in synaptic strength1, the NMDA receptor participates in the synaptic mechanisms that are thought to underpin learning and memory at a subcellular level. In response to depolarization of the postsynaptic neuron in parallel with presynaptic transmitter release, calcium enters via the NMDA receptor to initiate AMPA receptor insertion or removal2. These receptor dynamics underlie synapse plasticity: an increase in synaptic strength is long-term potentiation2,3 (LTP), while a decrease in synaptic strength is long-term depression4 (LTD). Therefore AMPA receptor movement is thought to be responsible for synaptic plasticity expression, while NMDA receptors are thought to control its induction.
Determining the precise mechanisms underlying synaptic transmission and plasticity requires studying small populations of synapses, ideally single synapses. While some synapses are highly suited for study at this level, e.g., the Calyx of Held5, for most synaptic populations this is extremely difficult due to the small and diffuse nature of the synaptic connections. Two major electrophysiology techniques have been developed to examine single synaptic connections: The first is minimal stimulation, where one presynaptic fiber is presumed stimulated extracellularly. The second technique is paired recordings, where two simultaneous whole cell recordings from synaptically connected neurons is performed. A major advantage of minimal stimulation is that it is rapid and relatively simple to perform, involving placement of an extracellular stimulating electrode into the axonal tract while simultaneously recording from a postsynaptic neuron. The primary concern when using this technique is that reliable stimulation of a single cell can rarely be guaranteed trial after trial.
Over the past fifteen years we have routinely used paired whole-cell recordings from two synaptically connected pyramidal neurons6-17. The major advantage of this technique is that only one presynaptic neuron is consistently and reliably stimulated. It also allows not only electrophysiological characterization but also pharmacological manipulation of the presynaptic neuron6,18. However, the probability of synaptic connectivity between neurons is low, making connected pairs difficult to obtain19. The use of organotypic brain slice cultures circumvents this obstacle as synaptic connectivity can re-establish in vitro and moreover the nature of the resulting connectivity is similar to that in native brain tissue20. In addition, organotypic cultures express LTP, LTD7-10,12-15,21 and additional forms of short-term synaptic plasticity including paired-pulse facilitation (PPF) and depression (PPD)6,22,23, enabling plasticity mechanisms to be studied in pairs of neurons. Here we describe the detailed methodology involved in successfully attaining paired recordings in this in vitro system. This information can readily be adapted to other experimental systems, including acute slices and other brain regions.
Protocol
Animal Ethics Statement:
The protocols described in this manuscript follow the animal care guidelines established by The University of Auckland and Stanford University. P7 rat pups were euthanized by rapid decapitation. Hippocampal dissection is then immediately performed as described below.
1. Organotypic Hippocampal Slice Culture
- Preparation
- Prepare dissection Medium (used only for dissecting the brain). Combine 200 ml Minimum Essential Medium, 2 ml penicillin-streptomycin solution (10,000 units of each, in 0.85% NaCl), 5 ml HEPES buffer solution, 2 ml 1 M Tris stock solution (pH 7.2), and filter sterilize with 0.22 μm filter. Carry out hippocampal dissection in ice-cold dissection medium.
- Cool the dissection medium. Place the dissection media in the freezer approximately 1 hr prior to beginning the dissection until the liquid is very cold. Do not allow large ice crystals to form. Store on ice until required.
- Prepare culture medium (used for everything except dissection of hippocampi). Combine 100 ml Minimum Essential Medium, (1x concentration, liquid) w/Hank’s salts, w/ L-glutamine, 2 ml penicillin-streptomycin solution (liquid, 10,000 units of each, in 0.85% NaCl ), 2.5 ml HEPES 1 M buffer solution, 50 ml Hank’s Balanced Salt Solution, 50 ml Horse Serum (defined, heat inactivated), and filter sterilize with 0.22 μm filter.
- Prepare the culture dishes. Place 1 ml of culture media per 35 mm culture dish, and add a membrane insert to each dish. Put up to seven of these dishes into one 150 mm Petri dish (referred to hereafter as a ‘plate’). Place the plate in a CO2 incubator for at least an hour before the dissection begins so that the culture medium in the dishes attains the proper temperature and pH.
- Dissection of the Hippocampi from Rat Pups at Postnatal Day 7 (P7)
- Pre-sterilize all dissection tools under UV light before the procedure.
- After rapid decapitation, remove the brain and place into chilled medium in one dish, and then remove it to a piece of moistened filter paper for the dissection. Tease the cortex away from the midbrain using blunt smooth plastic-coated miniature spatulas, exposing the hippocampus. Cut the fornix, and then gently work the spatula underneath the hippocampus to flip it out (Figure 1A).
NOTE: Successful slice cultures can be prepared using animals up to P10. - Trim the isolated hippocampus away from the rest of the brain. Transfer the hippocampi into a new dish containing chilled dissection media using a moistened soft paintbrush (e.g., white sable #4).
- Clean the underside (i.e. the flatter side) of the hippocampus of choroid plexus during dissection, as these spongy, meninge-like tissues make it difficult to separate hippocampal slices from one another later on.
NOTE: Leave these tissues in place if they cannot be teased away gently. - Perform the entire dissection as quickly as possible without damaging the hippocampi.
NOTE: A careful dissection is more important than a fast one, as long as the experimental conditions are chilled. Slice the hippocampi from three rats simultaneously. The slice health is not affected as long as the total time from start to when the plated slices go into the incubator is under 30 min.
- Slicing
- Slice the hippocampi transversely into 400 μm cross-sections using a manual tissue slicer. The method by which the slicing is conducted is not important, so long as it is not overly damaging to the tissue and produces slices of consistent thickness with readily visible laminar architecture (i.e. Ammon’s Horn is easily seen to be preserved in the tissue).
- Lie the hippocampi on the stage of the tissue chopper on top of a triple thickness of #2 filter paper. The hippocampi are sliced entirely without removing any slices individually (i.e. the entire hippocampus is left on the stage of the chopper until all cuts have been made; rather like loaves of sliced bread at this point; Figure 1B).
- Transfer the hippocampi into dissection medium using a soft paintbrush laid next to each hippocampus (white sable #2). Gently push each hippocampus to the side to break the adhesion between the hippocampus and the underlying filter paper. Roll the brush underneath each hippocampus to pick it up off the stage. Place all hippocampi into the same 35 mm Petri dish of chilled dissection medium. Separate the hippocampi into individual slices by manually agitating the dish in alternating clockwise and counterclockwise motions.
- Inspect the slices under a dissecting microscope and discard any which are damaged, small, or which do not possess clearly visible cell body layers.
NOTE: It will often be the case that not all the slices are successfully separated from their siblings in this manner. In this case, they can be separated by turning them on edge and prodding them with the tips of fine forceps. “Good” slices are then transferred to another clean Petri dish containing chilled dissection medium using a cut off and fire-polished Pasteur pipette (Figures 1C-E).
- Storage
- Place the individual slices onto the membrane filter inserts. Using a Pasteur pipette cut off and fire polished to give a larger bore, individually transfer slices to the culture inserts.
NOTE: The size of the bore created when cutting and fire-polishing the pipette is important. Too small and the slices will not easily leave the pipette for the membrane. Too large and excess fluid is placed on the surface of the membrane along with the slice. The best bore diameter is typically about half the diameter of the barrel of the pipette. When cutting, this diameter can be chosen by cutting at the proper place on the taper of the pipette. - Slice Transfer
- Fill the pipette with medium first, and then suck up a single slice with small suction forces. This keeps the slice near the opening of the pipette and prevents too much medium expulsion into the insert to place the slice.
- Apply a slight pressure on the bulb to form a small hanging droplet, allow the slice to settle into that droplet, and then touch that droplet to the membrane and let the slice “fall” onto the membrane. Generally three slices are placed onto each membrane insert.
NOTE: If the fluid droplet is too large, the droplets from the three slices tend to merge on the membrane surface, and the slices will thus all gather to the center of the membrane, making it more difficult to separate them for electrophysiological recording later.
- Fluid Removal
- Aspirate any excess medium from the top of the culture plate insert for all slices in that plate (3 slices per insert = 21 slices per plate), so that slices are not immersed in or surrounded by a pool of dissection medium.
- Perform this is with an uncut, but fire-polished Pasteur pipette. Allow the slice to settle and adhere to the membrane for a minute before pipetting. Take care not to suck the slice up into the pipette. Carry out fluid removal for the inserts that are plated first, to allow sufficient time for the slices to settle.
- Slice Storage
- Store slice cultures in a 5.0% CO2 incubator at 37 °C. Observe the final arrangement of the cultures in Figure 1E. The individual slices rest upon a culture plate insert which has a porous membrane for a bottom, and this insert fits inside a Petri dish filled with a small amount of medium; by this means, the slices are not immersed in the medium, but have access to it only through the membrane at the bottom of the culture plate insert.
- Optionally, remove the slices may for study either by removing the culture plate insert or by cutting out a section of insert membrane containing a slice.
- Place the individual slices onto the membrane filter inserts. Using a Pasteur pipette cut off and fire polished to give a larger bore, individually transfer slices to the culture inserts.
- Maintenance
- Exchange the medium in the Petri dishes the day after making cultures. Transfer the culture plate insert to a new petri dish containing 1 ml of fresh culture medium that is equilibrated in the incubator for at least an hour prior to transfer.
- Change the medium again in the same manner on the third day after making cultures. On the third day, transfer the cultures to an incubator set at 34 °C. Change the medium twice each week (every 3-4 days).
- Identify healthy cultures. Select healthy cultures for paired whole cell recordings.
NOTE: Healthy cultures have a well-defined edge and a clearly defined pyramidal cell layer. Cultures with dark (likely necrotic) areas present or a vacuolated appearance with flattened borders are rejected. Employing these criteria, usually two-thirds of slice cultures are sufficiently healthy for recording. - Utilize these cultures within a fairly small window of time. Do not used tissues that are cultured for more than two weeks since the neurons begin to display slightly epileptiform behavior which slowly worsens with time. The exact upper limit of neuronal survival is not known but it is possible to record from functioning pyramidal cells as late as 16 weeks in culture.
2. Paired Whole Cell Recordings
- ACSF and Intracellular Solutions
- Prepare presynaptic electrode solution by mixing (in mM) 120 potassium gluconate, 40 HEPES, 5 MgCl2, 2 NaATP, and 0.3 NaGTP (pH 7.2 with KOH; osmolarity: 290 mOsm). Whilst the same electrode solution is used for postsynaptic neurons, cesium gluconate (120 mM) is typically used as the major salt in the postsynaptic electrode, plus 5 mM QX314.
NOTE: This enables stable voltage clamping at positive potentials to record postsynaptic NMDAR-mediated currents8-10,13. It also prevents the potassium-induced hyperexcitability of the presynaptic neuron by the internal solution flowing from the postsynaptic recording electrode before a giga ohm seal is made. - Prepare postsynaptic electrode solution composing (in mM) 120 cesium gluconate, 40 HEPES, 5 MgCl2, 5 QX314, 2 NaATP, and 0.3 NaGTP (pH 7.2 with CsOH; osmolarity: 290 mOsm). Pull glass microelectrodes at 5-10 MΩ resistance and fill with filtered internal solution.
- Prepare artificial cerebrospinal fluid (ACSF) composing (in mM) 119 NaCl, 2.4 KCl, 1.3 MgSO4, 2.4 CaCl2, 1 Na2HPO4, 26.2 NaHCO3, 11 glucose, pH 7.4, saturated with 95% O2, 5% CO2. NOTE: If AMPAR-mediated responses need to be blocked for examination of NMDAR EPSCs, include 10 μM CNQX or NBQX in the ACSF.
- Prepare presynaptic electrode solution by mixing (in mM) 120 potassium gluconate, 40 HEPES, 5 MgCl2, 2 NaATP, and 0.3 NaGTP (pH 7.2 with KOH; osmolarity: 290 mOsm). Whilst the same electrode solution is used for postsynaptic neurons, cesium gluconate (120 mM) is typically used as the major salt in the postsynaptic electrode, plus 5 mM QX314.
- Obtain Paired Whole Cell Recordings
- To examine synaptic transmission between two individual neurons, designate one neuron as the presynaptic neuron and hold in current clamp to induce action potentials and initiate synaptic transmission.
- Designate the second neuron, as the postsynaptic neuron, and hold it in current or voltage clamp depending on the information required by the researcher. Obtain detailed examination of the glutamatergic currents, with AMPAR-mediated EPSCs examined at -65 mV and NMDAR-mediated EPSCs examined at +30 mV6-16 by holding the postsynaptic neuron in voltage clamp
- Establish a paired recording. To establish a paired recording, the presynaptic whole cell recording is always obtained first, enabling multiple sequential postsynaptic neurons to be obtained until one that is synaptically connected is obtained. If performing synaptic plasticity experiments, it is especially critical to obtain the presynaptic neuron first as the induction of LTP in the postsynaptic neuron must be initiated within 10 min of obtaining the whole cell recordings to prevent washout of cytoplasmic factors required for LTP8,19.
- Avoid movement-induced cell loss. A major challenge when performing paired whole cell recordings is avoiding vibration and movement-induced disruption of the first whole cell recording whilst obtaining the second recording.
- Obtain the first whole cell recording and then move the microscope lens 10-200 μm in the x-axis so that the established recording is located at the edge of the area visible on the monitor (Figure 2A).
- Raise the lens of the microscope ~5-10 mm while ensuring that the ACSF still maintains contact with the lens. Mount the second electrode and guide into the fluid meniscus (Figure 2B).
- Move the electrode in the x-y plane until it is directly under the light path through the lens but still significantly above the established recording electrode. Once the electrode is visible under the microscope, ensure the tip is at the far edge of the monitor away from the first electrode.
- Move the second electrode down in sequence with the microscope focus until both electrodes are in the same focal plane. It is important that the level of positive pressure is strong enough to avoid tip blockage but not too strong so as to disrupt the first whole cell recording. This translates to the positive pressure inducing movement in 2-3 neurons from the electrode when it first enters the slice.
- Locate a postsynaptic partner in a similar focal plane to the first presynaptic recording (Figure 2C). Recording is generally Obtain paired recordings between CA3 pyramidal neurons the second whole cell from a 0-200 mm neuron of the first recording (Figures 2C,D).
NOTE: Synaptic transmission between neuronal pairs are stable over time periods of up to 3-4 hr when using these standard whole cell recording techniques.
- Synaptic plasticity: Once Upon obtaining a successful synaptically-connected pyramidal cell pair (Figures 3A,B), examine the characteristics of synaptic transmission and plasticity.
- LTP induction.
- Induce LTP by pairing presynaptic action potentials (1 Hz) with postsynaptic depolarization to -10 to 0 mV for 1 min (Figure 3C)7-9,12-14. Initiate pairing within 10 min of break-in to the postsynaptic neuron.
NOTE: LTP can also be induced with both neurons held in current clamp by pairing presynaptic and postsynaptic action potentials at 1 Hz for 1 min, with postsynaptic action potentials elicited 10 msec following injection of current into the presynaptic neuron8.
- Induce LTP by pairing presynaptic action potentials (1 Hz) with postsynaptic depolarization to -10 to 0 mV for 1 min (Figure 3C)7-9,12-14. Initiate pairing within 10 min of break-in to the postsynaptic neuron.
- Further induce LTD by low frequency stimulation (LFS) at 1 Hz combined with a slight depolarization of the postsynaptic neuron to -55 mV for 5-10 min (Figure 3C)9.
- LTP induction.
Representative Results
Synaptic connectivity is evident by stimulating the presynaptic neuron to fire an action potential by passing a depolarizing current pulse (typically 20-50 pA for 20 msec) via the recording electrode. The postsynaptic current trace is then examined for the presence of a monosynaptic EPSC evoked at short (<5 msec) and consistent latencies after the peak of the presynaptic action potential (Figure 3A). In most experiments multiple postsynaptic neurons are tested before a synaptically-connected pair can be obtained. Overall, ~1/3 of presynaptic CA3 cells are monosynaptically coupled to the postsynaptic CA3 cell by active synaptic connections (Figure 3A), and approximately 20% of CA3 neurons are connected by all-silent synaptic connections6,8.
The amplitude of baseline AMPA receptor mediated currents is variable from trial to trial within a paired recording, and also between independent paired recordings (Figure 3B)6,8. Average AMPAR EPSC amplitude ranged from <10 pA to >800 pA, and likely arises from differences in the number of functional synapses between paired recordings. Failure rates have also been observed to vary significantly between paired recordings6,7, ranging from 100% AMPAR EPSC failure rates at silent synapses, to 0-95% failure rates in pairs connected by active synapses6,7. Within individual paired recordings, synaptic failures occurred, especially in recordings with smaller AMPAR EPSC amplitudes. The variability in AMPAR EPSC amplitude is evident from trial to trial in the raw data (Figure 3B) is likely due to fluctuation in quantal number released from trial to trial, as occurs at other synapses6.
Dual whole cell recordings also provide direct access to both the pre- and the postsynaptic neuronal cytoplasm, enabling pharmacological manipulation of either/both the presynaptic and postsynaptic cells via the recording electrodes6. Typically, presynaptic pharmacological manipulations are very rapid (within 10 min). This is not limited to small molecules (e.g., BAPTA), as fluorescently labeled dextrans can access presynaptic terminals ≤200 μm away from the recording electrode. Therefore analysis similar to that performed at the squid giant synapse25 could be extended to studies of synaptic vesicle release machinery in the hippocampal slice. The position of the specific synapses measured in the paired recordings are not identified in the recording process. Therefore the proximal versus distal sites of synapses cannot be assessed as readily as local stimulation with extracellular stimulating electrodes or focal glutamate application. However, dye filling of each neuron during the paired recording can enable morphological reconstruction of axonal arbors and potential synaptic sites6.
LTP and LTD are reliably induced at CA3-CA3 synapses in this culture system (Figure 3C), and both forms of plasticity last for the duration of whole cell recordings (over 2 hr). The LTP and LTD induction paradigms utilized are identical to those performed in acute slices and LTP and LTD exhibit the properties of NMDAR-dependence, associativity and pathway independence, and therefore this combination of culture system and paired recordings provides a valuable model system to examine synaptic plasticity7.
Polysynaptic inhibitory events are frequently observed in paired recordings between CA3 pyramidal cell pairs (Figure 3D). We do not pharmacologically block this GABAergic inhibition as this produces highly disruptive hyperactivity. However, due to the longer latency of these polysynaptic inhibitory events, they generally do not prevent measurement of the monosynaptic excitatory current. If the polysynaptic inhibitory events were observed to interfere with the monosynaptic current, obscuring the peak of the monosynaptic current, these pairs need to be excluded from the analysis. Polysynaptic excitatory connections are also sometimes observed, but are significantly less common and also are excluded from analysis. Rarely, a monosynaptic inhibitory synaptic current is obtained due to the presynaptic neuron being an inhibitory interneuron. This is readily identified by the lack of accommodation of action potential firing in response to longer (1 sec) current injection. This is not possible in postsynaptic neurons in which the internal solution is cesium gluconate-based. However, as neurons are identified visually prior to recording, in our experience this occurs <1% of the time. Interneurons within organotypic hippocampal slices are readily identified visually by their non-pyramidal cell body, especially in the stratum radiatum and oriens. Therefore this preparation also enables recordings of interneuron excitatory and inhibitory currents. However, very little is known about the inhibitory networks in organotypic slices, and whether they maintain connectivity similar to that in the brain is not clear.

Figure 1. Preparation of organotypic hippocampal slice cultures. (A) The hippocampus, in situ, outlined by the dotted line. To remove the hippocampus, the connections to the fornix are severed and the hippocampus gently rolled out of the brain. (B) Hippocampi are positioned on the stage of the slicer on top of filter paper. (C) Hippocampal slices are transferred via the wide bore of a Pasteur pipette to prevent damage. (D) Examples of ‘good’ versus ‘bad’ slices. (E) Cartoon of slice setup on membranes. Please click here to view a larger version of this figure.

Figure 2. Obtaining paired whole cell recordings from hippocampal CA3 neurons. Sequential images of attaining a paired recording, showing optimal electrode and neuronal positioning. (A) After obtaining the first whole cell recording the microscope is moved 10-200 mm in the x-axis so that the established recording (arrow) is located at the edge of the area visible on the monitor. Scale bar: 25 μm. (B) To enable room for the second electrode, the lens of the microscope is then raised ~5-10 mm, ensuring that the ACSF still maintains contact with the lens. To ensure the second electrode does not bump the other recording electrode, the second electrode is moved in the x-axis until it is directly under the light path through the lens but still significantly above the established recording electrode. (C) Established paired recording showing both electrodes (arrows) in the same focal plane. Scale bar: 25 μm. (D) Paired recording from adjacent CA3 neurons in a transgenic animal, showing the EGFP-positive neurons on the RHS. Scale bar: 20 μm. Please click here to view a larger version of this figure.

Figure 3. Synaptic transmission and plasticity between CA3-CA3 pyramidal cell pairs. (A) Example of pre- and postsynaptic traces from a paired recording between two CA3 pyramidal neurons. In response to a 20 sec current pulse to induce presynaptic action potential firing, the postsynaptic AMPAR-mediated current (recorded at -65 mV) occurs in direct response to the action potential. (B) Left: The amplitude of AMPAR-mediated EPSCs varies not only between paired recordings, but also from trial-to-trial within a paired recording. Each trial represents the amplitude of the EPSC measured at 0.1 Hz. In this example, AMPAR EPSC amplitude fluctuates from 5 pA to >60 pA. Right: NMDAR EPSC amplitudes measured at +40 mV in the presence of 10 μM CNQX. NMDAR EPSC amplitudes are typically significantly smaller in amplitude that AMPAR EPSCs, but still show significant variability. NMDAR EPSC amplitude typically averages between 10-20 pA in paired recordings between CA3 pyramidal cells, making failures readily identifiable. (C) Left: CA3 pyramidal cell pairs express LTP that persists for the length of the paired recording. LTP is readily evident by an increase in the amplitude of AMPAR EPSCs. Significant trial-to-trial variability in the AMPAR EPSC amplitude remains after the induction of LTP. Right: Expression of long-term depression (LTD) between CA3 pyramidal neurons. Note the decrease in average amplitude of the AMPAR EPSC and concomitant decrease in the trial to trial amplitude variability. (D) Example of postsynaptic polysynaptic inhibitory currents, which occur at a longer latency compared to the monosynaptic AMPAR EPSC. When the postsynaptic neuron is voltage clamped at -65 mV, the polysynaptic currents are visible as a second peak at a latency of >5 msec after the peak of the presynaptic action potential. Holding the postsynaptic neuron at -30 mV confirms that the polysynaptic current is inhibitory due to the reversal of the current direction. Please click here to view a larger version of this figure.
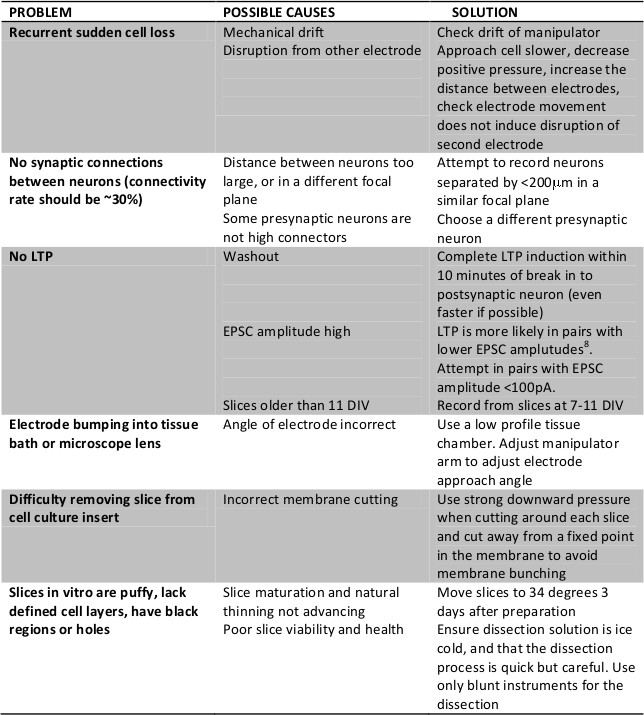
Table 1. Potential problems arising in paired recordings and preparation of organotypic slice cultures. This lists common potential problems that occur with paired whole cell recordings in organotypic slice cultures. In addition, we also advise that significant time is required to be spent ‘driving’ the electrophysiology setup for paired recordings, as most problems are related to movement disruptions that can readily be visualized and rectified.
Discussion
Here we have described the requirements for establishing successful paired whole cell recordings in organotypic hippocampal slice cultures. Paired recordings can also be performed in multiple preparations, including acute slices and dissociated culture systems26,27. While the focus here has been on the induction of longer forms of synaptic plasticity (namely LTP and LTD), it is important to highlight that paired whole cell recordings in organotypic, acute slice and dissociated cell preparations have provided important insights into quantal size, short-term plasticity, presynaptic function, as well as LTP and LTD2,4,8,26,27.
The major advantage of the organotypic slice system is the increased connectivity between neurons. Synaptic connectivity in the acute slice preparation is the low due to the severing of axonal connections during slice preparation. In our experience, paired recordings from synaptically connected CA3 neurons in acute slices were possible in only ~5% of paired recordings obtained. That is, 95% of paired recordings were unconnected. An alternative is to use primary dissociated hippocampal cultures where synaptic connectivity is significantly higher26. However, with dissociated cultured preparations come questions regarding neuron identity and whether their properties are similar to those of mature synapses in native tissue. The use of organotypic hippocampal slice cultures partially ameliorates these concerns because neuronal subtypes can readily by identified, and the cells maintain morphology and connectivity that are similar to native brain tissue20. However, as slices are maintained in vitro for over one week, it is important to note that significant axonal growth occurs during this time. The resulting increase in connectivity is greatly beneficial to performing paired recordings that examine synaptic function and plasticity, however it should be noted that studies examining network connectivity may not be suitable for this preparation.
Whilst the majority of our paired recordings have been conducted on CA3-CA3 pyramidal cell pairs, this technique can be readily adapted to CA3-CA1 and dentate gyrus-CA3 paired recordings. The incidence of synaptic connectivity between dentate gyrus and CA3 pyramidal neurons in this system is considerably lower. However, monosynaptic excitatory connections between CA3 and CA1 in this system have been reported to be as high as 76%28.
In addition, paired whole cell recordings are routinely performed in mouse hippocampal tissue, including transgenic mice (Figure 2D)29-31. The technique can be further refined to record synaptic transmission and plasticity between neurons with mosaic gene expression30. GFP labeling of wild-type neurons enables targeted paired whole cell recordings in neurons of known genotype. These experiments have identified alterations in synaptic connectivity between individual neurons, including hyperconnectivity or asymmetric presynaptic function30,31 that may contribute to behavioral deficits observed in these animals.
In summary, paired whole cell recordings increase the ability to interpret electrophysiological synaptic properties and to determine the subcellular mechanisms that neurons employ to change synaptic strength. Using this technique has allowed the predictions of pre- and postsynaptic models of synaptic plasticity to be directly tested. In addition, the phenotype of silent synapses, and specific roles of active-zone and PSD proteins have also been described in this preparation6-16. It has also enabled the discovery that synapses exist in distinct electrophysiologically-defined states, and that during synaptic plasticity synapses move between these states9,17.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We would like to thank the members of the Montgomery and Madison labs for helpful discussion. We acknowledge the funding received from the following sources in this research: NFNZ, AMRF, Marsden Fund, HRC, and NIH.
Materials
| Organotypic cultures | Paired recordings | ||
| Minimum Essential Medium | Stable motorized micromanipulators | ||
| Penicillin-Streptomycin solution | Shallow tissue bath | ||
| HEPES buffer solution | DIC camera | ||
| 1M Tris stock solution | Amplifier | ||
| Hank’s Balanced Salt Solution | Computer | ||
| Horse Serum | Vibration isolation table | ||
| plastic-coated miniature spatulas | Upright microscope | ||
| soft paintbrush | Data acquistion and analysis software | ||
| manual tissue chopper | Electrode puller | ||
| #2 filter paper | Faraday cage | ||
| #5 forceps | |||
| Membrane inserts | |||
| CO2 incubator | |||
| Dissection hood | |||
| Class II hood |
References
- Dingledine, R., Borges, K., Bowie, D., Traynelis, S. F. The glutamate receptor ion channels. Pharmacology Reviews. 51, 7-61 (1999).
- Malenka, R. C., Nicoll, R. A. Long-term potentiation – A decade of progress. Science. 285, 1870-1874 (1999).
- Bliss, T. V. P., Lomo, T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. Journal of Physiology. 232, 331-356 (1973).
- Dudek, S. M., Bear, M. F. Homosynaptic long-term depression in area CA1 of hippocampus and effects of N-methyl-D-aspartate receptor blockade. Proceedings of the National Academy of Sciences USA. 89, 4363-4367 (1992).
- Borst, J. G., Soria van Hoeve, J. The calyx of held synapse: from model synapse to auditory relay. Annual Reviews in Physiology. 74, 199-224 (2012).
- Pavlidis, P., Madison, D. V. Synaptic transmission in pair recordings from CA3 pyramidal cells in organotypic culture. Journal of Neurophysiology. 81, 2787-2797 (1999).
- Pavlidis, P., Montgomery, J. M., Madison, D. V. Presynaptic protein kinase activity supports long-term potentiation at synapses between individual hippocampal neurons. Journal of Neuroscience. 20 (12), 4497-4505 (2000).
- Montgomery, J. M., Pavlidis, P., Madison, D. V. Pair recordings reveal all-silent synaptic connections and the postsynaptic expression of long-term potentiation. Neuron. 29, 691-701 (2001).
- Montgomery, J. M., Madison, D. V. State-dependent heterogeneity in synaptic depression between pyramidal cell pairs. Neuron. 33, 765-777 (2002).
- Montgomery, J. M., Selcher, J. C., Hansen, J. E., Madison, D. V. Dynamin-dependent NMDAR endocytosis during LTD and its dependence on synaptic state. BMC Neuroscience. 6, 48 (2005).
- Waites, C. L., et al. Synaptic SAP97 isoforms regulate AMPA receptor dynamics and access to presynaptic glutamate. Journal of Neuroscience. 29 (14), 4332-4345 (2009).
- Emond, M., et al. AMPA receptor subunits define properties of state-dependent synaptic plasticity. Journal of Physiology. 588, 1929-1946 (2010).
- Li, D., et al. SAP97 directs NMDA receptor spine targeting and synaptic plasticity. Journal of Physiology. 589, 4491-4510 (2011).
- Genoux, D., Bezerra, P., Montgomery, J. M. Intra-spaced stimulation and protein phosphatase 1 dictate the direction of synaptic plasticity. European Journal of Neuroscience. 33 (10), 1761-1770 (2011).
- Selcher, J. C., Xu, W., Hanson, J. E., Malenka, R. C., Madison, D. V. Glutamate receptor subunit GluA1 is necessary for long-term potentiation and synapse unsilencing, but not long-term depression in mouse hippocampus. Brain Research. 1435, 8-14 (2012).
- Arons, M. H., et al. Autism-associated mutations in ProSAP2/Shank3 impair synaptic transmission and neurexin-neuroligin-mediated transsynaptic signaling. Journal of Neuroscience. 32 (43), 14966-14978 (2012).
- Montgomery, J. M., Madison, D. V. Discrete synaptic states define a major mechanism of synapse plasticity. Trends in Neuroscience. 27 (12), 744-750 (2004).
- Miles, R., Poncer, J. C. Pair recordings from neurones. Current Opinion in Neurobiology. 6 (3), 387-394 (1996).
- Malinow, R. Transmission between pairs of hippocampal slice neurons: quantal levels, oscillations and LTP. Science. 252 (5006), 722-724 (1991).
- Gähwiler, B. H., Capogna, M., Debanne, D., McKinney, R. A., Thompson, S. M. Organotypic slice cultures: a technique has come of age. Trends in Neuroscience. 20 (10), 471-477 (1997).
- Stoppini, L., Buchs, P. A., Muller, D. A simple method for organotypic cultures of nervous tissue. Journal of Neuroscience Methods. 37 (2), 173-182 (1991).
- Debanne, D., Guérineau, N. C., Gähwiler, B. H., Thompson, S. M. Paired-pulse facilitation and depression at unitary synapses in rat hippocampus: quantal fluctuation affects subsequent release. Journal of Physiology. 491, 163-176 (1996).
- Debanne, D., Gähwiler, B. H., Thompson, S. M. Cooperative interactions in the induction of long-term potentiation and depression of synaptic excitation between hippocampal CA3-CA1 cell pairs in vitro. Proceedings of the National Academy of Sciences U S A. 93 (20), 11225-11230 (1996).
- Malinow, R., Tsien, R. W. Presynaptic enhancement shown by whole cell recordings of long-term potentiation in hippocampal slices. Nature. 346 (6280), 177-180 (1990).
- DeBello, W. M., et al. SNAP-mediated protein-protein interactions essential for neurotransmitter release. Nature. 373 (6515), 626-630 (1995).
- Bekkers, J. M., Stevens, C. F. Origin of variability in quantal size in cultured hippocampal neurons and hippocampal slices. Proceedings of the National Academy of Sciences U S A. 87, 5359-5362 (1990).
- Malinow, R. Transmission between pairs of hippocampal slice neurons: quantal levels, oscillations, and LTP. Science. 252, 722-724 (1991).
- Debanne, D., Guérineau, N. C., Gähwiler, B. H., Thompson, S. M. Physiology and pharmacology of unitary synaptic connections between pairs of cells in areas CA3 and CA1 of rat hippocampal slice cultures. Journal of Neurophysiology. 73 (3), 1282-1294 (1995).
- Mitra, A., Blank, M., Madison, D. V. Developmentally altered inhibition in Ts65Dn, a mouse model of Down syndrome. Brain Research. 1440, 1-8 (2012).
- Hanson, J. E., Madison, D. V. Presynaptic FMR1 genotype influences the degree of synaptic connectivity in a mosaic mouse model of fragile X syndrome. Journal of Neuroscience. 27 (15), 4014-4018 (2007).
- Hanson, J. E., Blank, M., Valenzuela, R. A., Garner, C. C., Madison, D. V. The functional nature of synaptic circuitry is altered in area CA3 of the hippocampus in a mouse model of Down’s syndrome. Journal of Physiology. 579, 53-67 (2007).

