一种同时分析人胰腺组织中N-糖肽和磷酸肽的自旋尖端富集策略
Summary
翻译后修饰(PTM)改变蛋白质结构和功能。同时富集多个PTM类型的方法可以最大限度地提高分析的覆盖率。我们提出了一个使用双功能Ti(IV)固定化金属亲和色谱法,然后进行质谱分析的方案,用于同时富集和分析胰腺组织中的蛋白质N-糖基化和磷酸化。
Abstract
质谱法可以提供翻译后修饰(PTM)的深度覆盖,尽管由于与非修饰分析物相比,它们化学计量低,因此通常需要从复杂的生物基质中富集这些修饰。在自下而上的蛋白质组学工作流程中,PTMs对肽的大多数富集工作流程,其中蛋白质在分析所得肽之前被酶消化,仅富集一种类型的修饰。然而,PTM的整个补体导致了生物学功能,并且单一类型的PTM的富集可能会错过PTM的这种串扰.PTM在蛋白质糖基化和磷酸化之间观察到串扰,这是人类蛋白质中最常见的两种PTM,也是使用质谱工作流程研究最多的两种PTM。使用本文描述的同时富集策略,两个PTM都从死后人胰腺组织(一种复杂的生物基质)中富集。双功能Ti(IV)固定化金属亲和色谱用于以方便的基于自旋尖端的方法同时分离多种形式的糖基化和磷酸化,从而允许下游分析潜在的PTM串扰相互作用。这种糖肽和磷酸肽的富集工作流程可应用于各种样品类型,以实现对多个PTM的深度分析,并为未来的研究确定潜在的目标分子。
Introduction
蛋白质翻译后修饰(PTM)在调节蛋白质结构中起主要作用,从而调节其功能和下游生物过程。由于各种PTM提供的组合变异性,人类蛋白质组的多样性呈指数级增长,基因组预测的蛋白质从其规范序列中产生的不同变体被称为蛋白质形式,许多蛋白质形式来自PTM1。研究健康和疾病中的蛋白质形式多样性已成为近年来非常感兴趣的研究领域2,3。
通过开发基于质谱(MS)的蛋白质组学方法,对蛋白质形式和更具体的PTM进行深入研究变得更加容易。使用MS,分析物被电离,破碎,并根据片段的 m / z 进行鉴定。富集方法通常是必要的,因为与非修饰形式的蛋白质相比,PTM的相对丰度较低。虽然完整蛋白质及其PTM的分析(称为自上而下分析)已变得更加常规,但在自下而上分析中蛋白质的酶消化及其组分肽的分析仍然是PTM分析中使用最广泛的途径。两种研究最广泛的PTM,以及 体内最常见的两种PTM,是糖基化和磷酸化4。这两个PTM在细胞信号传导和识别中起着重要作用,因此是在疾病研究中表征的重要修饰。
各种PTM的化学性质通常为在分析之前在蛋白质和肽水平上富集这些PTM提供了途径。糖基化是一种亲水性PTM,因为每个单糖上都有丰富的羟基。该性质可用于在亲水相互作用色谱(HILIC)中富集糖肽,其可以从疏水性非修饰肽5中分离出更多的亲水性糖肽。磷酸化增加了磷酸盐部分,除酸性pH外,磷酸盐部分带负电。由于这种电荷,各种金属阳离子,包括钛,可用于吸引和结合磷酸肽,同时非磷酸化物质被洗掉。这就是固定化金属亲和色谱(IMAC)的原理。关于糖基化和磷酸化的这些和其他富集策略的进一步讨论可以在最近的综述6,7中找到。
由于肽上PTM的化学计量较低,富集方案通常需要相对大量的起始肽材料(0.5mg或更多)。在可能不容易获得这种数量的样品的情况下,例如肿瘤核心活检或脑脊液分析,使用简单的工作流程来产生最大的生物分子信息是有益的。我们实验室和其他实验室最近制定的策略强调了使用相同的PTM富集工作流程8,9,10,11,12同时和平行分析糖基化和磷酸化。虽然这两种PTM的化学性质可能不同,但由于采用了创新的分离技术和材料,这些PTM可以分多个步骤进行分析。例如,静电排斥-亲水相互作用色谱(ERLIC)覆盖基于分析物与流动相之间的亲水相互作用与分析物与固定相材料13,14,15,16之间的电荷 – 电荷相互作用的分离。在酸性pH下,磷酸化肽对固定相的吸引力可以改善它们与非修饰肽的保留和分离。由固定在亲水性微球上的Ti(IV)组成的材料可用于HILIC和基于IMAC的洗脱,以分离磷酸肽和中性,酸性和甘露糖-6-磷酸化糖肽17,18。这种策略被称为双功能钛(IV)-IMAC。使用这些策略在单个工作流程中丰富多个 PTM 可以使潜在的 PTM 串扰交互分析更容易获得。此外,当并行执行时,总样品量和时间要求低于传统的富集方法(即,在单独的样品等分试样上进行HILIC和IMAC)。
为了证明同时分析蛋白质糖基化和磷酸化的双功能Ti(IV)-IMAC策略,我们将其应用于分析死后人胰腺组织。胰腺产生消化酶和调节激素,包括胰岛素和胰高血糖素。胰腺功能在胰腺疾病中受损。在糖尿病中,血糖的调节受到影响,导致血液中葡萄糖水平升高。在胰腺炎中,炎症是由器官的自动消化引起的3.PTM谱的变化,包括糖基化和磷酸化,可能导致其他疾病,通常的情况是这样的。
在这里,我们描述了基于双功能Ti(IV)-IMAC策略的基于自旋尖端的同时富集方法的方案,用于从胰腺组织中提取的蛋白质衍生的N-糖肽和磷酸肽。该方案包括蛋白质提取和消化、富集、MS数据收集和数据处理,如图 1所示。本研究的代表性数据 可通过 蛋白质交换联盟获得,标识符为PXD033065。
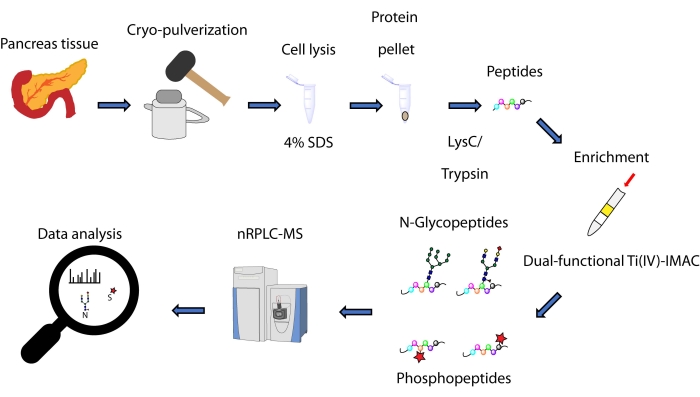
图 1:同时分析来自人胰腺组织的 N-糖肽和磷酸肽的工作流程。 在使用洗涤剂十二烷基硫酸钠(SDS)提取蛋白质之前,首先将组织冷冻粉碎成细粉。然后对蛋白质进行酶消化。在使用双功能Ti(IV)-IMAC富集之前,将所得肽等分。使用纳米级反相液相色谱- 质谱(nRPLC-MS)收集原始数据,并使用数据库搜索软件进行分析。 请点击此处查看此图的大图。
该协议旨在使PTM分析更易于访问,并在同一工作流程中对多个PTM进行更广泛的分析。该协议可应用于其他复杂的生物基质,包括细胞和生物流体。
Protocol
Representative Results
Discussion
双功能Ti(IV)-IMAC策略可用于在单个样品制备工作流程中同时分析来自同一样品的N-糖肽和磷酸肽。基于 ERLIC 的方法也被证明可以同时富集 PTM。这两种策略以前都用于PTM分析14,18的深度覆盖。通过使用自旋头使双Ti方法适应减少样品孵育时间,我们希望该协议已经变得不那么资源密集,从而更广泛地获得。
多个PTM的同时分析是通?…
Disclosures
The authors have nothing to disclose.
Acknowledgements
这项研究得到了NIH(R01DK071801,RF1AG052324,P01CA250972和R21AG065728)和青少年糖尿病研究基金会(1-PNF-2016-250-S-B和SRA-2016-168-B)的赠款资助的部分支持。这里提供的数据也部分是通过威斯康星大学临床和转化研究所获得NIH / NCATS UL1TR002373奖的支持而获得的。Orbitrap仪器是通过NIH共享仪器赠款(NIH-NCRR S10RR029531)和威斯康星大学麦迪逊分校研究和研究生教育副校长办公室的支持购买的。我们还要感谢威斯康星大学器官和组织捐赠组织的慷慨支持,该组织为人类胰腺的研究提供了帮助,Dan Tremmel,Sara D. Sackett博士和Jon Odorico教授为我们的实验室提供了样本。我们的研究团队特别感谢为这项研究捐赠纸巾的家庭。L.L.承认NIH拨款S10OD025084,这是威斯康星大学Carbone癌症中心(233-AAI9632)的胰腺癌试点补助金,以及维拉斯杰出成就教授职位和查尔斯·墨尔本约翰逊杰出讲座教授职位,由威斯康星州校友研究基金会和威斯康星大学麦迪逊分校药学院提供资金。
Materials
| Acetic Acid, Glacial (Certified ACS) | Fisher Scientific | A38S-500 | |
| Acetone (Certified ACS) | Fisher Scientific | A18-1 | |
| Acetonitrile, Optima LC/MS Grade | Fisher Scientific | A955-4 | |
| Ammonium Acetate (Crystalline/Certified ACS) | Fisher Scientific | A637-500 | |
| Ammonium Hydroxide (Certified ACS Plus) | Fisher Scientific | A669-212 | |
| Byonic software | Protein Metrics | n/a | Commercial software used for glycoproteomic analysis (https://proteinmetrics.com/byos/) |
| C18 BEH material | Waters | 186002353 | Material removed from column and used to pack nano capillaries (pulledto integrate tip used directly in line with instrument inlet) |
| CAE-Ti-IMAC, 100% | J&K Scientific | 2749380-1G | Material used for dual-functional Ti(IV)-IMAC; can also be used for conventional IMAC/conventional phosphopeptide enrichment |
| Cellcrusher kit | Cellcrusher | n/a | Used for grinding tissue samples into powder before extraction |
| Eppendorf 5424R Microcentrifuge | Fisher Scientific | 05-401-205 | For temperature-controlled centrifugation |
| cOmplete protease inhibitor cocktail tablets | Sigma | 11697498001 | |
| DTT, Molecular Grade (DL-Dithiothreitol) | Promega | V3151 | Protein reducing agent |
| Ethanol, 200 proof (100%), USP | Fisher | 22-032-601 | |
| Fisherbrand Analog Vortex Mixer | Fisher Scientific | 02-215-414 | |
| Fisherbrand Low-Retention Microcentrifuge Tubes (1.5 mL) | Fisher Scientific | 02-681-320 | |
| Fisherbrand Low-Retention Microcentrifuge Tubes (2 mL) | Fisher Scientific | 02-681-321 | |
| Fisherbrand Model 120 Sonic Dismembrator | Fisher Scientific | FB120110 | For sample lysis using ultrasonication |
| Formic Acid, 99.0+%, Optima LC/MS Grade | Fisher Scientific | A117-50 | |
| Fused silica capillary (75 μm inner diameter, 360 μm outer diameter) | Polymicro Technologies LLC | 100 m TSP075375 | For in-house pulled and packed columns with integrated emitter |
| Hydrofluoric acid (48 wt. % in H2O) | Sigma-Aldrich | 339261-100ML | Used for opening emitter of pulled capillary column |
| Iodoacetamide, BioUltra | Sigma | I1149-5G | Protein reducing reagent |
| MaxQuant software | n/a | n/a | Free software used for phosphoproteomic analysis (https://www.maxquant.org/) |
| Multi-therm Shaker with heating and cooling | Benchmark Scientific | H5000-HC | Heating block |
| Oasis HLB 1 cc Vac Cartridge, 10 mg Sorbent per Cartridge, 30 µm, 100/pk | Waters | 186000383 | Larger-scale cartridge desalting for tryptic digests (loading capacity approximately up to 1 mg each) |
| OMIX C18 pipette tips, 100 µL tip, 10 – 100 μL elution volume, 1 x 96 tips | Agilent | A57003100 | Smaller-scale packed pipette tip for desalting for enrichment elutions |
| P-2000 Micropipette Puller | Sutter Instrument Co. | P-2000/F | For pulling nano-capillary columns for LC-MS |
| PhosSTOP phosphatase inhibitor tablets | Sigma | 4906845001 | |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 | |
| Pierce Quantitative Colorimetric Peptide Assay | Thermo Fisher Scientific | 23275 | |
| PolySAX LP (12 μm, pore size 300 Å) | PolyLC | BMSX1203 | Material for strong anion-exchange chromatography used for ERLIC/conventional glycopeptide enrichment |
| Potassium Phosphate Monobasic (Crystalline/Certified ACS) | Fisher Scientific | P285-500 | |
| Pressure injection cell with integrated magnetic stirplate | Next Advance | PC77-MAG | For packing nano-capillary columns with stationary phase up to 2500 psi limit |
| Proteome Discoverer software | Thermo Fisher Scientific | n/a | Commercial software for proteomics anaysis (with integrated database searching software nodes) and data visualization (https://www.thermofisher.com/us/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html) |
| SpeedVac SC110 Vacuum Concentrator Model SC110-120 | Savant | n/a | Centrifugal vacuum concentrator for drying samples (under heat) |
| SDS Solution, 10% Sodium Dodecyl Sulfate Solution, Molecular Biology/Electrophoresis | Fisher Scientific | BP2436200 | |
| Sequencing Grade Modified Trypsin | Promega | V5111 | |
| Sodium Chloride (Crystalline/Certified ACS) | Fisher Scientific | S271-500 | |
| TopTip, Empty, 10-200 µL, Pack of 96 | Glygen Corporation | TT2EMT.96 | Empty pipette tip with micron-sized hole used that can be used to pack chromatographic materials for enrichments, bundled with tube adapters |
| Triethylammonium bicarbonate buffer (TEAB, 1 M, pH 8.5 (volatile)) | Sigma | 90360-100ML | |
| Trifluoroacetic acid, Reagent Grade, 99% | Fisher Scientific | 60-017-61 | |
| Tris Base (White Crystals or Crystalline Powder/Molecular Biology) | Fisher Scientific | BP152-500 | |
| Trypsin/Lys-C Mix, Mass Spec Grade | Promega | V5071 | |
| Urea (Certified ACS) | Fisher Scientific | U15-500 | |
| Water, Optima LC/MS Grade | Fisher Scientific | W64 |
References
- Smith, L. M., Kelleher, N. L. Proteoform: a single term describing protein complexity. Nature Methods. 10 (3), 186-187 (2013).
- Pan, S., Brentnall, T. A., Chen, R. Glycoproteins and glycoproteomics in pancreatic cancer. World Journal of Gastroenterology. 22 (42), 9288-9299 (2016).
- Tabang, D. N., Ford, M., Li, L. Recent advances in mass spectrometry-based glycomic and glycoproteomic studies of pancreatic diseases. Frontiers in Chemistry. 9, 707387 (2021).
- Khoury, G. A., Baliban, R. C., Floudas, C. A. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Scientific Reports. 1, 90 (2011).
- Alpert, A. J. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. Journal of Chromatography A. 499, 177-196 (1990).
- Riley, N. M., Bertozzi, C. R., Pitteri, S. J. A pragmatic guide to enrichment strategies for mass spectrometry-based glycoproteomics. Molecular & Cellular Proteomics. 20, 100029 (2020).
- Low, T. Y., et al. Widening the bottleneck of phosphoproteomics: Evolving strategies for phosphopeptide enrichment. Mass Spectrometry Reviews. 40 (4), 309-333 (2021).
- Cho, K. C., Chen, L., Hu, Y., Schnaubelt, M., Zhang, H. Developing workflow for simultaneous analyses of phosphopeptides and glycopeptides. ACS Chemical Biology. 14 (1), 58-66 (2019).
- Zhou, Y., et al. An integrated workflow for global, glyco-, and phospho-proteomic analysis of tumor tissues. Analytical Chemistry. 92 (2), 1842-1849 (2020).
- Tang, R., et al. Facile preparation of bifunctional adsorbents for efficiently enriching N-glycopeptides and phosphopeptides. Analytica Chimica Acta. 1144, 111-120 (2021).
- Wang, Z., Wang, J., Sun, N., Deng, C. A promising nanoprobe based on hydrophilic interaction liquid chromatography and immobilized metal affinity chromatography for capture of glycopeptides and phosphopeptides. Analytica Chimica Acta. 1067, 1-10 (2019).
- Glover, M. S., et al. Characterization of intact sialylated glycopeptides and phosphorylated glycopeptides from IMAC enriched samples by EThcD fragmentation: Toward combining phosphoproteomics and glycoproteomics. International Journal of Mass Spectrometry. 427, 35-42 (2018).
- Alpert, A. J. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Analytical Chemistry. 80 (1), 62-76 (2008).
- Cui, Y., et al. Counterion optimization dramatically improves selectivity for phosphopeptides and glycopeptides in electrostatic repulsion-hydrophilic interaction chromatography. Analytical Chemistry. 93 (22), 7908-7916 (2021).
- Cui, Y., et al. Finding the sweet spot in ERLIC mobile phase for simultaneous enrichment of N-Glyco and phosphopeptides. Journal of the American Society for Mass Spectrometry. 30 (12), 2491-2501 (2019).
- Tabang, D. N., et al. Analysis of pancreatic extracellular matrix protein post-translational modifications via electrostatic repulsion-hydrophilic interaction chromatography coupled with mass spectrometry. Molecular Omics. 17 (5), 652-664 (2021).
- Huang, J., et al. Dual-functional Titanium(IV) immobilized metal affinity chromatography approach for enabling large-scale profiling of protein Mannose-6-Phosphate glycosylation and revealing its predominant substrates. Analytical Chemistry. 91 (18), 11589-11597 (2019).
- Huang, J., et al. Dual-functional Ti(IV)-IMAC material enables simultaneous enrichment and separation of diverse glycopeptides and phosphopeptides. Analytical Chemistry. 93 (24), 8568-8576 (2021).
- Jami-Alahmadi, Y., Pandey, V., Mayank, A. K., Wohlschlegel, J. A. A robust method for packing high resolution C18 RP-nano-HPLC columns. Journal of Visualized Experiments: JoVE. (171), e62380 (2021).
- Bern, M., Kil, Y. J., Becker, C. Byonic: advanced peptide and protein identification software. Current Protocols in Bioinformatics. , (2012).
- Tyanova, S., Temu, T., Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols. 11 (12), 2301-2319 (2016).
- Perez-Riverol, Y., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research. 47, 442-450 (2019).
- Zhou, Y., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications. 10 (1), 1523 (2019).
- Zacharias, L. G., et al. HILIC and ERLIC enrichment of glycopeptides derived from breast and brain cancer cells. Journal of Proteome Research. 15 (10), 3624-3634 (2016).
- Yang, W., et al. Comparison of enrichment methods for intact N- and O-linked glycopeptides using strong anion exchange and hydrophilic interaction liquid chromatography. Analytical Chemistry. 89 (21), 11193-11197 (2017).
- Toghi Eshghi, S., Shah, P., Yang, W., Li, X., Zhang, H. GPQuest: A spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides. Analytical Chemistry. 87 (10), 5181-5188 (2015).
- Liu, M. -. Q., et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nature Communications. 8 (1), 438 (2017).
- Lu, L., Riley, N. M., Shortreed, M. R., Bertozzi, C. R., Smith, L. M. O-Pair Search with MetaMorpheus for O-glycopeptide characterization. Nature Methods. 17 (11), 1133-1138 (2020).
- Caval, T., Heck, A. J. R., Reiding, K. R. Meta-heterogeneity: Evaluating and describing the diversity in glycosylation between sites on the same glycoprotein. Molecular & Cellular Proteomics. 20, 100010 (2021).
- Lee, J. S., Smith, E., Shilatifard, A. The language of histone crosstalk. Cell. 142 (5), 682-685 (2010).
- Leutert, M., Entwisle, S. W., Villén, J. Decoding post-translational modification crosstalk with proteomics. Molecular & Cellular Proteomics. 20, 100129 (2021).
- Hart, G. W., Slawson, C., Ramirez-Correa, G., Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annual Review of Biochemistry. 80 (1), 825-858 (2011).

