Transcriptome-Wide Profiling of Protein-RNA Interactions by Cross-Linking and Immunoprecipitation Mediated by FLAG-Biotin Tandem Purification
Summary
Here we present a modified CLIP-seq protocol called FbioCLIP-seq with FLAG-biotin tandem purification to determine the RNA targets of RNA-binding proteins (RBPs) in mammalian cells.
Abstract
RNA and RNA-binding proteins (RBPs) control multiple biological processes. The spatial and temporal arrangement of RNAs and RBPs underlies the delicate regulation of these processes. A strategy called CLIP-seq (cross-linking and immunoprecipitation) has been developed to capture endogenous protein-RNA interactions with UV cross-linking followed by immunoprecipitation. Despite the wide use of conventional CLIP-seq method in RBP study, the CLIP method is limited by the availability of high-quality antibodies, potential contaminants from the copurified RBPs, requirement of isotope manipulation, and potential loss of information during a tedious experimental procedure. Here we describe a modified CLIP-seq method called FbioCLIP-seq using the FLAG-biotin tag tandem purification. Through tandem purification and stringent wash conditions, almost all the interacting RNA-binding proteins are removed. Thus, the RNAs interacting indirectly mediated by these copurified RBPs are also reduced. Our FbioCLIP-seq method allows efficient detection of direct protein-bound RNAs without SDS-PAGE and membrane transfer procedures in an isotope-free and protein-specific antibody-free manner.
Introduction
RNAs and RNA-binding proteins (RBPs) control diverse cellular processes including splicing, translation, ribosome biogenesis, epigenetic regulation, and cell fate transition1,2,3,4,5,6. The delicate mechanisms of these processes depend on the unique spatial and temporal arrangement of RNAs and RBPs. Therefore, an important step towards understanding RNA regulation at the molecular level is to reveal the positional information about the binding sites of RBPs.
A strategy referred to as cross-linking and immunoprecipitation (CLIP-seq) has been developed to capture protein-RNA interactions with UV cross-linking followed by immunoprecipitation of the protein of interest7. The key feature of the methodology is the induction of covalent cross-links between an RNA-binding protein and its directly bound RNA molecules (within ~1 Å) by UV irradiation8. The RBP footprints can be determined by CLIP tag clustering and peak calling, which usually have a resolution of 30−60 nt. Alternatively, the reverse transcription step of CLIP can lead to indels (insertions or deletions) or substitutions to the cross-linking sites, which allows identification of protein binding sites on the RNAs at a single-nucleotide resolution. Pipelines like Novoalign and CIMS have been developed for the analysis of the high-throughput sequencing results of CLIP-seq8. Several modified CLIP-seq methods have also been proposed, including individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP), enhanced CLIP (eCLIP), irCLIP, and photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP)9,10,11,12.
Despite the wide use of traditional CLIP-seq methods in the study of RBPs, the CLIP methods have several drawbacks. First, the tedious denatured gel electrophoresis and membrane transfer procedure may lead to loss of information, and cause limited sequence complexity. Second, the protein specific antibody-based CLIP method may pull down a protein complex instead of a single target protein, which may lead to false positive protein-RNA interactions from the copurified RBPs. Third, the antibody-based strategy requires a large amount of high-quality antibodies, which makes the application of these methods inadequate for the study of RBPs without high-quality antibodies available. Fourth, the traditional CLIP method requires radiolabeled ATP to label the protein-bound RNAs.
The high affinity of streptavidin to biotinylated proteins makes it a very powerful approach to purify specific proteins or protein complexes. The efficient biotinylation of proteins carrying an artificial peptide sequence by ectopically expressed bacterial BirA biotin ligase in mammalian cells makes it an efficient strategy to perform biotin purification in vivo13. We developed a modified CLIP-seq method called FbioCLIP-seq (FLAG-Biotin-mediated Cross-linking and Immunoprecipitation followed by high-throughput sequencing) using FLAG-biotin tag tandem purification14 (Figure 1). Through tandem purification and stringent wash conditions, almost all the interacting RBPs are removed (Figure 2). The stringent wash conditions also allow circumventing the SDS-PAGE and membrane transfer, which is labor intensive and technically challenging. And similar to eCLIP and irCLIP, the FbioCLIP-seq method is isotope-free. Skipping the gel running and transfer steps avoids the loss of information, keeps authentic protein-RNA interactions intact, and increases the library complexity. Moreover, the high efficiency of the tagging system makes it a good choice for RBPs without high-quality antibodies available.
Here we provide a step-by-step description of the FbioCLIP-seq protocol for mammalian cells. Briefly, cells are cross-linked by 254 nm UV, followed by cell lysis and FLAG immunoprecipitation (FLAG-IP). Next, the protein-RNA complexes are further purified by biotin affinity capture and RNAs are fragmented by partial digestion with MNase. Then, the protein-bound RNA is dephosphorylated and ligated with a 3’ linker. A 5’ RNA linker is added after the RNA is phosphorylated with PNK and eluted by proteinase K digestion. After reverse transcription, the protein-bound RNA signals are amplified by PCR and purified by agarose gel purification. Two RBPs were chosen to exemplify the FbioCLIP-seq result. LIN28 is a well-characterized RNA-binding protein involved in microRNA maturation, protein translation, and cell reprogramming15,16,17. WDR43 is a WD40 domain-containing protein thought to coordinate ribosome biogenesis, eukaryotic transcription, and embryonic stem cell pluripotency control14,18. Consistent with previously reported results for LIN28 with CLIP-seq, FbioCLIP-seq reveals binding sites of LIN28 on “GGAG” motifs in the microRNA mir-let7g and mRNAs16,19 (Figure 3). WDR43 FbioCLIP-seq also identified the binding preference of WDR43 with 5' external transcribed spacers (5'-ETS) of pre-rRNAs20 (Figure 4). These results validate the reliability of the FbioCLIP-seq method.
Protocol
1. Cell line construction
- Clone the gene of interest into a PiggyBac vector pPiggyBac-FLAG-bio-[cDNA of interest]-(Hygromycin-resistant) plasmid that carries a FLAG-biotin epitope13,21 to express a FLAG-biotin tag fused RBP (FBRBP).
- Cotransfect the FBRBP expressing vector with the pBase vector into a cell line expressing the BirA enzyme22.
NOTE: In this study, the FBRBP gene was transfected into mouse embryonic stem cells (mESCs) carrying an integrated BirA expression vector21. Alternatively, the FBRBP expressing vector can be cotransfected into cells with pBase and pPiggyBac-BirA-V5 together. - Grow the cells in mESC on gelatinized dishes and maintain in mES culture medium (Table of Materials) in 5% CO2 at 37 °C. Select the transfected cells with hygromycin b (100 µg/mL) for 1 week.
- After drug selection, harvest cells by SDS loading buffer (Table 1) and use a Western blot23 to confirm the tagging and expression of the gene with a FLAG antibody and streptavidin-HRP, respectively.
2. Cross-linking
- Plate ~3 x 106 mES cells in a 10 cm plate 1 day before the experiment so that the cells will grow to 70−90% confluence when harvested (~5−10 million cells per sample).
- Remove the medium with a vacuum. Treat the cells with 2 mL of 0.25% trypsin-EDTA for 2 min at room temperature (RT) and quench the trypsin by 4 mL of fresh mESC medium. Transfer the cells to a 15 mL centrifuge tube. Spin down the cells by centrifugation at 300 x g for 3 min at 4 °C and remove the supernatant.
- Suspend the cells with 4 mL of cold PBS and plate the cells back to the 10 cm plate for cross-linking. Cross-link the cells by radiation with 254 nm UV light (set the UV cross-linker with a parameter of 400 mJ/cm2).
NOTE: For cell monolayers, the cells can be cross-linked with UV light directly on the plate before trypsin treatment. - Transfer the cross-linked cells to a 15 mL tube. Spin down by centrifugation at 300 x g for 3 min at 4 °C and remove the supernatant.
NOTE: The cross-linked cell pellet can be stored at -80 °C until use.
3. Cell lysate preparation
- Lyse the cells with 500 µL of wash buffer A (Table 1) freshly supplemented with 1 mM DTT, 1 mM PMSF, 1/500 protease inhibitor cocktail, and 400 U/mL RNase inhibitor. Transfer the cells to an RNase-free 1.5 mL tube. Incubate the cells for 30−60 min on a rotor with gentle rotation at 4 °C.
- Treat the lysate with 30 µL of DNase I at 37 °C for 10 min. Stop the reaction by adding 4 µL of 0.5 M EDTA. Spin down the insoluble pellet by 12,000 x g for 20 min at 4 °C and transfer the supernatant to a new prechilled 1.5 mL tube.
NOTE: For immediate use, keep on ice. If the supernatant will not be used immediately, store it at -80 °C until use.
4. FLAG beads preparation
- Add 40 µL of slurry FLAG beads per sample to a fresh 1.5 mL tube.
- Quickly wash the beads with 0.5 mL of ice-cold wash buffer A and spin down by 3,000 x g for 2 min at 4 °C.
- Repeat step 4.2 once. Spin down the beads with 3,000 x g for 2 min at 4 °C. Carefully remove the buffer with a narrow-end pipette tip. Avoid removing the beads.
5. Immunoprecipitation
- Transfer the cell lysate from step 3.2 to the pre-equilibrated FLAG beads. Rotate all the samples on a roller shaker at 4 °C gently. Incubate the FLAG beads with lysate for 2−4 h or overnight.
- Centrifuge the beads for 2 min at 3,000 x g and remove the supernatant.
- Wash the beads with 0.5 mL of prechilled wash buffer A by incubation for 5 min in a rotator at 4 °C, spin down the beads with 3,000 x g for 2 min and remove the supernatant. Repeat the wash 1x.
- Repeat the wash 2x as described in step 5.3 with 0.5 mL of prechilled wash buffer B (Table 1).
6. Elution with 3x FLAG peptide
- Prepare 3x FLAG elution solution by dissolving 1 mg of 3x FLAG peptide with 5 mL of wash buffer A to a final concentration of 200 ng/mL.
- Spin down the FLAG beads from step 5.4 with 3,000 x g for 2 min. Remove the supernatant carefully. Make sure most of the supernatant is removed by using a narrow end pipette tip.
- Add 200 µL of 3x FLAG elution solution to each sample. Incubate the samples with gentle rotation for 30 min at 4 °C. Spin down the FLAG beads for 2 min at 3,000 x g. Transfer the supernatants to fresh tubes.
- Repeat the elution 2x as described in steps 6.2 and 6.3 and pool the eluents together. Save 5% of the eluents for Western blot analysis or silver staining to check the efficiency of FLAG-IP.
7. Streptavidin beads preparation
- Prepare 50 µL of a streptavidin bead slurry for each sample. Collect the beads with a magnetic stand for 30 s and remove the supernatant with a pipette tip.
- Quickly wash the beads with 0.5 mL of ice-cold wash buffer A and collect the beads with a magnetic stand for 30 s.
- Repeat the wash 1x. Remove the supernatant after the wash.
8. Biotin affinity purification
- Transfer the pooled eluents from step 6.4 to the streptavidin beads from step 7.3 and gently rotate the samples at 4 °C for 1−3 h or overnight.
- Collect the beads with a magnetic stand for 30 s and remove the supernatant. Wash the beads 2x with 500 µL of wash buffer C (Table 1) by gentle rotation at RT for 5 min.
- Collect the beads with the magnetic stand and remove the supernatant. Wash the beads 2x with 500 µL of wash buffer D (Table 1) by rotating at RT for 5 min.
- Collect the beads with the magnetic stand and remove the supernatant. Quickly wash the beads 2x with 500 µL of ice-cold PNK buffer (Table 1).
9. Partial RNA digestion
- Make a 105-fold dilution of MNase with MNase reaction buffer (Table 1). Add 100 µL of MNase solution to each sample. Vortex the beads at 37 °C with a thermal mixer set at 1,200 rpm (5 s run, 30 s stop) for 10 min, collect the beads with a magnetic stand for 30 s and remove the supernatant.
NOTE: The concentration of MNase should be optimized for different RNA-binding proteins. The MNase concentration that can digest RNAs into 30−50 nt fragments (determined by the size of insert of the final library) is optimal. - Quickly wash the beads 2x with 500 µL of ice-cold PNK+EGTA buffer (Table 1). Collect the beads with a magnetic stand and remove the supernatant between each wash step.
- Wash the beads 2x with 500 µL of wash buffer C by gentle rotation for 5 min at RT. Then quickly wash the beads 2x with 500 µL of ice-cold PNK buffer.
10. Dephosphorylation of RNA
- Make a calf intestine phosphatase (CIP) reaction mix with 8 µL of 10x CIP buffer (Table of Materials), 3 µL of CIP enzyme, and 69 µL of water. The total volume is 80 µL per reaction.
- Collect the beads with a magnetic stand and remove the buffer. Add the CIP reaction mix to the beads. Vortex the beads at 37 °C with a thermal mixer set at 1,200 rpm (5 s run, 30 s stop) for 10 min.
- Quickly wash the beads 2x with 500 µL of ice-cold PNK+EGTA buffer. Quickly wash the beads 2x with 500 µL of ice-cold PNK buffer.
11. 3’ linker ligation
- Make a 3’ linker mix with 4 µL of 20 µM 3’ linker, 4 µL of 10x T4 RNA ligase buffer, 4 µL of 50% PEG8000, 2 µL of T4 RNA ligase 2 (truncated), and 26 µL of water. The total volume is 40 µL per reaction.
- Collect the beads from step 10.3 with a magnetic stand and remove the buffer. Add 40 µL of the 3’ linker ligation mix to the beads. Vortex the beads at 16 °C with a thermal mixer set at 1,200 rpm (5 s run, 30 s stop) for 3 h or overnight.
- Quickly wash the beads 2x with 500 µL of ice-cold PNK+EGTA buffer. Quickly wash the beads 2x with 500 µL of ice-cold PNK buffer.
12. PNK treatment
- Make a PNK mix with 4 µL of 10x PNK buffer, 2 µL of T4 PNK enzyme, 1 µL of 10 mM ATP, and 33 µL of water. The total volume is 40 µL per reaction.
- Collect the beads with a magnetic stand and remove the buffer. Add 40 µL of PNK mix to the beads. Vortex the beads gently at 37 °C with a thermal mixer set at 1,200 rpm (5 s run, 30 s stop) for 10 min.
- Quickly wash the beads with 500 µL of ice-cold PNK+EGTA buffer.
- Quickly wash the beads with 500 µL of ice-cold PNK buffer. Save 5% of the beads for Western or silver staining analysis to confirm the efficiency of immunoprecipitation.
13. RNA isolation
- Collect the beads from step 12.4 with a magnetic stand and remove the buffer. Add 200 µL of proteinase K digestion buffer (Table 1). Vortex the beads for 30 min at 37 °C with a thermal mixer set at 1,200 rpm (5 s run, 30 s stop). Magnetically isolate the beads and take the supernatant for the experiment.
- Add 400 µL of RNA isolation reagent (Table of Materials) to the supernatant from step 13.1, mix thoroughly, and incubate on ice for 5 min. Add 80 µL of chloroform and mix thoroughly and then incubate on ice for 5 min. Spin down with 12,000 x g for 10 min 4 °C.
NOTE: This step should be performed in a chemical hood. - Take the supernatant (~500 µL) and add two volumes of isopropanol:ethanol mix (1:1), 1/10 volume of 3 M sodium acetate (pH = 5.5), and 1 µL of glycogen. Freeze at -20 °C for 2 h. Centrifuge at 4 °C with 12,000 x g for 20 min.
- Wash the pellet 2x with ice-cold 70% ethanol. Spin down the pellet at 4 °C with 12,000 x g for 5 min. Remove the supernatant and dry the pellet. Dissolve the RNAs in 5 µL of RNase-free H2O.
14. 5’ RNA linker ligation
- Make a 5’ RNA ligation mix with 1 µL of 10x T4 RNA ligase buffer, 0.5 µL of BSA (1 µg/µL), 1 µL of 10 mM ATP, 1 µL of RNase inhibitor, and 0.5 µL of T4 RNA ligase. The total volume is 4 µL per reaction.
- Add 1 µL of 5’ 20 µM RNA linker to the RNA from step 13.4, denature the RNA by heating at 70 °C for 2 min, and chill on ice immediately.
- Add the 5’ RNA ligation mix to the RNA from step 14.2. Incubate at 16 °C overnight.
15. Reverse transcription
- Purify the RNAs as described in section 13 and dissolve the RNA with 11.5 µL of RNase-free water.
- Add 1 µL of 10 µM reverse transcription primer to the purified RNA, heat at 70 °C for 2 min, and chill on ice immediately.
- Make a reverse transcription mix with 4 µL of 5x reverse transcription buffer, 1 µL of 10 mM dNTPs, 1 µL of 0.1 M DTT, 1 µL of RNase inhibitor, and 0.5 µL of reverse transcriptase (Table of Materials). The total volume is 7.5 µL per reaction.
- Add 7.5 µL of reverse transcription mix to the RNA, incubate at 50 °C for 30 min, and stop the reaction by incubation at 70 °C for 10 min. Leave at 4 °C.
16. PCR amplification
- Make a PCR amplification mix with 15 µL of 2x PCR master mix (Table of Materials), 5 µL of cDNA from step 15.4, 0.6 µL of 10 µM forward primer 1 (Table 2), 0.6 µL of 10 µM reverse primer 1 (Table 2), and 8.8 µL of H2O. The total volume is 30 µL.
- Amplify the cDNA with the following settings: 98 °C 30 s, 98 °C 10 s, 60 °C 30 s, 72 °C 30 s (repeat steps 2−4 for 15−20 cycles), 72 °C 2 min, and hold at 4 °C. Run the PCR product on a 2−3% agarose gel. Cut out the DNA of ~100−150 bp, extract DNA with gel extraction kit (Table of Materials), and elute the DNA with 20 µL of water.
- Take 3 µL of the purified PCR product, amplify with forward primer 2 (Table 2) and reverse primer 2 (index primer; Table 2) as described in step 16.2 for five cycles to introduce index sequences. Purify the PCR product as described in step 16.2.
- Sequence the library with a high-throughput sequencing platform.
17. Bioinformatics analysis
- Perform data analysis using commercial programs as previously described8.
Representative Results
The schematic representation of the FbioCLIP-seq procedure is shown in Figure 1. Compared with FLAG-mediated or streptavidin-mediated one-step affinity purification, FLAG-biotin tandem purification removed almost all the copurified proteins, avoiding the contamination of indirect protein-RNA interactions (Figure 2). Representative results for FbioCLIP-seq for LIN28 and WDR43 are depicted in Figure 3 and Figure 4. We performed LIN28 or WDR43 FbioCLIP-seq with mESCs. Figure 3A shows the track view of LIN28 and WDR43 FbioCLIP-seq in pre-let-7g. The reported GGAG motif in pre-let-7g and the cross-linked sites identified by FbioCLIP-seq are shown in Figure 3B. The classification of the mutation sites called by CIMS algorithm showed that LIN28 prefers to bind and cross-link to G nucleotide (Figure 3C). The enriched RNA motifs in LIN28 FbioCLIP-seq binding sites are shown in Figure 3D. More representative tracks of FbioCLIP-seq on LIN28 and HNRNPU are shown in Figure 3E. Comparison of WDR43 and LIN28 FbioCLIP-seq showed their different binding patterns in Rn45 pre-rRNA locus (Figure 4). Figure 5 shows the representative results of FLAG and biotin tag validation after cell line construction. Figure 6 shows representative results of efficient (Figure 6A) or unsuccessful (Figure 6B) FLAG-IP and FLAG-biotin tandem purification.
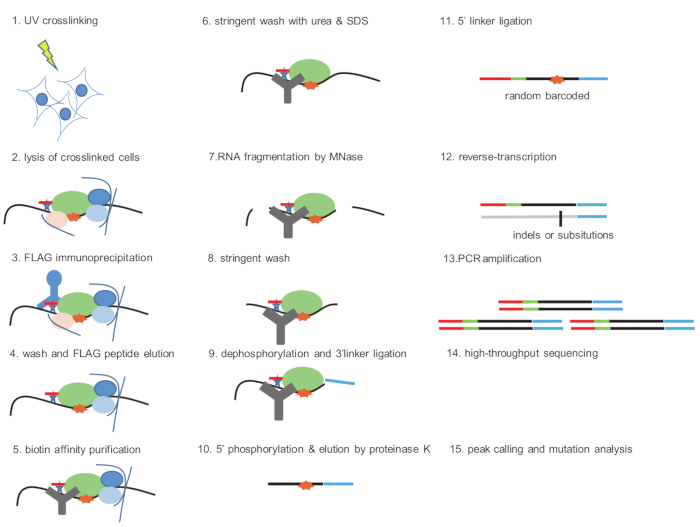
Figure 1: Schematic representation of FbioCLIP. UV-cross-linked cells (step 1) are lysed in lysis buffer (step 2). The FBRBP-RNA complex is immunoprecipitated using anti-FLAG resin (steps 3−4). The eluted protein-RNA complex is further purified with streptavidin beads and stringent wash conditions are used to remove protein-protein interactions (steps 5−6). RNAs are partially digested by MNase and non-protein-bound RNA fragments are removed by further wash (steps 7−8). The RNAs are then dephosphorylated and ligated with 3’ linker (step 9). Then the RNAs are phosphorylated and eluted by proteinase K treatment (step 10). Purified RNAs are ligated with a 5’ RNA linker containing a 6 nt random barcode (step 11). After reverse transcription (step 12), the cDNA library is amplified with PCR and high-throughput sequencing is performed (steps 13−15). Please click here to view a larger version of this figure.
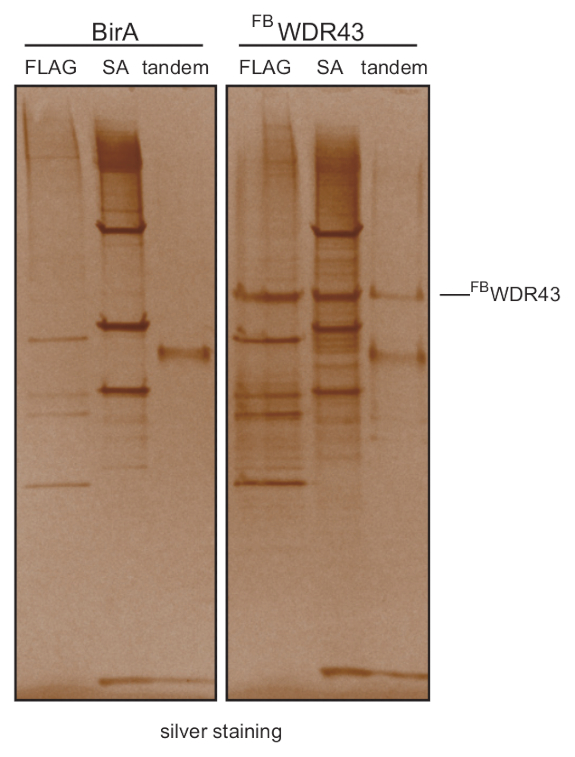
Figure 2: FLAG-biotin tandem purification removes copurified proteins. Silver staining of FBWDR43 showed that almost all the interacting proteins presented in FLAG- or biotin-mediated one-step purification were eliminated after stringent wash in tandem purification. FLAG: FLAG-mediated affinity purification, SA: streptavidin-mediated biotin purification, tandem: FLAG-biotin tandem purification. Please click here to view a larger version of this figure.
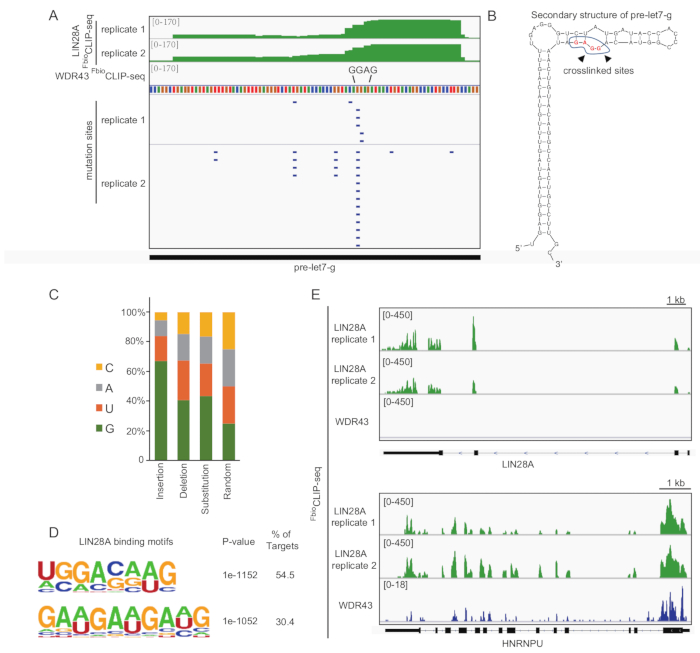
Figure 3: Representative results of LIN28 FbioCLIP-seq. (A) Track view of LIN28 FbioCLIP-seq tags and mutation reads called by CIMS in pre-let-7g RNA. The tags shown are unique reads of FbioCLIP-seq. In total, ~7.7 million unique reads for LIN28 FbioCLIP-seq and ~2.2 million unique reads for WDR43 FbioCLIP-seq were retrieved after removing redundant reads. (B) Cross-linked sites on the GGAG motif of pre-let7-g RNA by LIN28 FbioCLIP-seq. The arrows indicate the cross-linked sites. (C) Percentage of mutated nucleotides in different types of mutations. G is the most frequent cross-linked and mutated nucleotide. Random: random distribution of the four nucleotides. (D) Predicted LIN28 binding motifs by FbioCLIP-seq with HOMER algorithm. (E) Track views of LIN28 FbioCLIP-seq on LIN28 and HNRNPU mRNAs. Please click here to view a larger version of this figure.
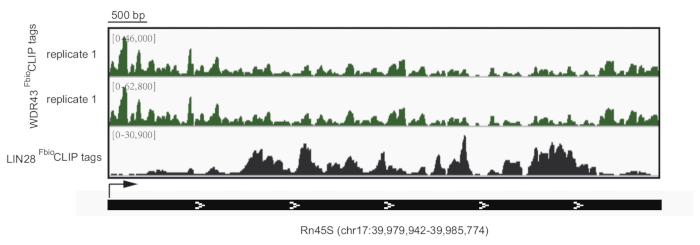
Figure 4: Representative results of WDR43 and LIN28 FbioCLIP-seq on Rn45S locus. Track view of WDR43 and LIN28 FbioCLIP-seq on Rn45S locus. WDR43 and LIN28 showed different binding patterns on pre-rRNA. Please click here to view a larger version of this figure.
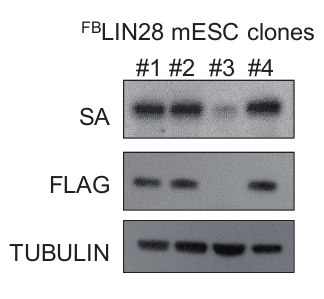
Figure 5: Representative results of FLAG and biotin tag validation. FLAG or biotin Western blot validates the tagging of the cell lines. Clones #1, #2, and #4 showed efficient expression and biotinylation of the tag while #3 showed poor expression of the tagged protein. Please click here to view a larger version of this figure.
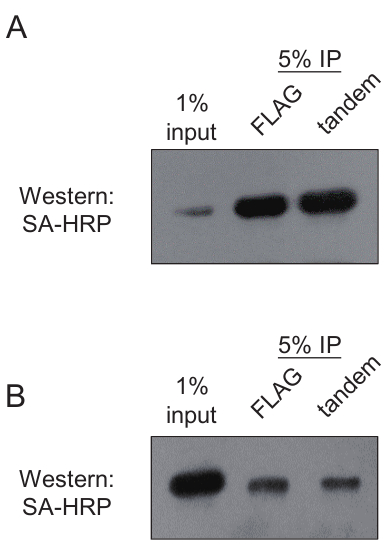
Figure 6: Representative results of FLAG immunoprecipitation and tandem purification efficiency validation. (A) Example of efficient purification of tagged protein by FLAG and biotin affinity purification. (B) Example of inefficient purification of tagged protein by FLAG and biotin affinity purification. Please click here to view a larger version of this figure.
| Buffer | Composition |
| SDS loading buffer | 50 mM Tris pH 6.8, 2% SDS, 0.1% bromophenolblue, 10% glycerol, 100 mM DTT |
| Wash buffer A | 1x PBS, 0.5% NP-40, 0.5% sodium deoxycholate, 0.1% SDS |
| Wash buffer B | 5x PBS, 0.5% NP-40, 0.5% sodium deoxycholate, 0.1% SDS |
| Wash buffer C | 50 mM Tris pH 7.4, 2% SDS |
| Wash buffer D | 5x PBS, 0.5% NP-40, 0.5% SDS, 1 M urea |
| PNK buffer | 50 mM Tris pH 7.4, 0.5% NP-40, 10 mM MgCl2 |
| MNase reaction buffer | 10 mM Tris pH 8.0, 1 mM CaCl2 |
| PNK+EGTA buffer | 50 mM Tris pH 7.4, 0.5% NP-40, 10 mM EGTA |
| Proteinase K digestion buffer | 50 mM Tris pH 7.4, 10 mM EDTA, 50 mM NaCl, 0.5% SDS, 20 µg of proteinase K |
Table 1: Composition of buffers used in this study.
| Oligo name | Sequence | Notes | ||||
| 3' linker | rAppAGATCGGAAGAGCACACGTCT-NH2 | |||||
| 5' RNA linker | GUUCAGAGUUCUACAGUCCGACGUCNNNNN | |||||
| RT primer | AGACGTGTGCTCTTCCGATCT | |||||
| Forward primer 1 | GTTCAGAGTTCTACAGTCCGACGATC | |||||
| Reverse primer 1 | AGACGTGTGCTCTTCCGATCT | |||||
| Forward primer 2 | AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGAC | |||||
| Reverse primer 2 | CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC-s-T | The red and underlined sequences represent Illumina index sequence. | ||||
Table 2: Oligonucleotides used in this study.
Discussion
Here we introduce a modified CLIP-seq method called FbioCLIP-seq, taking advantage of the FLAG-biotin double tagging system to perform tandem purification of protein-RNA complexes. The FLAG-biotin double tagging system has been shown to be powerful in identifying protein-protein and protein-DNA interactions13,21. Here we demonstrate the high specificity and convenience of this system in identifying the RNAs interacting with proteins. Through tandem purification and stringent wash conditions, we skipped the challenging SDS-PAGE and membrane transfer step, so that more protein-RNA interactions were preserved and to avoid contaminated RNA signals mediated by RBPs in the same complex. Besides, bypass of these steps avoids labeling of the RNA with radiolabeled ATP. This makes the procedure much easier. Note that during the preparation of our publication, a similar method called uvCLAP has also been proposed24.
Several steps are important for the success of the protocol. First, the efficiency of the biotinylation of the tagged protein should be confirmed by Western blot before the experiment (Figure 5). Second, the efficient elution of purified protein by 3 x FLAG peptide is required for the successful amplification of the RNA signals. The beads from step 12.4 should be analyzed with Western blot or silver staining to guarantee that a decent amount of target protein is retrieved (Figure 6). To increase the efficiency, either increase the 3 x FLAG peptide concentration in step 6 up to 500 ng/mL or repeat the elution step multiple times to get better production. Third, it is optimal to titrate the MNase concentration for each protein at the first trial. Overdigestion or insufficient treatment may lead to RNA fragments of inappropriate sizes. Last, the protocol contains two rounds of affinity purification and highly stringent wash. Only a small amount of purified RNAs is retrieved. The reagent should be RNase-free and avoid RNA degradation after MNase treatment step.
Despite the ease of this method, there are still some aspects that can be improved. For example, ligation of a 5’ adaptor to the RNA directly may limit the recovery of the signals because a significant portion of reverse transcription will be terminated by residual cross-linked peptides. Our current study is mainly based on ectopic expression of tagged proteins, which may lead to some artifacts due to protein overexpression. It is worth improving the method by introducing the tag into the endogenous locus. The CRISPR/Cas9 system makes cell line construction much easier and doable. Some studies have applied the eCLIP-seq experiment to mouse tissues25. Derivation of knock-in mice with the double tag is also a potential and promising direction in the improvement and application of the FbioCLIP-seq method. Furthermore, ectopically expressed bacterial BirA ligase may lead to unexpected biotinylation events in vivo. Tagging of the protein may also affect its biological functions.
A large number of studies have shown that the gene expression and epigenome of cells are heterogeneous instead of fully homogeneous, suggesting that it is valuable to study the interaction network at a single-cell level. Several strategies have been developed to study the transcriptome and epigenome of single cells. However, no strategies have been reported to analyze the protein-RNA interactome at a single-cell level. Without the denatured gel running and membrane transfer steps, FbioCLIP-seq can preserve more signals, which makes it a potential strategy to study protein-RNA interactions at a single-cell level.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Grant support is from the National Basic Research Program of China (2017YFA0504204, 2018YFA0107604), the National Natural Science Foundation of China (31630095), and the Center for Life Sciences at Tsinghua University.
Materials
| Equipment | |||
| UV crosslinker | UVP | HL-2000 HybrilLinker | |
| Affinity Purification Beads | |||
| ANTI-FLAG beads | Sigma-Aldrich | A2220 | |
| Streptavidin beads | Invitrogen | 112.06D | |
| Reagents | |||
| 10x PBS | Gibco | 70013032 | |
| 3 M NaOAc | Ambion | AM9740 | |
| 3 x FLAG peptide | Sigma-Aldrich | F4799 | |
| ATP | Sigma-Aldrich | A6559 | |
| Calcium chloride (CaCl2) | Sigma-Aldrich | C1016 | |
| CIP | NEB | M0290S | CIP buffer is in the same package. |
| DTT | Sigma-Aldrich | D0632 | |
| EDTA | Sigma-Aldrich | E9884 | |
| EGTA | Sigma-Aldrich | E3889 | |
| Gel purification kit | QIAGEN | 28704 | |
| Glycogen | Ambion | AM9510 | |
| Magnesium chloride (MgCl2) | Sigma-Aldrich | 449172 | |
| MNase | NEB | M0247S | |
| NP-40 | Amresco | M158-500ML | |
| PMSF | Sigma-Aldrich | 10837091001 | |
| Porteinase K | TAKARA | 9033 | |
| Protease inhibitor cocktail | Sigma-Aldrich | P8340 | |
| Q5 High-Fidelity 2X Master Mix | NEB | 0492S | |
| reverse trancriptase (SupperScriptIII) | Invitrogen | 18080093 | |
| RNA isolation reagent (Trizol) | Invitrogen | 15596018 | |
| RNase Inhibitor | ThermoFisher | EO0381 | |
| RNaseOUT | Invitrogen | 10777019 | |
| RQ1 Dnase | Promega | M6101 | |
| SDS | Sigma-Aldrich | 1614363 | |
| Sodium chloride | Sigma-Aldrich | S9888 | |
| Sodium deoxycholate | Sigma-Aldrich | D6750 | |
| T4 PNK | NEB | M0201S | PNK buffer is in the same package. |
| T4 RNA ligaes | ThermoFisher | EL0021 | T4 RNA ligase buffer and BSA are in the same package. |
| T4 RNA ligase2, truncated | NEB | M0242S | T4 RNA ligase buffer and 50% PEG are in the same package. |
| Trypsin-EDTA | ThermoFisher | 25200072 | |
| Urea | Sigma-Aldrich | 208884 | |
| mESC culture medium | |||
| DMEM (80%) | Gibco | 11965126 | |
| 2-Mercaptoethanol | Gibco | 21985023 | |
| FCS (15%) | Hyclone | ||
| Glutamax (1%) | Gibco | 35050061 | |
| LIF | purified recombinant protein; 10,000 fold dilution | ||
| NEAA (1%) | Gibco | 11140050 | |
| Nucleoside mix (1%) | Millipore | ES-008-D | |
| Penicillin-Streptomycin (1%) | Gibco | 15140122 | |
| Kit | |||
| DNA gel extraction kit | QIAGEN | 28704 |
References
- Kim, B., Jeong, K., Kim, V. N. Genome-wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates. Molecular Cell. 66 (2), 258-269 (2017).
- Guallar, D., et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nature Genetics. 50 (3), 443-451 (2018).
- Holmqvist, E., Li, L., Bischler, T., Barquist, L., Vogel, J. Global Maps of ProQ Binding In Vivo Reveal Target Recognition via RNA Structure and Stability Control at mRNA 3′ Ends. Molecular Cell. 70 (5), 971-982 (2018).
- Wei, C., et al. RBFox2 Binds Nascent RNA to Globally Regulate Polycomb Complex 2 Targeting in Mammalian Genomes. Molecular Cell. 62 (6), 875-889 (2016).
- Kim, D. S., et al. Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. Molecular Cell. 75 (6), 1270-1285 (2019).
- Yao, R. W., et al. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Molecular Cell. 76 (5), 767-783 (2019).
- Licatalosi, D. D., et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 456 (7221), 464-469 (2008).
- Moore, M. J., et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nature Protocols. 9 (2), 263-293 (2014).
- Konig, J., et al. iCLIP–transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. Journal of Visualized Experiments. (50), e2638 (2011).
- Zarnegar, B. J., et al. irCLIP platform for efficient characterization of protein-RNA interactions. Nature Methods. 13 (6), 489-492 (2016).
- Van Nostrand, E. L., et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nature Methods. 13 (6), 508-514 (2016).
- Hafner, M., et al. PAR-CliP–a method to identify transcriptome-wide the binding sites of RNA binding proteins. Journal of Visualized Experiments. (41), e2034 (2010).
- de Boer, E., et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 100 (13), 7480-7485 (2003).
- Bi, X., et al. RNA Targets Ribogenesis Factor WDR43 to Chromatin for Transcription and Pluripotency Control. Molecular Cell. 75 (1), 102-116 (2019).
- Heo, I., et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 138 (4), 696-708 (2009).
- Cho, J., et al. LIN28A Is a Suppressor of ER-Associated Translation in Embryonic Stem Cells. Cell. 151 (4), 765-777 (2012).
- Yu, J., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 318 (5858), 1917-1920 (2007).
- Zhao, C., et al. Tissue specific roles for the ribosome biogenesis factor Wdr43 in zebrafish development. PLoS Genetics. 10 (1), 1004074 (2014).
- Wilbert, M. L., et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Molecular Cell. 48 (2), 195-206 (2012).
- Hunziker, M., et al. UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nature Communications. 7, 12090 (2016).
- Kim, J., Cantor, A. B., Orkin, S. H., Wang, J. L. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nature Protocols. 4 (4), 506-517 (2009).
- Li, Z., Michael, I. P., Zhou, D., Nagy, A., Rini, J. M. Simple piggyBac transposon-based mammalian cell expression system for inducible protein production. Proceedings of the National Academy of Sciences of the United States of America. 110 (13), 5004-5009 (2013).
- Hnasko, T. S., Hnasko, R. M. The Western Blot. Methods in Molecular Biology. 1318, 87-96 (2015).
- Aktas, T., et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 544 (7648), 115-119 (2017).
- Xu, Q., et al. Enhanced Crosslinking Immunoprecipitation (eCLIP) Method for Efficient Identification of Protein-bound RNA in Mouse Testis. Journal of Visualized Experiments. (147), e59681 (2019).

