Characterization of Immune Cells and Proinflammatory Mediators in the Pulmonary Environment
Summary
This protocol describes the use of flow cytometry to identify the changes in immune cell composition, cytokine profile, and chemokine profile in the pulmonary environment following transient middle cerebral artery occlusion, a murine model of ischemic stroke.
Abstract
Immune cell expansion, activation, and trafficking to the lungs, which are controlled by the expression of multiple cytokines and chemokines, may be altered by severe brain injury. This is evidenced by the fact that pneumonia is a major cause of mortality in patients who have suffered from ischemic stroke. The goal of this protocol is to describe the use of multicolor flow cytometric analysis to identify 13 types of immune cells in the lungs of mice, including alveolar macrophages, interstitial macrophages, CD103+ or CD11b+ dendritic cells (DCs), plasmacytoid DCs, eosinophils, monocytes/monocyte-derived cells, neutrophils, lymphoid-derived T and B cells, NK cells, and NKT cells, following ischemic stroke induction by transient middle cerebral artery occlusion. Moreover, we describe the preparation of lung homogenates using a bead homogenization method, to determine the expression levels of 13 different cytokines or chemokines simultaneously by multiplex bead arrays coupled with flow cytometric analysis. This protocol can also be used to investigate the pulmonary immune response in other disease settings, such as infectious lung disease or allergic disease.
Introduction
The lungs are a barrier organ, exposed to the external environment and, therefore, are constantly receiving immunological challenges such as pathogens and allergens1. The activation of lung-resident immune cells and the infiltration of immune cells from the periphery are required to clear pathogens from the pulmonary environment. Additionally, lung-resident immune cells maintain tolerance to commensal bacteria, suggesting that these cells play a role in pathogen clearance and maintaining homeostasis1. Alveolar and interstitial macrophages are among the lung-resident sentinel immune cells that sense pathogens via pattern recognition receptors and clear these pathogens by phagocytosis2. Lung-resident dendritic cells bridge the innate and adaptive immune response through antigen presentation3. In addition, activated local innate immune cells produce cytokines and chemokines that amplify the inflammatory response and stimulate the infiltration of immune cells such as monocytes, neutrophils, and lymphocytes into the lungs1. Ischemic stroke has been shown to modify systemic immunity and lead to increased susceptibility to pulmonary infection; however, few studies have evaluated the pulmonary compartment following ischemic stroke, though some studies have examined it during inflammatory conditions4,5,6,7,8,9. The goal of the methods described herein is to simultaneously determine lung pathology, immune cell composition, and the levels of cytokine and chemokine expression in the lungs to evaluate alterations to the pulmonary compartment and assess potential alterations to the pulmonary immune response following ischemic attack.
Described here is a protocol for obtaining single cell suspensions from the lungs of the mice to identify 13 types of immune cells. This protocol is based on tissue digestion with collagenase D without the need of an automated tissue dissociator. Additionally, we developed a protocol to prepare tissue homogenates that can be used to determine the expression levels of 13 different cytokines or chemokines using flow cytometry-based multiplex bead arrays. This protocol was successfully used to investigate the effects of ischemic stroke on pulmonary immunity and can be used in other disease models as well.
Protocol
All protocols and procedures performed were approved by the Institutional Animal Care and Use Committee (IACUC) of West Virginia University. The mice were housed under specific-pathogen-free conditions in the vivarium at West Virginia University.
1. Preparation of solutions
- Prepare perfusion buffer (phosphate buffered saline, PBS). Use approximately two 10 mL aliquots of ice-cold PBS per mouse.
- Prepare lung cell medium/FACS buffer. FACS buffer contains PBS supplemented with 1% fetal bovine serum (FBS). Keep the medium cold for the entire process of lung excision and transfer. Prepare approximately 8 mL per lung sample. Prepare fresh medium/FACS buffer prior to the experiments.
- Prepare dissociation buffer for single cell isolation. Buffer contains 1 mg/mL collagenase D and 200 µg/mL DNase I in Hank’s buffered salt solution (HBSS). Prepare fresh from the stock solution (100 mg/mL collagenase D and 10 mg/mL DNase I) prior to the experiments. Approximately 6 mL per lung sample is needed. Ensure buffer reaches room temperature prior to the use.
- Prepare homogenization buffer for lung tissue homogenization. Buffer contains PBS and 1x proteinase and phosphatase inhibitor cocktail (stock = 100x). Freshly prepare prior to the experiments. Keep the buffer cold during the entire homogenization process. Approximately 200 µL of the buffer is sufficient for homogenizing a single right lobe of the lungs.
2. Transient middle cerebral artery occlusion (tMCAO)
NOTE: Procedures for tMCAO via monofilament insertion to the middle cerebral artery were documented in detail previously10. In these experiments, 8- to 12-week-old male C57BL/6J mice, weighing 25-30 g, were used.
- In brief, subcutaneously administer 2 mg/kg meloxicam to the mice before surgery. Deeply anesthetize the mice in an induction chamber using 5% isoflurane. Confirm deep anesthetization using the toe-pinch method. Maintain anesthesia with 1-2% isoflurane during the surgery using a nose cone. Maintain the body temperature at 36.5 – 38 °C throughout the procedure by placing a warm blanket underneath the mice. All surgical tools should be autoclave-sterilized a day prior to the surgery.
- Shave and prepare the skin on the ventral neck using 70% ethanol followed by chlorhexidine scrub. Subcutaneously administer 2 mg/kg bupivacaine over the incision area prior to making the first incision. Use eye ointment to cover the eyes to prevent dryness. Make a midline neck incision. Gently pull aside soft tissues around the trachea.
- Identify the left common carotid artery and the external carotid artery.
- Apply a temporary suture (size 6/0) to the common carotid artery to stem the flow of the blood. Make an incision in the external carotid artery.
- Insert a silicone rubber-coated monofilament (size 6-0) into the external carotid artery, then advance the monofilament to the middle cerebral artery. Leave the monofilament in the middle cerebral artery for 60 min (occlusion).
- Monitor the rate of blood flow using Laser Doppler Flowmetry. A reduction in the flow rate to the middle cerebral artery that is > 80% indicates a successful occlusion.
- Following the 60 min of occlusion, remove the monofilament to allow for the reperfusion to occur. Close the incision.
- In sham-operated mice, perform steps 2.1-2.6 without the insertion of the monofilament. In these mice, the rate of blood flow should remain steady during the entire procedure.
- Monitor the mice daily and measure the neurological deficits using standard scoring criteria11. 0 – no neurological deficit; 1 – retracts contralateral forepaw when lifted by the tail; 2- circles to the contralateral when lifted by the tail; 3- falling to the contralateral while walking; 4 – does not walk spontaneously or is comatose; 5 – dead.
- Provide subcutaneous injections of normal saline and analgesic (meloxicam, 2 mg/kg) every 24 hours for 3 days following the surgery.
- Isolate the lungs 24 and 72 h following tMCAO for the downstream analysis.
3. Harvesting the lung tissues
- Deeply anesthetize the mouse by intraperitoneal (i.p.) injection of 100 mg/kg of ketamine and 10 mg/kg of xylazine. Confirm deep anesthetization using the toe-pinch method.
- To perform the whole body transcardial perfusion12, pin the mouse to the surgical platform and wet fur with 70% ethanol to reduce fur distribution into the body cavity. Use fine dissection scissors to make a transverse incision in the caudal abdomen and then a midline incision of the peritoneum, stopping at the thoracic cavity prior to piercing the diaphragm.
- Do not cut through the muscle/hide into the peritoneal cavity and do not continue incision past abdomen.
- Stop the incision as soon as the thoracic cavity has been reached before puncturing the diaphragm. Take care not to damage the heart and lungs which may compromise perfusion quality or cell recovery
- Using fine dissection scissors make a midline incision of the peritoneum stopping at the thoracic cavity prior to piercing the diaphragm.
- Grasp the mouse by the distal end of the sternum using forceps and lift upward while cutting through the diaphragm being certain not to damage the heart, lungs, or vasculature. Once organs are visible make an incision through the rib cage and separate and pin back the rib case so that the heart and lungs are fully exposed.
- Carefully make a small incision in the right atrium near the aortic arch. This will serve as the outflow for the perfusion.
- Immediately after, gently insert a 25 G x 5/8 needle attached to a syringe filled with 10 mL of cold PBS into the distal superior surface of the left ventricle. Use care not to insert through the ventricular septum into the right ventricle. Needle should be barely inserted into the left ventricle.
NOTE: If ventricular septum is pierced, fluid will rush out from the mouse’s nose. This will decrease the perfusion quality and result in cell loss. - If desired, a clamp may be used to hold the needle securely in place in the left ventricle during the perfusion.
- Steadily but gently push 10 mL of cold PBS into the heart. Mouse tissue should begin to perfuse and clear off the blood as fluid exits from the right atrium.
- Push a second aliquot of 10 mL of PBS until sufficient blood clearance has occurred. Liver will have a light brown appearance and lungs will transition from a reddish pink to mostly white in color.
- Using forceps and fine dissection scissors, carefully remove all surrounding tissues including the heart, trachea, esophagus, thymus, connective tissue, lymph nodes, and large bronchials.
- Separate individual lung lobes and place the lobes into a Petri dish on ice containing a 1-2 mL aliquot of cold lung cell medium.
4. Homogenization of lung tissue for multiplex bead arrays using bead homogenizer
- Pre-chill a 2 mL conical screw cap tube containing 3 sterile 2.3 mm zirconia/silica beads. Add 200 µL of cold homogenization buffer to the tube.
- Weigh the lobe, transfer the lobe to the pre-chilled conical tube containing the beads and homogenization buffer.
NOTE: Approximately 50-100 mg of tissue homogenized in 200 µL of buffer will be sufficient for detecting cytokine and chemokine expression in the sample.- Homogenize the lobe using a bead-based homogenizer (see Table of Materials) at 4,000 rpm for 2 min.
- Ensure the tissue is completely homogenized. Increase the time if needed.
- Following homogenization, centrifuge the tube in a pre-chilled (4 °C) micro-centrifuge at 15,870 x g for 3 min.
- Transfer the supernatants to a pre-chilled 1.5 mL microtube.
- Place the sample directly on ice for immediate use or store the sample at -80 °C for future use. Avoid multiple freeze-thaw cycles.
5. Lung tissue dissociation and single cell isolation
- Pour the lung cell medium off, of the remaining lung lobes to avoid dilution of the dissociation buffer. Then, carefully insert a 1 mL syringe with 25G and 5/8 needle containing 1 mL of the dissociation buffer into lung tissue.
- Inject small fractions (approximately 1/4th to 1/5th) of the dissociation buffer into each lung lobe and gently inflate.
NOTE: Most of the buffer will rush out soon after inflating as the lobe has been excised. - Repeat the injection of dissociation buffer 1-2x.
- Incubate the lung lobes with dissociation buffer for 2 min at room temperature.
- Finely mince the lung lobes into small pieces using fine dissection scissors.
- Transfer the minced lung pieces with the dissociation buffer to a 15 mL centrifuge tube. Supplement with additional dissociation buffer to a total of 6 mL. Vortex vigorously for 1 min.
- Incubate the sample for 45 min at 37 °C. Vortex vigorously every 7-8 min.
- After 45 min of incubation, vortex the sample vigorously for 1 min for optimal results.
- Further dissociate the sample by passing through a 100 µm cell strainer to remove residual, undesired connective, and interstitial tissue.
- Centrifuge the sample for 10 min at 380 x g, discard the supernatant.
- Add 5 mL of cold lung cell medium to the sample. Resuspend the cell pellet by gentle vortexing.
- Centrifuge the sample for 10 min at 380 x g, discard the supernatant.
- Add 1 mL of cold lung cell medium. Resuspend the cell pellet by gentle vortexing.
- Pass through a 100 µm cell strainer, and count cells.
6. Flow cytometric analysis for the lung immune cell niche
- Seed 1-2 x 106 cells/antibody set into a 96 well round bottom plate (3 sets in total). Centrifuge the cells for 3 min at 830 x g. Discard supernatants.
- Resuspend the cell pellet in 50 µL of cold PBS containing fixable live/dead stain. Prepare the stain according to the manufacturer’s instructions.
- Incubate the cells at 4 °C for 20 min protected from light. Centrifuge the cells for 3 min at 830 x g. Discard supernatants.
- Resuspend the cell pellet with 25 µL of cold FACS buffer containing 5 µg/mL of anti-mouse CD16/32 antibody. Incubate at 4 °C for 10-15 min.
- Add 25 µL of cold FACS buffer containing antibody combinations listed in Table 1 to the cells. Mix by gentle pipetting.
NOTE: Total volume of FACS buffer for the antibody staining is 50 µL. Therefore, when preparing a master mix of antibodies in 25 µL, the concentration of antibodies should be double the final concentration. - Incubate the cells at 4 °C for 20 min protected from light. Centrifuge the cells for 3 min at 830 x g. Discard supernatants.
- Resuspend the cells with 100 µL of cold FACS buffer. Centrifuge the cells for 3 min at 830 x g. Discard supernatants.
- Resuspend the cells with 100 µL of cold FACS buffer. Transfer to a polystyrene round-bottom 5 mL FACS tube containing 150 µL of additional FACs buffer. Analyze the sample immediately using a flow cytometer.
NOTE: Cell fixation using the following steps allows samples to be analyzed later. - Following step 6.7., resuspend the sample with 100 µL of 1% paraformaldehyde in PBS.
- Incubate the sample at 4 °C for 15 min protected from light. Centrifuge the cells for 3 min at 830 x g. Discard supernatants.
- Resuspend the cells with 100 µL of cold FACS buffer. Centrifuge the cells for 3 min at 830 x g. Discard supernatants.
- Resuspend the cells with 100 µL of cold FACS buffer. Store the cells at 4 °C, protected from light. Analyze the sample within 72 h.
7. Multiplex bead arrays for cytokine and chemokine detection
NOTE: Commercially available proinflammatory chemokine and inflammation multiplex panels (see Table of Materials) were used to determine the expression of chemokines and cytokines in the lungs following tMCAO induction according to manufacturer protocol, which has been described in detail13.
- In brief, thaw the analytes on ice if they were stored at -80 °C after tissue homogenization. Centrifuge the sample at 590 x g for 2 min at 4 °C before use to remove any residual tissue.
- Generate a standard curve by serially diluting the standard cocktail (C7) which is at a known concentration of 10 ng/mL into 7 standards (C1-6) with the last standard being assay buffer alone (C0).
- Once all reagents have warmed to room temperature, add 25 µL of each standard and 25 µL of assay buffer to a V bottom 96 well plate.
- To each sample well, add 25 µL of lung homogenate (analyte) and 25 µL of assay buffer to a V bottom 96 well plate
- Vortex the pre-coated beads vigorously, then add 25 µL of pre-coated beads to each of the standard and sample wells.
- Seal the plate and protect from light by covering with foil. Shake the plate at 110 rpm for 2 h at room temperature.
- Centrifuge at 230 x g for 5 min. Add 200 µL of wash buffer at 1x (stock solution is 20x, dilute to 1x with water). Incubate for 1 min.
- Centrifuge at 230 x g for 5 min. Resuspend the standard and sample with 25 µL detection antibodies.
- Seal and cover the plate to protect from light. Shake plate at 110 rpm for 1 h at room temperature.
- Add 25 µL of streptavidin-phycoerythrin (SA-PE) to each standard and sample well.
- Seal and cover the plate to protect from light. Shake at 110 rpm for 30 min at room temperature.
- Spin the plate at 230 x g for 5 min. Resuspend the standard and samples in 1x wash buffer.
- Transfer to a well-labeled polystyrene round-bottom 5 mL FACS tube containing an additional 150 µL of 1x wash buffer.
- Determine the PE signal fluorescence intensity for the standards and samples using a flow cytometer.
- Construct a standard curve for each chemokine and cytokine using the mean fluorescence intensity (MFI) of PE against the pre-determined concentrations.
- Determine the concentration of each chemokine and cytokine using the standard curve and the MFI from each sample. The multiplex panel provides data analysis software that can be used to determine the concentration of each analyte. The weight of the apical/superior lobe is used to determine the concentration of cytokines or chemokine per milligram of tissue.
Representative Results
We recently reported that ischemic stroke induction in mice alters the immune cell composition of the lungs11. Specifically, transient cerebral ischemia increased percentages of alveolar macrophages, neutrophils, and CD11b+ DCs, while diminishing percentages of CD4+ T cells, CD8+ T cells, B cells, NK cells, and eosinophils in the pulmonary compartment. Moreover, cellular alteration corresponded to significantly diminished levels of multiple chemokines in the lungs. Described here is a method for the isolation and identification of different immune cell populations in the pulmonary compartment. Representative results shown here were from mice that had undergone tMCAO induction and a sham operation.
We identified 13 different populations of immune cells in the lungs (L1-L13) using 3 sets of antibody combinations with each set containing 5-7 antibodies (Figure 1). Dead cells were excluded using a LIVE/DEAD stain in each set. Antibodies and markers for distinguishing different immune cell types are listed in Table 1. Alveolar and interstitial macrophages, CD103+ DCs, CD11b+ DCs, and eosinophils were identified in Set 1 (Figure 1A,B). Proinflammatory monocytes and neutrophils were identified in Set 2 (Figure 1C). During inflammatory responses, monocytes migrate to the site of inflammation, where these cells differentiate into monocyte-derived antigen presenting cells (mo-APCs)14. Downregulation of Ly6C and CCR2 are characteristic of monocyte differentiation, which can be evaluated using Set 215. CD4+ T cells, CD8+ T cells, B cells, plasmacytoid DCs, NK cells, and NKT cells were identified using Set 3 (Figure 1D).
To determine the quality of our single cell isolation protocol, we compared the number of viable cells and CD45+ immune cells isolated using the manual method with the cells isolated using a commercially available tissue dissociator (see Table of Materials), which is often used to isolate cells from tissues16,17,18,19,20. In the latter protocol, lung lobes were transferred into a dissociator-specific tube (see Table of Materials) following injection of the dissociation buffer, and the tissue was digested using the 37C_m_LDK_1 program. The total number of viable cells, percentage of CD45+ cells, and the total number of CD45+ cells obtained were comparable between the two methods (Figure 2A-C). The percentage of cell death among CD45+ cells using both protocols was ~ 10% (Figure 2D). These results suggest that the protocol presented here allows cell recovery with high yield and quality without the aid of an automated tissue dissociator.
A commercially available multiplex assay coupled with flow cytometric analysis was used to determine the concentration of 13 chemokines using 25 µL of sample (Figure 3). Two different sizes of beads were first identified by FSC/SSC (Figure 3A). Each bead is coated with 6-7 primary antibodies, which could be distinguished by fluorescence intensity in the APC channel (Figure 3B). The level of chemokines in the sample is proportional to the fluorescence intensity in the PE channel, which could be determined by MFI (Figure 3C). By comparing the MFI value of each chemokine with the standard curve constructed with known concentrations of chemokine, the concentration in the sample (per mg of tissue) can be determined.
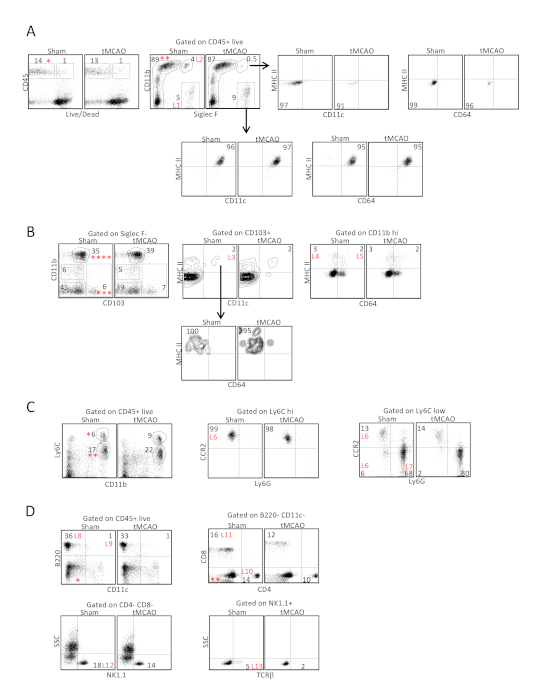
Figure 1: Identification of 13 immune cell types from the lungs following tissue digestion with collagenase D following ischemic stroke. Lung tissues were excised 24 h following tMCAO or sham operation, immune cells in the lungs were analyzed by flow cytometry, defined by surface markers listed on Table 1. (A) In antibody set 1, CD45+ viable cells (*) were first gated on Siglec F and CD11b to identify the alveolar macrophages (L1), which expressed CD11c and MHC II, and eosinophils (L2), which did not express CD11c and MHC II. (B) Cells within the Siglec F- population (**) in A were then gated to determine the expression of CD103 and CD11b. CD103+ CD11b- (***). Cells were further gated to determine the expression of CD11c and MHC II. CD103+ DCs expressed both CD11c and MHC II (L3) but did not express CD64. The CD11b hi population (****) was further gated to determine the expression of CD64 and MHC II. CD11b+ DCs expressed MHC II but not CD64 (L4), whereas interstitial macrophages (L5) expressed both markers. (C) In antibody set 2, CD45+ viable cells in A were gated to determine the expression of CD11b and Ly6C. Ly6C hi cells (*) represented the undifferentiated monocytes that maintained a high level of CCR2 expression (L6, middle plot), whereas Ly6C low cells (**) contained a mixed population of differentiating monocytes that were Ly6G- (L6, right plot), and Ly6G+ neutrophils (L7). (D) In antibody set 3, CD45+ viable cells in A were gated to determine the expression of CD11c and B220 to identify B cells (L8) and plasmacytoid DCs (L9). The CD11c- and B220- population (*) was then gated to determine the expression of CD4 and CD8 to identify CD4+ T cells (L10) and CD8+ T cells (L11). The CD4- CD8- population (**) was further gated to determine the expression of NK1.1 and TCRb to identify NK cells (L12) and NKT cells (L13). Shown are representative plots from 12 C57BL/6J mice following tMCAO and sham operation. Parts of the figure have been reprinted from previously published literature11 with permission. Please click here to view a larger version of this figure.
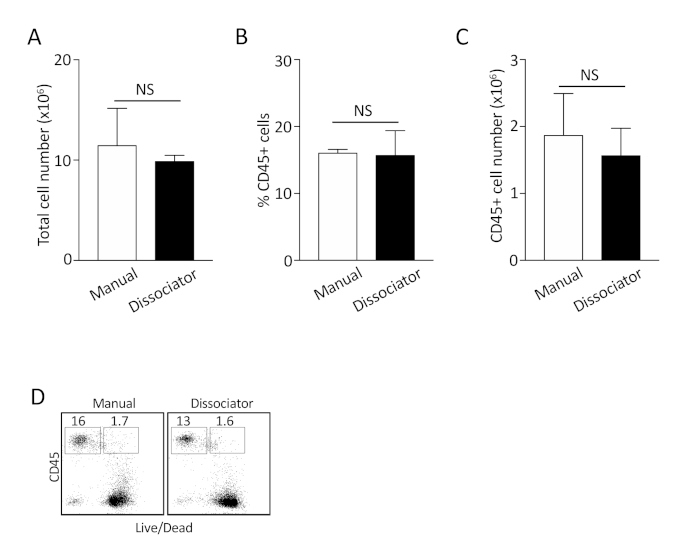
Figure 2: Comparison between manual dissociation method and the use of tissue dissociator for isolating single cells from the lungs. (A-C) The total number of cells, the percentage of CD45+ cells, and the total number of CD45+ cells were compared. Shown are combined results from 3 independent experiments. NS: not statistically significant. (D) Representative plots to determine the percentage of dead CD45+ cells following isolation. Shown are representative plots from 3 independent experiments. Please click here to view a larger version of this figure.
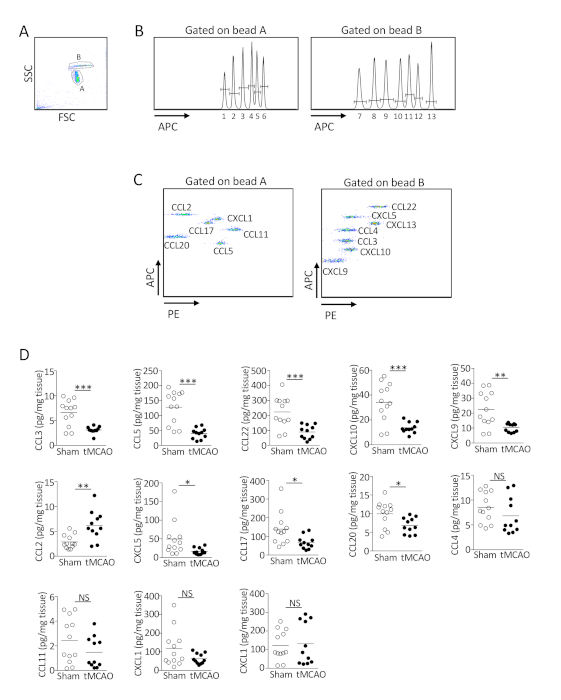
Figure 3: Ischemic stroke suppresses the production of multiple chemokines in the lungs. (A-C) Representative plots showing the determination of the level of 13 chemokines in the lungs by multiplex bead array. (A) FSC/SSC gate was used to identify beads A and B with different size. (B) Primary antibodies coated on the beads could be distinguished by fluorescence intensity in the APC channel. (C) The level of chemokines in the sample was proportional to the fluorescence intensity in the PE channel. Shown are representative plots from 12 C57BL/6J mice following sham operation. (D) Lung tissues were homogenized 24 h following tMCAO or sham operatio. The level of 13 chemokines in the lungs of individual animals was determined by multiplex bead array. Data shown are combined results from three independent experiments with n = 11-12 animals per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001. NS, not statistically different. Parts of the figure have been reprinted from previously published literature11 with permission. Please click here to view a larger version of this figure.
| Antibody | Clone | Immune Cell Type | Population | Surface Marker Expression |
| CD45-FITC | 30-F11 | Alveloar macrophages | L1 | CD45+ Siglec F+ CD11b- |
| Siglec F-PE | E50-2440 | Eosinophils | L2 | CD45+ Siglec F+ CD11b+ |
| CD11c-Percp/Cy5.5 | N418 | CD103+ DCs | L3 | CD45+ Siglec F- CD11b- CD103+ CD11c+ MHC II+ |
| CD11b-PE/Cy7 | M1/70 | CD11b+ DCs | L4 | CD45+ Siglec F- CD11b hi CD103- CD64- MHC II+ |
| CD64-APC | X54-5/7.1 | Interstitial macrophages | L5 | CD45+ Siglec F- CD11b hi CD103- CD64+ MHC II+ |
| CD103-BV421 | 2E7 | |||
| MHC II-BV510 | M5/114.15.2 | |||
| Live/dead-APC/Cy7 | ||||
| CD45-FITC | 30-F11 | Monocytes/moDCs | L8 | CD45+ CD11b hi Ly6C hi/int CCR2+/- Ly6G- |
| Ly6C-PE | HK1.4 | Neutrophils | L9 | CD45+ CD11b hi Ly6C int CCR2- Ly6G+ |
| CD11b-PE/Cy7 | M1/70 | |||
| CCR2-BV421 | SA203G11 | |||
| Ly6G-BV510 | 1A8 | |||
| Live/dead-APC/Cy7 | ||||
| CD45-FITC | 30-F11 | Plasmacytoid DCs | L6 | CD45+ B220+ CD11c+ |
| CD8-PE | 53-6.7 | B cells | L7 | CD45+ B220+ CD11c- |
| NK1.1-Percp/Cy5.5 | PK136 | CD4+ T cells | L10 | CD45+ B220- CD11c- CD4+ CD8- |
| CD11c-PE/Cy7 | N418 | CD8+ T cells | L11 | CD45+ B220- CD11c- CD4- CD8+ |
| APC-B220 | RA3-6B2 | NK cells | L12 | CD45+ B220- CD11c- CD4- CD8- NK1.1+ TCRb- |
| CD4-BV421 | GK1.5 | NKT cells | L13 | CD45+ B220- CD11c- CD4- CD8- NK1.1+ TCRb+ |
| TCRb-BV510 | H57-597 | |||
| Live/dead-APC/Cy7 |
Table 1: Surface markers and antibody combinations for determining immune cells isolated from the lungs following tMCAO.
Discussion
The protocols described here allow for the identification of lung immune cell types and the expression of chemokines or cytokines in the same mouse. If a histopathology study is desired, an individual lobe can be removed and fixed for that purpose prior to proceeding to the single cell isolation steps. One limitation of this method is that this approach may not be suitable in some disease settings if the change in the immune cell composition and the expression of chemokines and/or cytokines are anticipated to be unequally distributed between different lobes of the lungs. For example, some bacteria, such as Mycobacterium tuberculosis, show a predilection for infecting certain lobes of the lung21. In this case, a comparison between lobes may be required.
The concentration, incubation time, and temperature of the single cell isolation protocol from the lungs critically impact the recovery of the immune cells from the lungs. The quality of collagenase D is critical for obtaining optimal results and should be tested if a different source of collagenase D is used. Over-digestion of the tissues results in an increase of cell death; whereas, under-digestion of the tissues causes low yield of immune cells, especially macrophages and DCs.
We used 3 antibody combinations to determine 13 immune cell populations in the lungs, with each set containing 5-7 antibodies and a LIVE/DEAD stain. Antibodies in each set can be combined if the number of cells obtained from the samples is limited. However, one major issue that arises when the number of antibodies is increased is that the compensation of the fluorescence signal on the flow cytometer can be challenging, especially when distinguishing cells from myeloid linage under inflammatory conditions. Additional antibody cocktails can be used to identify other innate immune cells within the single-cell suspension, such as innate lymphoid cells and γδ T cells, that contribute to the immune response in the lungs22,23. Since one lobe of the lung is taken for the multiplex array, the exact number of each cell type in the lungs cannot be definitively determined and compared, and this constitutes a limitation of this method. To address this, a defined number of cells can be isolated from the lungs of sham and tMCAO-induced mice. In this case, the absolute number of each immune cell type can be accurately compared between the two groups. Additionally, this method does not allow the localization of the immune cells in the lung to be determined. This can be accomplished by performing immunohistochemistry on lung sections.
In conclusion, this protocol was used to investigate the effect of ischemic stroke in pulmonary immunity, but it can also be used to study other disease models, such as infection and allergies.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by NIH grant P20 GM109098 and the Innovation Award Program from Praespero to Edwin Wan. Flow Cytometry experiments were performed in the WVU Flow Cytometry & Single Cell Core Facility, which is supported by NIH grants S10 OD016165, U57 GM104942, P30 GM103488, and P20 GM103434.
Materials
| B220-APC, clone RA3-6B2 | Biolegend | 103212 | 1:200 dilution |
| Beadbug 3 position bead homogenizer | Benchmark Scientific | D1030 | Tissue homogenizer |
| CCR2-BV421, clone SA203G11 | Biolegend | 150605 | 1:200 dilution |
| CD103-BV421, clone 2E7 | Biolegend | 121422 | 1:200 dilution |
| CD11b-PE/Cy7, clone M1/70 | Biolegend | 101216 | 1:400 dilution |
| CD11c-PE/Cy7, clone N418 | Biolegend | 117318 | 1:200 dilution |
| CD11c-Percp/Cy5.5, clone N418 | Biolegend | 117328 | 1:200 dilution |
| CD4-BV421, clone GK1.5 | Biolegend | 100443 | 1:200 dilution |
| CD45-FITC, clone 30-F11 | Biolegend | 103108 | 1:200 dilution |
| CD64-APC, clone X54-5/7.1 | Biolegend | 139306 | 1:200 dilution |
| CD8-PE, clone 53-6.7 | Biolegend | 100708 | 1:800 dilution |
| Collagenase D | Sigma Aldrich | 11088882001 | Component in the dissociation buffer |
| Conical screw cap tube | ThermoFisher | 02-681-344 | Tube for tissue homogenization |
| DNase I | Sigma Aldrich | 10104159001 | Component in the dissociation buffer |
| Fc block CD16/32 antibody | Biolegend | 101320 | 1:100 dilution |
| genlteMACS dissociator | Miltenyi Biotec | 130-093-235 | Comparsion of lung digestion with or without mechanical dissociator |
| gentleMACS C tubes | Miltenyi Biotec | 130-093-237 | Tube for tissue disscoiation with genlteMACS dissociator |
| Halt protease and phosphatase inhibitor cocktial | ThermoFisher | 78442 | Component in the homogenization buffer |
| Laser doppler monitor | Moor | MOORVMS-LDF | Blood flow monitoring during tMCAO |
| LEGENDplex proinflammatory chemokine panel | Biolegend | 740451 | Multiplex bead array |
| LIVE/DEAD fixable near-IR stain | ThermoFisher | L34976 | Use for dead cell exclusion during flow cytometric analysis |
| Ly6C-PE, clone HK1.4 | Biolegend | 128008 | 1:800 dilution |
| Ly6G-BV510, clone 1A8 | Biolegend | 127633 | 1:200 dilution |
| MCAO suture L56 reusable 6-0 medium | Doccol | 602356PK10Re | tMCAO |
| MHC II-BV510, clone M5/114.15.2 | Biolegend | 107636 | 1:800 dilution |
| NK1.1-Percp/Cy5.5, clone PK136 | Biolegend | 108728 | 1:200 dilution |
| Siglec F-PE, clone E50-2440 | BD Biosciences | 552126 | 1:200 dilution |
| Silk suture thread, size 6/0 | Fine Science Tools | 18020-60 | tMCAO |
| SomnoSuite anesthesia system | Kent Scientific | SS-01 | Mouse anaesthetization for tMCAO |
| TCRb-BV510, clone H57-897 | Biolegend | 109234 | 1:200 dilution |
| Zirconia/silica beads, 2.3 mm | Biospec | 11079125z | Beads for tissue homogenization |
References
- Lloyd, C. M., Marsland, B. J. Lung Homeostasis: Influence of Age, Microbes, and the Immune System. Immunity. 46 (4), 549-561 (2017).
- Allard, B., Panariti, A., Martin, J. G. Alveolar Macrophages in the Resolution of Inflammation, Tissue Repair, and Tolerance to Infection. Frontiers in Immunology. 9, 1777 (2018).
- Hartl, D., et al. Innate Immunity of the Lung: From Basic Mechanisms to Translational Medicine. Journal of Innate Immunity. 10 (5-6), 487-501 (2018).
- Prass, K., et al. Stroke-induced immunodeficiency promotes spontaneous bacterial infections and is mediated by sympathetic activation reversal by poststroke T helper cell type 1-like immunostimulation. Journal of Experimental Medicine. 198 (5), 725-736 (2003).
- Smith, C. J., et al. Interleukin-1 receptor antagonist reverses stroke-associated peripheral immune suppression. Cytokine. 58 (3), 384-389 (2012).
- McCulloch, L., Smith, C. J., McColl, B. W. Adrenergic-mediated loss of splenic marginal zone B cells contributes to infection susceptibility after stroke. Nature Communications. 8 (1), 15051 (2017).
- Dames, C., et al. Immunomodulatory treatment with systemic GM-CSF augments pulmonary immune responses and improves neurological outcome after experimental stroke. Journal of Neuroimmunology. 321, 144-149 (2018).
- Jin, R., Liu, S., Wang, M., Zhong, W., Li, G. Inhibition of CD147 Attenuates Stroke-Associated Pneumonia Through Modulating Lung Immune Response in Mice. Frontiers in Neurology. 10, 853 (2019).
- Yu, Y. R., et al. A Protocol for the Comprehensive Flow Cytometric Analysis of Immune Cells in Normal and Inflamed Murine Non-Lymphoid Tissues. PLoS One. 11 (3), 0150606 (2016).
- Rousselet, E., Kriz, J., Seidah, N. G. Mouse model of intraluminal MCAO: cerebral infarct evaluation by cresyl violet staining. Journal of Visualized Experiments. (69), e4038 (2012).
- Farris, B. Y., et al. Ischemic stroke alters immune cell niche and chemokine profile in mice independent of spontaneous bacterial infection. Immunity, Inflammation and Diseases. 7 (4), 326-341 (2019).
- Gage, G. J., Kipke, D. R., Shain, W. Whole animal perfusion fixation for rodents. Journal of Visualized Experiments. (65), e3564 (2012).
- Lehmann, J. S., Zhao, A., Sun, B., Jiang, W., Ji, S. Multiplex Cytokine Profiling of Stimulated Mouse Splenocytes Using a Cytometric Bead-based Immunoassay Platform. Journal of Visualized Experiments. (129), e56440 (2017).
- Shi, C., Pamer, E. G. Monocyte recruitment during infection and inflammation. Nature Reviews Immunology. 11 (11), 762-774 (2011).
- Monaghan, K. L., Zheng, W., Hu, G., Wan, E. C. K. Monocytes and Monocyte-Derived Antigen-Presenting Cells Have Distinct Gene Signatures in Experimental Model of Multiple Sclerosis. Frontiers in Immunology. 10, 2779 (2019).
- Zhai, X., et al. A novel technique to prepare a single cell suspension of isolated quiescent human hepatic stellate cells. Science Reports. 9 (1), 12757 (2019).
- Platzer, B., et al. Dendritic cell-bound IgE functions to restrain allergic inflammation at mucosal sites. Mucosal Immunology. 8 (3), 516-532 (2015).
- Shinoda, K., et al. Thy1+IL-7+ lymphatic endothelial cells in iBALT provide a survival niche for memory T-helper cells in allergic airway inflammation. Proceedings of the National Academy of Sciences U. S. A. 113 (20), 2842-2851 (2016).
- Nakahashi-Oda, C., et al. Apoptotic epithelial cells control the abundance of Treg cells at barrier surfaces. Nature Immunology. 17 (4), 441-450 (2016).
- Barrott, J. J., et al. Modeling synovial sarcoma metastasis in the mouse: PI3′-lipid signaling and inflammation. Journal of Experimental Medicine. 213 (13), 2989-3005 (2016).
- Bouté, M., et al. The C3HeB/FeJ mouse model recapitulates the hallmark of bovine tuberculosis lung lesions following Mycobacterium bovis aerogenous infection. Veterinary Research. 48 (1), 73 (2017).
- Bal, S. M., et al. IL-1β, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nature Immunology. 17 (6), 636-645 (2016).
- Nakasone, C., et al. Accumulation of gamma/delta T cells in the lungs and their roles in neutrophil-mediated host defense against pneumococcal infection. Microbes Infection. 9 (3), 251-258 (2007).

