Transplantation of Chemogenetically Engineered Cortical Interneuron Progenitors into Early Postnatal Mouse Brains
Summary
Here we present a protocol, designed to use chemogenetic tools to manipulate the activity of cortical interneuron progenitors transplanted into the cortex of early postnatal mice.
Abstract
Neuronal development is regulated by a complex combination of environmental and genetic factors. Assessing the relative contribution of each component is a complicated task, which is particularly difficult in regards to the development of γ-aminobutyric acid (GABA)ergic cortical interneurons (CIs). CIs are the main inhibitory neurons in the cerebral cortex, and they play key roles in neuronal networks, by regulating both the activity of individual pyramidal neurons, as well as the oscillatory behavior of neuronal ensembles. They are generated in transient embryonic structures (medial and caudal ganglionic eminences - MGE and CGE) that are very difficult to efficiently target using in utero electroporation approaches. Interneuron progenitors migrate long distances during normal embryonic development, before they integrate in the cortical circuit. This remarkable ability to disperse and integrate into a developing network can be hijacked by transplanting embryonic interneuron precursors into early post-natal host cortices. Here, we present a protocol that allows genetic modification of embryonic interneuron progenitors using focal ex vivo electroporation. These engineered interneuron precursors are then transplanted into early post-natal host cortices, where they will mature into easily identifiable CIs. This protocol allows the use of multiple genetically encoded tools, or the ability to regulate the expression of specific genes in interneuron progenitors, in order to investigate the impact of either genetic or environmental variables on the maturation and integration of CIs.
Introduction
The function of neuronal networks relies in the existence of a balanced complement of excitatory projection neurons and inhibitory interneurons. Although cortical interneurons (CIs) only represent 20% of all neurons in the mammalian cortices, deficits in their number or function are thought to play a key part in the pathogenesis of neurodevelopmental disorders1,2. The study of CI development is challenging because CIs are generated in transient embryonic structures that are hard to access, and they follow a long tangential migration before they reach the pallium and develop their mature anatomical and physiological properties3. Both genetic and environmental mechanisms are known that regulate CI development4, but it has proven difficult to study the relative contribution of multiple factors.
Many insights in CI development were obtained using in vitro culture systems after isolation of progenitors from the ganglionic eminences5,6. One of the great advantages of these methods is the potential to label and genetically modify the isolated progenitors and follow their differentiation in detail, to detect cell-autonomous changes. However, these methods are unable to offer information regarding the interactions between developing interneurons and an active network. We have adapted these protocols, by transplantation of the modified precursors into early post-natal cortex. Interneuron progenitors isolated from embryonic ganglionic eminences are able to survive, disperse and integrate into the host network upon transplantation into the cortex7,8. This method has been used to reduce the severity of epileptic seizures in genetic mouse models, and has been proposed as a possible new therapy for different neurodevelopmental disorders9,10. A previous protocol describes a procedure to transduce these precursors with viral vectors prior to transplantations11. The protocol we describe here also allows the genetic modification of interneurons, but does not require the creation of a viral vector, requiring only plasmid DNA, which greatly increases its flexibility. Some studies reported success in using in utero electroporation to genetically modify interneuron progenitors in the caudal ganglionic eminences (CGE)12, but this method has proven very difficult to reproduce.
In the representative results section, we illustrate the use of this method to express designer receptors exclusively activated by designer drugs (DREADDs13) in the transplanted CIs, a method we used in a recent publication14. We expressed hM3D(Gq), an engineered receptor based on the human cholinergic receptor CHRM3, which does not affect neuronal function unless it binds its specific ligand clozapine-N-oxide (CNO). CNO administration selectively triggers activation of hM3D(Gq) expressing cells. We used this method to show that cell autonomous and transient depolarization is sufficient to prevent apoptosis of CIs during development14. Combined with different genetically encoded tools, this protocol has the potential to up- or down-regulate gene expression, and visualize or manipulate cell activity during different stages of interneuron differentiation.
Protocol
Animals were bred and housed in accordance with the United Kingdom Animals (Scientific Procedures) Act (1986).
NOTE: For the generation of the pCAGGs-hM3D(Gq)-IRES-RFP construct, a SalI-StuI fragment, containing the hM3D(Gq) sequence, has been isolated from plasmid 50463 (Addgene), and inserted into the expression vector pCAGGs-RFP (gift from F. Guillemot) digested with XhoI-EcoRV.
1. Preparation of Mouse Embryo Cortical Slices
- Sterilize lab equipment (e.g., stereo microscopes) and surfaces (e.g., benches) with an appropriate detergent solvent and 70% ethanol (EtOH) solution in water (H2O).
- Use dissection tools that have been autoclaved and keep them all the time in 100% EtOH.
- Prepare three 20 mL aliquots of 4% low-gelling temperature agarose in 1x phosphate buffer solution (PBS), in 50 mL tubes, and keep them at 55 °C.
- Sacrifice pregnant mice by cervical dislocation at 13.5 or 14.5 days of gestation (embryonic day E13.5-E14.5).
- With a pair of dissection scissors, open the abdomen of the pregnant mice, remove the uterine horns (with the embryos) and place them in ice-cold Krebs solution (Table 1), in a 90 mm Petri dish.
- With a pair of straight student fine forceps, open the amniotic sacs, remove the embryos and transfer them in fresh ice-cold Krebs solution, in a new 90 mm Petri dish. Dissect the mouse embryo brain (forebrain, midbrain, hindbrain) from the rest of the embryo body.
- Holding the dissected brains from the hindbrain, transfer them in ice-cold Krebs solution in a new 90 mm Petri dish, and keep them on ice.
- With a waterproof pen, draw a straight line at the outside surface, in the middle of the bottom part of six 35 mm Petri dishes.
- Place one 4% low-gelling agarose/PBS 20 mL aliquot at 37 °C for 5 min. Immediately afterwards poor 10 mL in two 35 mm Petri dishes and embed the dissected brains.
NOTE: The olfactory bulbs should face downwards (bottom of the Petri dish). Embed 3-4 brains per Petri dish, align them in the straight line previously drawn. Leave 3−5 mm space between each two brains. - Repeat steps 1.8 and 1.9 until all dissected brains have been embedded.
- Place the embedded brains at 4 °C, in order for the agarose to solidify, and subsequently carve the three brains into a single block of appropriate size and orientation.
NOTE: Leave approximately 3 mm around the edges of the brain samples. Change the orientation of the brains, with the olfactory bulbs on the top. - Glue the block on the surface of a microtome base and using a surgical blade cut all the way through the bottom of the block, between each two brains, in order to create 3 independent blocks (each mini block containing one brain).
- Section the blocks, in ice-cold Krebs solution, into 250 μm thick slices, using a vibrating blade microtome.
- With a bended flat-ended micro-spatula collect only the slices containing the medial or caudal ganglionic eminences (MGE or CGE; Figure 1) and individually transfer them onto filter membranes (13 μm diameter, 8.0 µm pore size), floating on minimal essential medium (MEM, Table 1) in polystyrene center-well organ culture dishes (60 mm x 15 mm).
- Place the dishes in a CO2 tissue culture incubator, at 37 °C, for 1 h, and prepare for the focal electroporation.
2. Acute Brain Slice Electroporation
- Before beginning the electroporation procedure, prepare 50 mL of 1% agarose gel in a 100 mm Petri dish. Leave the agarose gel to solidify at room temperature (RT) for approximately 30 min.
- Prepare tiny agarose columns (1 mm diameter and 10 mm length), punched with a glass pipette (225 mm length; 2 mL capacity) and transfer them in ice-cold Krebs solutions.
NOTE: A rubber dropper bulb suitable for 2 mL pipettes is attached to the pipette, and by pressing it the column can be released from the glass pipette to the Krebs solution. - With a surgical blade, cut small agarose blocks of two sizes: a small one that will fit on the surface of the electrode (see below) and a bigger one, which will be used as a base for performing the focal DNA injections into the brain slices. Transfer the agarose blocks in ice-cold Krebs solution as well.
- Prepare the set-up for the focal injections and the acute electroporation (Figure 2).
NOTE: For the injections, the following equipment is needed: 1) a bright field dissection stereo-microscope, 2) a pneumatic pico-pump injector, 3) a micromanipulator, 4) a magnetic stand, and 5) a steel base plate. For the acute slice electroporation, the following equipment is needed: 1) one square platinum 10 mm Petri dish electrode, 2) one square platinum 10 mm cover electrode, 3) a micromanipulator, and 4) an electroporator. - Focal DNA injections
- Prepare a DNA mixture of expression vectors: pCAGGs-IRES-GFP (control vector) + pCAGGs-hM3D(Gq)-IRES-RFP, at a concentration of 1 µg/µL for each vector and add fast green solution (stock 25 mg/mL) in a 1/10 dilution.
- Fill a pulled glass micropipette (0.5 mm inner diameter and 1 mm outer diameter) with 10 µL of the DNA mixture and inject small amounts (on the 25-50 nL range) into the selected region (MGE/CGE) of the slice (Figure 1 and Figure 2).
- Acute electroporation
NOTE: The electroporation should be performed immediately after the focal DNA injection.- Place the small agarose block on the Petri dish electrode and attach the agarose column to the mobile cover electrode with the help of a flat-ended micro-spatula.
- Transfer the slice with its supporting membrane onto the agarose block and place the top electrode with the agarose column on top of the selected region (MGE/CGE) of the slice.
NOTE: Charging voltages of 125 V (two pulses of 5 ms each, interval 500 ms) will yield a successful electroporation (Figure 3).
- After the electroporation, place the slice with its supporting membrane into the dish and transfer the dish in a CO2 tissue culture incubator, at 37 °C.
- After 1 h, exchange the MEM medium for a basic medium appropriate for primary neuronal cultures (neuron basic medium; Table 1) and incubate the slices overnight, for approximately 18-24 h.
3. Preparation of Cell-grafts
- Check the efficiency of the electroporation in all slices. Select only the slices where an acceptable number of fluorescent cells is observed (Figure 3).
NOTE: Electroporate approximately 30 slices/per experiment in order to acquire the appropriate number of cells for grafting (see below). - Dissect the selected region (MGE/CGE) from each slice and cut the tissue into small pieces in ice cold Krebs solution, under a fluorescent dissection stereo-microscope.
- Meanwhile, place a 1.5 mL tube with 900 µL neuron basic medium in a water bath at 37 °C.
- Transfer the tissue pieces with a P1000 micropipettor to a 1.5 mL tube containing 500 µL of L15/DNase medium prepared by adding 100 µL of 1 mg/mL DNase stock in DMEM/F12 medium into 900 µL of L15 medium. Wash the tissue pieces by tapping.
- Add 100 µL of DNase (stock 1 mg/mL in DMEM/F12 medium) into the 900 µL neuron basic medium.
- Discard the L15/DNase medium and resuspend the tissue pieces in 200 µL neuron basic/DNase medium prepared in step 3.5.
- Set a P200 micropipettor to 180 µL and mechanically dissociate the tissue pieces by pipetting up and down gently, 20-30 times, until a smooth and "creamy" suspension is obtained.
- Add 200 µL of neuron basic/DNase medium (final total volume 400 µL) and resuspend.
- Take a 4 µL aliquot of cells, dilute appropriately and mount on a haemocytometer.
- Under a bright field microscope, check the efficiency of the dissociation and count the number of cells.
NOTE: If the dissociation is successful, bright (alive) single cells and not cell-aggregates will be observed. - Centrifuge cell suspension from step 3.8, at 1,000 rpm, at RT, for 5 min, and subsequently, remove the supernatant from the tube and add the appropriate L15/DNase medium (usually 5-7 µL) so that the final concentration of cells will be 8 x 105-1.2 x 106 cells/µL.
NOTE: During the resuspension, it is extremely important to avoid air bubbles. - Place the cell aliquot on ice and have additional L15/DNase medium for the injections.
4. Intracranial Injections
NOTE: The following procedures take place in a procedure room within the Animal House Facility. Since the cells will be injected directly to the brain of newborn pups without exposing the brain, aseptic conditions are kept by sterilizing the working space with 70% EtOH solution and using autoclaved glass needles. The following equipment is needed for the intracranial injections: 1) a bright field dissection stereo-microscope, 2) a micro-injector, and 3) a heating pad for mouse recovery.
- Prepare a glass needle by pulling needles according to the manufacturer’s recommendations. Glass needles with an 80 µm outer diameter, 40 µm inner diameter, and a 30° bevel are used in this protocol.
NOTE: As mentioned above, needles should be autoclaved before use. - Backfill manually the needle with biologically inert oil using a 30 G, 2 inch needle and a syringe.
- Assemble and insert the needle to the injector unit according to the manufacturer's instructions.
- Determine the injection settings at maximum volume (69 nL) and a relative slow rate (23 nL/s).
- Empty the needle until the plunger is fully extended.
- Fill the needle.
- Cut a small piece from a grafting tape and place it under a bright field dissection stereo-microscope. With a P10 micropipette, transfer 5 µL from the sample (cell aliquot) onto the tape, so that a spherical drop is formed.
- Place the tip of the needle into the sample and fill the needle (the plunger retracts and draws the sample with it).
NOTE: Since the sample should be quite viscous, fill the needle in small steps so that the sample will equilibrate, and at a slow rate that prevents bubbles from forming. The sample should be smooth and homogeneous within the needle. If the sample is too viscous and it is not possible to fill the needle, add as much L15/DNase medium as it is required to obtain the correct viscosity. Nevertheless, this will change the concentration of the sample, and ideally should be avoided.
- Anaesthetize new born pups (postnatal day 0-2 [P0-P2]) on ice for 2-5 min.
NOTE: Make sure that the pup is not moving. - Place the anaesthetized pup under the bright field dissection stereo-microscope.
- Perform 3-4 injections of 69 nL each in each hemisphere.
NOTE: The injection sites are located approximately 1 mm lateral to the midline, and between 1 mm caudal to bregma and 1 mm rostral to the interaural line. The tip of the needle should be placed approximately 1 mm deep to the pial surface. After each injection, the needle is left in place for approximately 30 s and withdrawn in periods. - Immediately after the injections, place the pup on a heating pad with the heating at its lowest setting (37 °C). When the pup recovers transfer it to the cage with its mother.
NOTE: Never leave the mother with no pups in the cage. The whole procedure (from removing the pup from its littermates, until it is returned) should last less than 10 min.
5. Clozapine-N-oxide Injections
- Prepare the DREADD ligand, CNO stock solution by diluting 1 mg of CNO in 50 µL of dimethyl sulfoxide (DMSO) until the solution is translucent. Top-up to 10 mL with saline so that the final concentration of CNO is 0.1 mg/mL.
NOTE: DMSO is toxic. Avoid using it to a concentration higher than 0.1%. Use as a control a saline solution containing the same DMSO concentration as the CNO containing solution. - From postnatal day 14 and for 3 constitutively days (P14-P16), in each mouse perform two intraperitoneal (I.P.) injections of CNO (CNO concentration: 1 mg/kg) or 0.05% DMSO in saline, per day, 12 h apart.
- On the last day (P17), perform a single injection and sacrifice mice by cervical dislocation within a time window of 30 min-1 h.
Representative Results
Using the procedure presented here, we tested whether the survival of cortical interneurons during early postnatal stages is regulated by activity in a cell autonomous manner. We performed 3 brain slice electroporation experiments (12-16 embryos [E14.5 embryos] per experiment) with the pCAGGs-IRES-GFP (control) and pCAGGs-hM3D(Gq)-IRES-RFP expression vectors, at a concentration of 1 µg/µL for each construct. In our electroporation experiments, only a fraction (approximately 50%; Figure 3) of the GFP+ neurons co-expressed hM3D(Gq) (RFP+) and therefore the GFP+RFP– population served as an internal control for the effect of DREADD ligands. Transfected cortical embryonic interneurons were mechanically dissociated and the resulting cell suspension (8 x 105 cells/µL) grafted in the cortex of P0-P1 wild type mice. We had performed 6 injections per brain. In each experiment, a minimum of 6 new born pups were injected. Administration of CNO selectively increased the activity of transfected RFP+ cells, as demonstrated by the expression of the activity-dependent protein cFos (Figure 4). CNO treatment according to the described protocol (administer twice daily P14-P17) resulted in an increase in the proportion of GFP+RFP+ relative to GFP+RFP– cells, in comparison to vehicle (0.5% DMSO in saline) administered littermates (Figure 5).
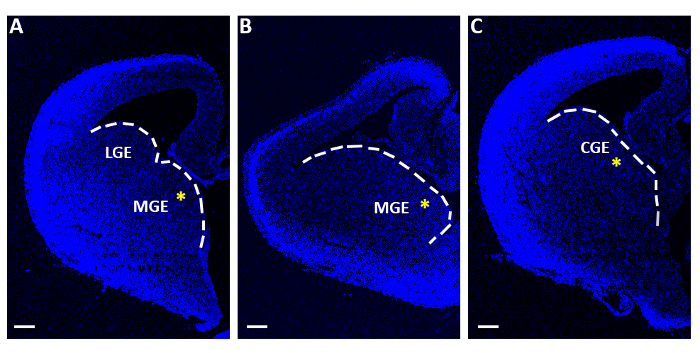
Figure 1: Representative telencephalic slices used for acute electroporation experiments. (A-C) Telencephalic slices obtained at three distinct sequential rostro-caudal levels, stained with 4′,6-diamidino-2-phenylindole (DAPI). LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence; CGE: caudal ganglionic eminence. Scale bars = 200 µm. The yellow asterisks indicate the electroporation site in each slice. The white line marks the edge of the ganglionic eminence. Please click here to view a larger version of this figure.
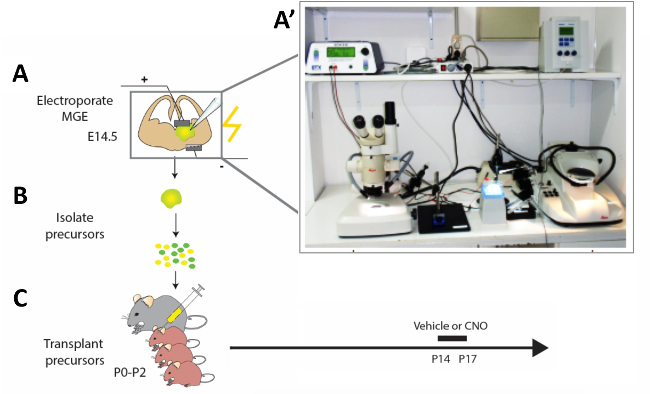
Figure 2: Schematic representation of the experimental workflow. (A) Mouse brain slices are electroporated with appropriate constructs, and (B) after 12 h modified cortical interneuron (CI) precursors are isolated and (C) transplanted in the pallium of newborn mouse pups (P0−P2). In order to modify the activity of immature CIs, P14 pups that had received cell transplantations were injected with CNO or vehicle for four constitutive days according to the presented protocol. (A') Photograph of the acute mouse brain slice electroporation set-up. Please click here to view a larger version of this figure.
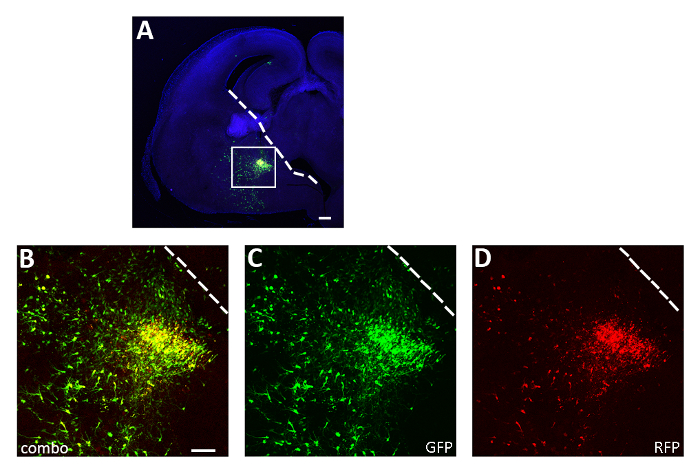
Figure 3: Representative successful acute slice electroporation experiment. (A) Representative coronal section from an E14.5 embryo brain transfected in the CGE with both pCAGGs-IRES-GFP (GFP) and pCAGGs-hM3D(Gq)-IRES-RFP (RFP) plasmids and cultured for 12 h. The section has been immunostained for GFP (A, B, C) and RFP (A, B, D). The boxed area in panel A is magnified to show the expression of both fluorescent reporters (B), GFP (C) and RFP only (D). The white line marks the edge of the ganglionic eminence. B-D: same photo, different channels or combination of the two different channels. Scale bars = 200 µm (A), 100 µm (B-D). This figure has been modified from Denaxa et al.14. Please click here to view a larger version of this figure.
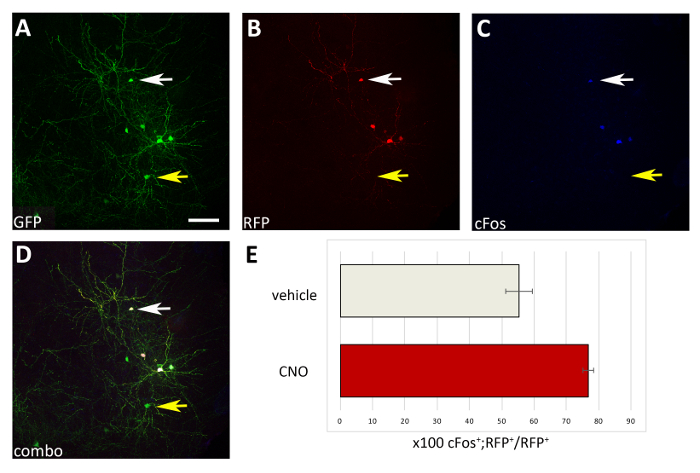
Figure 4: Cell autonomous increase in the activity of M3D(Gq)-expressing transplanted CIs upon CNO administration. (A-D) Representative confocal images of a coronal section of a P17 mouse transplanted at P1 with CI precursors transfected with both pCAGGs-IRES-GFP (GFP) and pCAGGs-hM3D(Gq)-IRES-RFP (RFP) plasmids and treated with CNO. The section has been immunostained for GFP (A), RFP (B), and cFos (C). (D) The combined image of A, B and C immunofluorescence (combo). Note that only CIs co-expressing both plasmids (white arrows in A-D) are also cFos+ compared to CIs expressing only the control-GFP plasmid (yellow arrows in A-D). (E) Quantification of cFos+RFP+ cells found in the pallium of P17 mice transplanted at P1 (normalized to the total RFP+ population) and treated with vehicle or CNO (N = 2). A-D: same photo, different channels, or combination of the three different channels. Scale bars = 50 µm. Please click here to view a larger version of this figure.
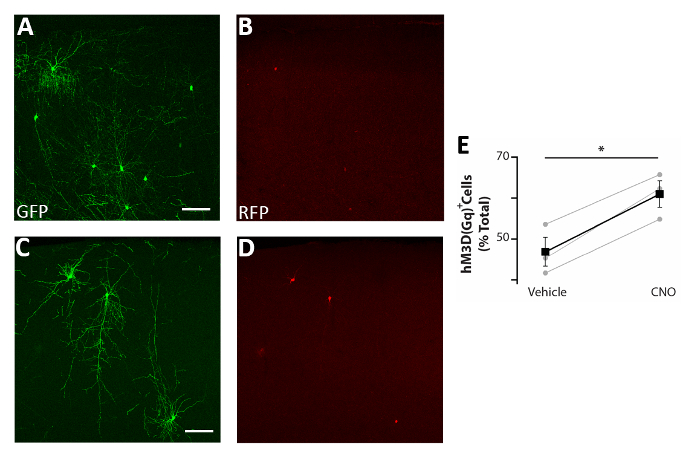
Figure 5: Cell autonomous increase in the activity of CIs enhances survival. (A-D) Representative confocal images of somatosensory cortex coronal slices of P17 mice transplanted at P0-P2 with CI precursors transfected with both pCAGGs-IRES-GFP (GFP) and pCAGGs-hM3D(Gq)-IRES-RFP (RFP) plasmids and treated with vehicle (A-B) or CNO (C-D). (E) Quantification of RFP+ cells found in the forebrain of P17 mice transplanted at P0-P2 (normalized to the total GFP+ population). RFP+(vehicle) = 47% ± 3%, CNO = 61% ± 3%, p = 0.01, Student’s paired sample t test, n = 3 vehicle and 3 CNO, a minimum of 150 cells counted per brain. A and B: same photo, different channels. C and D: same photo, different channels. Scale bars = 50 µm. This figure has been modified from Denaxa et al.14. Please click here to view a larger version of this figure.
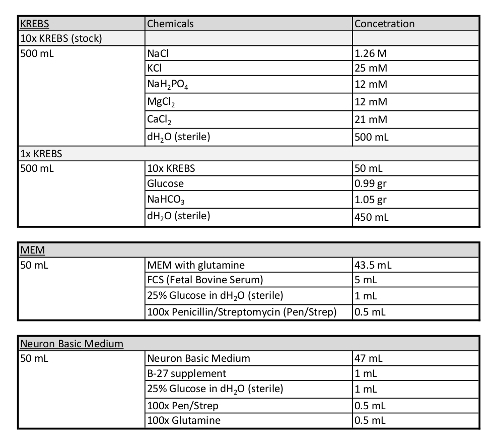
Table 1: Additional information concerning media used in this protocol.
Discussion
Here we describe a widely accessible methodology to genetically modify the activity of CI precursors to study the impact of intrinsic activity on CI maturation, and/or the effect of activity modulated CIs on the assembly/function of the integrated cortical circuits.
In the past, several labs, including ours, had performed in utero electroporation experiments in order to genetically modify projection neurons6. However, in utero electroporation into ganglionic eminences that include CI progenitors is very difficult, due to electrical conduction path problems. In order to solve this problem, a small number of labs are performing ultrasound guided injections followed by electroporation, which is a demanding technique that requires expensive equipment. This protocol provides an alternative to these methodologies which is accessible to the majority of the scientific community.
One of the most challenging aspect of this protocol is to maximize the number of cells that survive in the host cortex to mature stages, when phenotypic analysis is usually performed (very dependent on the experiment design, but typically older than P17). There are three key steps that the investigator should pay attention: (1) The efficiency of the electroporation. This can be maximized by ensuring the purity of DNA plasmids. Only high-quality DNA plasmids (an A260/A280 ratio of 1.9-2.0) should be used for this procedure. We obtain such high-quality DNA preparations by employing cesium chloride DNA purification. Another crucial factor is the promoter that drives the expression of the gene of interest. We found that the pCAGGs vector, which consists of the chicken b-actin promoter, is extremely powerful and can dramatically increase the electroporation efficiency. (2) The number of starting donor embryos. It is important to make sure that a large number (12-16) of embryos of the same stage are electroporated. This number can be increased, if more experimenters are performing embryo dissections and sectioning together, as it important that embryonic cortical slices are obtained, electroporated and transferred to the incubator as soon as possible. (3) It is important to make sure that a great number of cells are injected in each pup to ensure a high chance of transplanted cell survival until mature stages. In addition, this will dramatically improve the likelihood of successful transplants since low density cell preparations will result in uneven mixing of the cells with the medium, which will produce significant variability in the transplanted brains15.
The protocol described here was tailored for investigating the role of activity in regulating CI survival in a cell-autonomous manner. The P14-P17 time window for performing the CNO injections was specifically chosen according to published data, which show that the peak of transplanted CI progenitors' cell death occurs during this period16. Therefore, this time frame or the frequency of CNO injections might not hold true for other cell types or brain regions, and the investigator should adjust these parameters according to the specific experimental purposes. Finally, the methodology described here for the intracranial injections of CIs is only feasible for P0-P5 pups (depending also on the mouse line background). In principle, any injections over P5 will require thinning or removing of the skull15.
One of the key advantages of this protocol is the ability to use new genetically encoded tools to visualize or manipulate the activity of CIs during different stages of differentiation as they integrate into a developing network. With the pace of discovery of new genetically encoded voltage and calcium sensors, as well as new chemogenetic and optogenetic tools, this protocol allows researchers to use them within weeks of release into plasmid repositories, such as Addgene.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by an ERC Starter Grant (282047), a Wellcome Trust Investigator Award (095589/Z/11/Z), an FP7 EC DESIRE grant, and a Lister Institute Prize to JB. Work in V.P.'s laboratory is supported by the BBSRC (BB/L022974/1), the UK Medical Research Council (MRC), and the Francis Crick Institute (which receives funding from the MRC, Cancer Research UK, and the Wellcome Trust). The research in M.D. lab was made possible through the grant from the Stavros Niarchos Foundation to the B.S.R.C. "Alexander Fleming", as part of the Foundation’s initiative to support the Greek research.
Materials
| Medium/Supplements | |||
| B-27 | GIBCO (ThermoFisher Scientific) | 175040-044 | |
| DMEM/F12 | GIBCO (ThermoFisher Scientific) | 21331-020 | |
| DNAse | SIGMA | DN15-100MG | |
| FBS | GIBCO (ThermoFisher Scientific) | 10270-098 | |
| 100x Glutamine | GIBCO (ThermoFisher Scientific) | 35050-061 | |
| L15 | GIBCO (ThermoFisher Scientific) | 11415-049 | |
| MEM alpha, GlutaMAX | GIBCO (ThermoFisher Scientific) | 32561-029 | |
| Neurobasal medium | GIBCO (ThermoFisher Scientific) | 21103-049 | Neuron basic medium |
| 100x P/S | GIBCO (ThermoFisher Scientific) | 15140-122 | |
| Equipment | |||
| Electroporator | BTX | ECM 830 generator | |
| Injector for acute slice electroporation | Eppendorf | FemtoJet Microinjector | |
| Injector for cell transplantation (I) | Visual Sonics | Vevo Injector System | |
| Injector for cell transplantation (II) | WPI | NANOLITER2010 | |
| Magnetic Stand | WPI | M10L Magnetic Stand | |
| Kite Manual Micromanipulator | WPI | KITE-M3-R | |
| Platinum Elecrode (I) | Protech International Inc. | CUY-700-1 | |
| Platinum Elecrode (II) | Protech International Inc. | CUY-700-2 | |
| Steel Base Plate | WPI | 5479 | |
| Vibratome | Leica | VT1200S | |
| Other Material | |||
| Glass capillaries for electroporation | VWR | 1B100-4 | |
| Glass capillaries for cell transplantation | Visual Sonics | provided by Visual Sonics | |
| Nuclepore 8 µm whatman membrane | SLS | 110414 | |
| Organ tissue culture dishes | BD Biosciences (Falcon) | 353037 |
References
- Marin, O. Interneuron dysfunction in psychiatric disorders. Nature Review Neuroscience. 13 (2), 107-120 (2012).
- Glausier, J. R., Lewis, D. A. GABA and schizophrenia: Where we stand and where we need to go. Schizophrenia Research. 181, 2-3 (2017).
- Fishell, G., Rudy, B. Mechanisms of inhibition within the telencephalon: “where the wild things are”. Annual Review Neuroscience. 34, 535-567 (2011).
- Wamsley, B., Fishell, G. Genetic and activity-dependent mechanisms underlying interneuron diversity. Nature Review Neuroscience. 18 (5), 299-309 (2017).
- Xu, Q., Cobos, I., De La Cruz, E., Rubenstein, J. L., Anderson, S. A. Origins of cortical interneuron subtypes. Journal of Neuroscience. 24 (11), 2612-2622 (2004).
- Denaxa, M., et al. Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Reports. 2 (5), 1351-1362 (2012).
- Wichterle, H., Garcia-Verdugo, J. M., Herrera, D. G., Alvarez-Buylla, A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nature Neuroscience. 2 (5), 461-466 (1999).
- Alvarez-Dolado, M., et al. Cortical inhibition modified by embryonic neural precursors grafted into the postnatal brain. Journal of Neuroscience. 26 (28), 7380-7389 (2006).
- Baraban, S. C., et al. Reduction of seizures by transplantation of cortical GABAergic interneuron precursors into Kv1.1 mutant mice. Proccedings of the National Academy of Sciences of the United States of America. 106 (36), 15472-15477 (2009).
- Southwell, D. G., et al. Interneurons from embryonic development to cell-based therapy. Science. 344 (6180), 1240622 (2014).
- Vogt, D., et al. Viral-mediated Labeling and Transplantation of Medial Ganglionic Eminence (MGE) Cells for In Vivo Studies. Journal of Visualized Experiments. (98), e52740 (2015).
- De Marco Garcia, N. V., Karayannis, T., Fishell, G. Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature. 472 (7343), 351-355 (2011).
- Urban, D. J., Roth, B. L. DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annual Review of Pharmacology and Toxicology. 55, 399-417 (2015).
- Denaxa, M., et al. Modulation of Apoptosis Controls Inhibitory Interneuron Number in the Cortex. Cell Reports. 22 (7), 1710-1721 (2018).
- Quatrocolo, G., et al. Homochronic Transplatation of Interneuron Precursors into Early Postnatal Mouse Brains. Journal of Visualized Experiments. (136), e57723 (2018).
- Southwell, D. G., et al. Intrinsically determined cell death of developning cortical interneurons. Nature. 491 (7422), 103-113 (2012).

