噬菌体转导:将氨基青霉素耐药性从捐赠者转移到受体大肠杆菌的方法
English
Share
Overview
资料来源:亚历山大·金1,托尼娅·科尔皮茨1
1波士顿大学医学院微生物学系,国家新兴感染疾病实验室,波士顿,马萨诸塞州
转导是细菌之间的一种基因交换形式,它利用噬菌体或噬菌体,一种完全感染原核生物的病毒。这种形式的DNA转移,从一种细菌到另一种细菌通过噬菌体的方式,发现于1951年由诺顿·津德和约书亚·莱德勒格,谁称这个过程”转导”(1)。1915年,英国细菌学家弗雷德里克·特奥尔首次发现噬菌体,1917年由法裔加拿大微生物学家费利克斯·德赫勒(Felix d’Herelle)再次独立发现。自那时以来,这些噬菌体的结构和功能被广泛使用特征(3),将这些噬菌体分为两类。第一类是赖舍噬菌体,感染后在宿主细菌内繁殖,破坏细菌代谢,使细胞变流,并释放后代噬菌体(4)。由于这种抗菌活性和抗生素耐药细菌的日益流行,这些溶性噬菌体最近被证明是有用的抗生素替代治疗。第二类是溶源噬菌体,它可以通过溶源循环在宿主中繁殖,或者进入一种静止状态,其中它们的基因组被集成到宿主的基因组中(图1),这个过程称为溶质,具有噬菌体的能力生产在后世的诱导(4)。
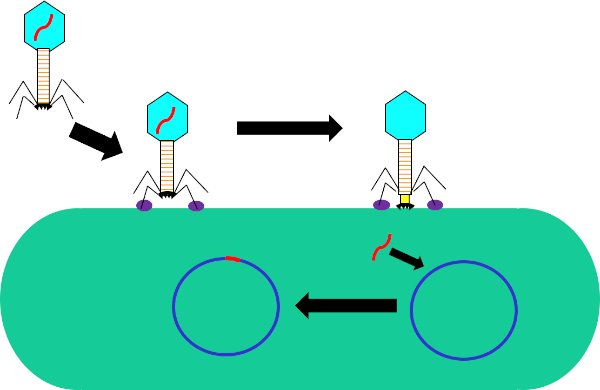
图1:噬菌体感染宿主细胞。通过尾纤维和受体(紫色)之间的相互作用,通过噬菌体吸附到细菌细胞壁。一旦在细胞表面,噬菌体是不可逆转地连接到细菌细胞使用底板(黑色),这是由收缩护套(黄色)移动到细胞壁。噬菌体基因组(红色)然后进入细胞并集成到宿主细胞基因组中。
虽然细菌转导是一个自然发生的过程,但利用现代技术,它纵在实验室环境中将基因转移到细菌中。通过将感兴趣的基因插入基源噬菌体的基因组(如噬菌体)中,能够将这些基因转移到细菌的基因组中,从而在这些细胞中表达这些基因。虽然其他基因转移方法,如转化,使用质粒进行基因转移和表达,但将噬菌体基因组插入受体细菌中不仅有可能赋予这种细菌新的特性,而且还允许自然发生的突变和细胞环境的其他因素,以改变转移基因的功能。
与其他水平基因转移方法(如共偶)相比,转导在供体和受体细胞所需的标准上相当灵活。任何可以适应所使用噬菌体基因组的遗传元素都可以从任何供体细菌菌株转移到任何受体细菌菌株,只要两者都允许噬菌体,需要表达必要的噬菌体受体。细胞表面。一旦这个基因从供体基因组中移出并打包到噬菌体中,就可以转移到受体。转导后,有必要为含有感兴趣的基因的受体细菌选择。这可以通过使用基因标记,如FLAG标记或多血氨酸标记,来标记感兴趣的基因,或基因的内在功能,在编码抗生素耐药性的基因的情况下。此外,PCR还可用于进一步确认成功的转导。通过使用感兴趣的基因中的一个区域的引体并将信号与阳性对照、具有感兴趣基因的细菌和阴性对照的细菌进行比较,细菌经历了与无噬菌体的转导反应相同的步骤。虽然细菌转导是分子生物学的有用工具,但它在细菌的进化中已经并将继续发挥重要作用,特别是在最近抗生素耐药性的上升方面。
在本实验中,细菌转导用于通过P1噬菌体(5)将抗生素氨基青霉素的抗药性基因编码从大肠杆菌的W3110菌株转移到J53菌株。这个实验包括两个主要步骤。首先,从供体菌株制备含有甲异素抗性基因的P1噬菌体。第二,通过P1噬菌体转导将该基因转移到受体菌株(图1)。一旦进行,环青素抗性基因的成功转移可以通过qPCR确定(图2)。如果转导成功,大肠杆菌的J53菌株对氨基青霉素具有抗药性,而该基因可由qPCR检测出这种抗性。如果不成功,将不会检测到阿霉素耐药性基因,而阿霉素仍将作为对抗J53菌株的有效抗生素。
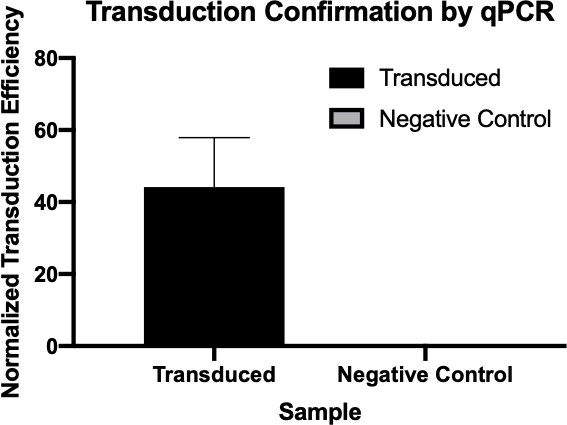
图2:qPCR成功转导的确认。通过比较从转导反应和阴性对照反应中检测到的感兴趣的基因的Cq值,并将这些值与内务管理基因规范化,从而证实细菌转导是成功的。
Procedure
Applications and Summary
The transfer of genes to and from bacteria by bacteriophage, while a natural process, has proved extremely useful for a multitude of research purposes. While other methods of gene transfer such as transformation and conjugation are possible, transduction uniquely uses bacteriophages; not only allowing for gene integration into the host genome, but also for gene delivery to multiple bacteria that are not susceptible to other methods. This process, while especially useful in the laboratory, has also been used in the recently emerging field of gene therapy, more specifically in alternative gene therapy, a therapeutic strategy that utilizes bacteria to deliver therapeutics to target tissues, many of which are not susceptible to other delivery methods and have much clinical relevance (8,9).
References
- Lederberg J, Lederberg E.M., Zinder, N.D., et al. Recombination analysis of bacterial heredity. Cold Spring Harbor symposia Quantitative Biol. 1951;16:413-43.
- Duckworth DH. "Who Discovered Bacteriophage?". Bacteriology Reviews. 1976;40:793-802.
- Yap ML, Rossman, M.G. Structure and Function of Bacteriophage T4. Future Microbiol. 2014;9:1319-27.
- Sulakvelidze A, Alavidze, Z., Morris, J. G. Bacteriophage Therapy Antimicrobial Agents and Chemotherapy 2001;45(3):649-59.
- Moore S. Sauer:P1vir phage transduction 2010 [Available from: https://openwetware.org/wiki/Sauer:P1vir_phage_transduction].
- Kobayashi A, et al. Growth Phase-Dependent Expression of Drug Exporters in
- Escherichia coli and Its Contribution to Drug Tolerance. Journal of Bacteriology. 2006;188(16):5693-703.
- Rocha D, Santos, CS, Pacheco LG. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685-93.
- Pálffy R. et al. Bacteria in gene therapy: bactofection versus alternative gene therapy. Gene Ther. 2006 13:101-5.
- O'Neill JM, et al. Intestinal delivery of non-viral gene therapeutics: physiological barriers and preclinical models. Drug Discovery Today. 2011;16:203-2018.
Transcript
Bacteria can adapt quickly to a fast-changing environment by exchanging genetic material and one way they can do this is via transduction, the exchange of genetic material mediated by bacterial viruses. A bacteriophage, often abbreviated to phage, is a type of virus that infects bacteria by first attaching to the surface of the host and then injecting its DNA into the bacterial cell. It then degrades the host cell’s own DNA and replicates its viral genome, whilst hijacking the cell’s machinery to synthesize many copies of its proteins. These phage proteins then self-assemble and package the phage genomes to form multiple progeny. However, due to the low fidelity of the DNA packaging mechanism, occasionally, the phage packages fragments of bacterial DNA into the phage capsid. After inducing the lysis of the host, the phage progeny are released and, once such a phage infects another host cell, it transfers the DNA fragment of its previous host. This can then recombine and become permanently incorporated into the new host’s chromosome, thereby mediating gene transfer between the two bacteria.
To carry out phage transduction in the laboratory requires a donor strain that contains a gene of interest, a recipient strain that lacks it, a phage that can infect both the strains, and a method to select the transduced bacteria. In most cases, this will be a selective solid growth media that supports the growth of transduced bacteria but inhibits the growth of non-transduced ones. To begin, the donor strain that contains the gene of interest is cultured in a liquid growth medium. When all the bacteria are actively dividing in the log phase of their growth, the culture is inoculated with the target phage. After three to four hours of incubation, when nearly all the bacteria have lysed and released the phage particles, the donor phage lysate is inoculated into a freshly grown culture of the recipient bacterial strain. After a brief incubation of one hour, the culture should now contain a mixture of transduced and non-transduced bacterial cells and this is screened for the transduced cells by spreading a fraction of the suspension onto an appropriate selective solid growth media. Upon further incubation, the transduced cells should grow and multiply to yield visible colonies. These colonies can then be selected for further analysis using a variety of methods to further confirm successful transduction, such as colony PCR, DNA sequencing, or quantitative PCR.
Before starting the procedure, put on any appropriate personal protective equipment, including a lab coat and gloves. Next, sterilize the workspace with 70% ethanol and wipe down the surface.
After this, prepare three one-milliliter aliquots of LB salt solution. Now, prepare a donor strain culture by adding 100 microliters of E. coli to a 15 milliliter conical vial containing five milliliters of LB growth medium with 500 micrograms of ampicillin. Then, grow the culture overnight at 37 degrees Celsius with aeration and shaking at 220 rpm. The next day, wipe down the bench top with 70% ethanol before removing the culture from the shaking incubator. Next, dilute the overnight culture one to 100 by adding 10 microliters of donor strain to 990 microliters of fresh LB supplemented with salt solution.
Allow the bacterial dilution to grow at 37 degrees Celsius for two hours with aeration and shaking at 220 rpm. Once the cells have reached early log phase, remove the culture from the incubator, add 40 microliters of P1 phage to the culture and incubate again. Continue to monitor the cells for one to three hours until the culture has lysed. Next, add 50 to 100 microliters of chloroform to the lysate and mix by vortexing. Then, centrifuge the lysate to remove debris and transfer the supernatant to a fresh tube. Add a few drops of chloroform to the supernatant and store it at four degrees Celsius for no more than one day.
To begin the transduction procedure, obtain a one milliliter culture of recipient strain. Next, transfer 100 microliters of donor phage lysate into a 1.5 milliliter microcentrifuge tube and incubate it at 37 degrees Celsius with the cap open for 30 minutes to allow any remaining chloroform to evaporate. While the donor phage lysate incubates, pellet the recipient strain cells via gentle centrifugation. Discard the supernatant and resuspend the cell pellet in 300 microliters of fresh LB containing 100 millimolar magnesium sulfate and five millimolar calcium chloride.
Next, set up the transduction reaction by combining 100 microliters of the recipient strain and 100 microliters of the donor phage lysate in a microcentrifuge tube. Then, set up the negative control by combining 100 microliters of the recipient strain and 100 microliters of the LB with magnesium sulfate and calcium chloride. After incubation, add 200 microliters of one molar sodium citrate and one milliliter of LB to both tubes, and mix by gently pipetting up and down. Then, after the tubes have been incubated for an hour, gently pellet the cells via centrifugation.
After centrifuging, discard the supernatant and resuspend the pelleted cells in 100 microliters of LB with 100 millimolar sodium citrate. Vortex the solutions and pipette the entire transduced sample onto an LB agar plate with 1X ampicillin. Finally, pipette the entire volume of the negative control cell mixture onto an LB agar plate without ampicillin. After incubating the plates overnight at 37 degrees Celsius, use a sterile pipette tip to pick three to four colonies from the transduction plate and streak them onto a new LB agar plate containing 1X ampicillin and 100 microliters of one molar sodium citrate. Repeat this plating method for the negative control on another LB agar plate containing only 100 microliters of one molar sodium citrate. Then, incubate the plates at 37 degrees Celsius overnight to allow colonies free of phage to grow.
The next day, wipe down the bench top with 70% ethanol before removing your plates from the incubator. Using a sterile pipette tip, pick three colonies from the transduction plate and add them each to a separate tube containing five milliliters of LB media. Then, select three colonies from the negative control plate and add them to another tube containing five milliliters of LB media. Grow the cultures overnight at 37 degrees Celsius with aeration and shaking at 220 rpm. After sterilizing the bench top as previously demonstrated, use a DNA miniprep kit to isolate DNA from 4.5 milliliters of each culture according to the manufacturer’s instructions. Then, elute the DNA with 35 microliters of nuclease-free water and measure the resulting concentration by lab spectrophotometer. Finally, prepare glycerol stocks by adding the remaining 0.5 milliliters of both bacterial solutions to 0.5 milliliters of 100% glycerol.
To confirm transduction, first prepare two qPCR master mixes for 24 qPCR reactions. For the first master mix, add 150 microliters of qPCR buffer mix to a microcentrifuge tube and 12 microliters each of a forward and reverse primer designed to amplify the ampicillin resistance gene. Next, prepare a second qPCR master mix by adding 150 microliters of qPCR master mix to a microcentrifuge tube and then adding 12 microliters each of a forward primer and reverse primer designed to amplify a housekeeping gene.
For each qPCR reaction, combine 100 micrograms of experimental DNA from each reaction with 14.5 microliters of qPCR master mix. Now, prepare the remaining reactions as previously demonstrated. Transfer the reactions to a thermocycler preheated to 94 degrees Celsius and then initiate the program. Finally, use the cycle quantification, or Cq, values generated by qPCR to calculate the normalized transduction efficiency of the ampicillin resistance gene.
The cycle quantitation, or Cq, values for the genes of interest were tabulated for each of the negative controls and transduced samples. Low Cq values, typically below 29 cycles, like the transduced samples in this example indicate high amounts of the target sequence.
A housekeeping gene, also tabulated here, is used as a loading control to normalize the amount of DNA in each reaction and as a positive control to ensure the qPCR is working. Provided the same amounts of the housekeeping gene are loaded, it is found at relatively the same rate in each sample.
Next, to calculate the delta Cq value for each sample, subtract the Cq value of the housekeeping gene for each sample from the Cq value of its corresponding target gene. For example, the delta Cq of the first negative control is 13.54. Then, use this value to calculate the normalized transduction efficiency of each sample using the formula shown here. Finally, the average normalized transduction efficiency for each sample group can be calculated.
