A Protocol for Immunohistochemistry and RNA In-situ Distribution within Early Drosophila Embryo
Summary
Here, we describe a protocol for detection and localization of Drosophila embryo protein and RNA from collection to pre-embedding and embedding, immunostaining, and mRNA in situ hybridization.
Abstract
Calcium induced calcium release signaling (CICR) plays a critical role in many biological processes. Every cellular activity from cell proliferation and apoptosis, development and ageing, to neuronal synaptic plasticity and regeneration have been associated with Ryanodine receptors (RyRs). Despite the importance of calcium signaling, the exact mechanism of its function in early development is unclear. As an organism with a short gestational period, the embryos of Drosophila melanogaster are prime study subjects for investigating the distribution and localization of CICR associated proteins and their regulators during development. However, because of their lipid-rich embryos and chitin-rich chorion, their utility is limited by the difficulty of mounting embryos on glass surfaces. In this work, we introduce a practical protocol that significantly enhances the attachment of Drosophila embryo onto slides and detail methods for successful histochemistry, immunohistochemistry, and in-situ hybridization. The chrome alum gelatin slide-coating method and embryo pre-embedding method dramatically increases the yield in studying Drosophila embryo protein and RNA expression. To demonstrate this approach, we studied DmFKBP12/Calstabin, a well-known regulator of RyR during early embryonic development of Drosophila melanogaster. We identified DmFKBP12 in as early as the syncytial blastoderm stage and report the dynamic expression pattern of DmFKBP12 during development: initially as an evenly distributed protein in the syncytial blastoderm, then preliminarily localizing to the basement layer of the cortex during cellular blastoderm, before distributing in the primitive neuronal and digestion architecture during the three-gem layer phase in early gastrulation. This distribution may explain the critical role RyR plays in the vital organ systems that originate in from these layers: the suboesophageal and supraesophageal ganglion, ventral nervous system, and musculoskeletal system.
Introduction
Calcium induced calcium release signaling (CICR) plays critical role in many biological processes, such as skeletal/smooth muscle and cardiac vascular function, cell proliferation and apoptosis, development, aging, neuronal synaptic plasticity, and regeneration1,2,3,4,5,6. Ryanodine receptors (RyRs) and inositol 1,4,5-trisphosphate receptors (IP3Rs) are major players in the calcium signaling pathway controlled by their regulators protein kinase A (PKA), Ca2+/calmodulin-dependent protein kinase II (CaMKII), FK506 binding proteins (FKBPs), calsequestrin (CSQ), triadin, and junctin1,2,3,4,5,6. Abnormal human expression and mutations in these proteins can lead to pathological physiology such as arrhythmias7 and oncogenic proliferation8,9.
FKBPs regulate the calcium release from the endoplasmic reticular (ER) by RyRs. This process is essential for the mechanism of contraction, and thus responsible for all mechanical movement generated by myosin-actin contraction through calcium-induced calcium release along with embryonic RyRs1,2. In mouse models, the lack of RyR2 and its regulator FKBP12/Calstabin is invariably lethal, either during embryonic development or in the early postnatal period10,11,12. FKBP12/Calstabin knockout mice exhibit critical cardiac defects with irregular excitation- contraction coupling (EC) and cerebral edema during embryonic development. This indicates that FKBP12/Calstabin plays an essential role in regulating RyR2 channel expression, which is important both to cardiac and cerebral development10.
RyR conducted calcium sparks were initially discovered in the zygote formation phase of fertilized Medaka eggs13,14. However, few investigations have been performed on the function of calcium signaling in early embryonic development. In Drosophila melanogaster, results obtained from DmFKBP12 S107 mutants provide strong evidence supporting the importance of this gene for larval development and a healthy life span, which is attributed to its function against oxidative stress15,16. Recently, we identified the dynamic localization of FKBP12/Calstabin protein and messenger RNA during early Drosophila melanogaster development17. Using the approaches described in this methodology, we were able to track the expression of FKBP12/Calstabin in D. melanogaster during the syncytial blastoderm (0-2 h), cellular blastoderm (2-3 h), early gastrula (3-12 h), and late gastrula (12-24 h). In this paper, we present the detailed protocols of every approach in the previous study, including pre-embryo embedding for classic paraffin sectioning, pre-coating slide treatment for embryonic sections, histo-chemistry staining and immunostaining, and mRNA in-situ hybridization for identification of gene expression.
Protocol
1. Preparation of grape juice agar plates
- Add 5 g of agar and 5 g of sucrose to 150 mL of distilled water. Boil it using a microwave until the agar and sucrose are fully dissolved. Mix 50 mL of 100% grape juice and the solution together.
- Add 1 mL of 100% propionic acid to make the final concentration to 0.5% propionic acid. Pour 25 mL of the prepared solution into each plate. After the agar has solidified, store the media in 4 °C.
2. Coating slides
- Pretreat slides with detergent. Wash the slides 3 times with tap water and once with distilled water. Then dry the slides in a 45 °C oven for 2 h.
- Immerse the slides in about 450 mL of potassium dichromate sulfuric acid solution (20 g of potassium dichromate, 40 mL of distilled water, and 400 mL of concentrated sulfuric acid) for 24 hours. Wash the slides 3 times with tap water and once with distilled water.
- Dry the slides in a 45 °C oven for two hours. Immerse in 95% ethanol for more than 2 h.
- Add 0.5 g of chromium potassium sulfate and 5 g of gelatin to 1 L of distilled water. Dissolve in a 60 °C water bath before use. The chrome alum gelatin can be stored at 4 °C for long periods.
- Drop 10 µL of the chrome alum gelatin on the slide and fully and evenly cover the entire surface of the slide with the solution. Place the slide with the gelatin side up in an oven at 42 °C for 48 h. The prepared slides can be stored at 4 °C for later use.
NOTE: This is a critical step. The complete coating is necessary. For fruit fly and its embryo, this slide coating approach appears more efficient than poly-lysine coating.
3. Drosophila embryo embedding
- Drosophila embryo collection
NOTE: We collected Drosophila embryos in four periods of 0-2 hours (syncytial blastoderm), 2-3 hours (cellular blastoderm), 3-12 hours (early gastrula) and 12-24 hours (late gastrula)18. The proportion of the main embryo types in each period is shown in Table 1.- Place 400-500 flies in a plastic collection bottle and cover the bottle with the grape juice agar plate containing yeast extract. Drosophila embryos are laid on the grape juice agar at room temperature.
- Collect four periods of embryos, gently adding 2 mL of PBS (137 mM NaCl, 2.7 mM KCL, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) to each plate to float the embryos. Transfer about 200 embryos to a 50 mL centrifuge tube and keep it on ice for 2 min to settle down the embryos.
- Pre-embedding
- Transfer 200 Drosophila embryos to a 1.5 mL centrifuge tube. Keep the tube on ice for 2 min to settle down the embryos, and then discard the supernatant with pipette carefully. Add 1 mL of 50% bleach in the tube and shake it gently for 2 min.
NOTE: The bleach should prepare fresh, and the amount of the bleach could vary depending on the amount of embryos collected. - Gently collect embryos by pour on filter paper and wash them 3 times with 1 mL of PBS at each time.
- Transfer the embryos with filter paper to about 5 mL of n-heptane.
- Transfer the mixture to a fresh 1.5 mL centrifuge tube. Add 1 mL of methanol and shake it gently. Collect embryos after sedimentation.
- Remove supernatant and wash the embryos in 1 mL of PBS 3-5 times.
- Fix the embryos with 400 µL of Bouin's solution (71% saturated picric acid, 24% formalin, 5% acetic acid) for 30 min. Wash the fixed embryos 3 times in 1 mL of PBS for 5 min each time.
- To aggregate embryos, transfer the samples into a rubber stopper and remove the PBS.
- Prepare 1.2% agarose-PBS solution and keep solution at 60 °C. Add 200 µL of 1.2% agarose-PBS solution onto embryos to fix them into an agar block.
- Take the block out from stopper and store the block in PBS.
NOTE: During pre-embedding approach, pro-warming is critical for successful. We strongly suggest that the centrifuge tubes and micropipette tips, which will be used in this step should be completely pre-warm before operating.
- Transfer 200 Drosophila embryos to a 1.5 mL centrifuge tube. Keep the tube on ice for 2 min to settle down the embryos, and then discard the supernatant with pipette carefully. Add 1 mL of 50% bleach in the tube and shake it gently for 2 min.
- Paraffin embedding
- Immerse the pre-embedded agar block in 35% ethanol, 50% ethanol, 75% ethanol, and 85% ethanol for 15 min each. Then dip 2 times in 95% ethanol and 100% ethanol.
NOTES: About 5-10x of block volume of each followed by a graded fixative for 15-30 minutes is necessary depending on number of embryos in the block. - Immerse the embryos block in 100% ethanol and xylene (1:1 ratio) solution for 15 min.
- Transfer the block to xylene for 1 min to make the embryo tissue transparent.
- Submerge the block in xylene and paraffin (1:1 ratio) for 30 min.
- Submerge the block in paraffin I, paraffin II, and paraffin III for 2 h each time.
- Fill the embedding box with paraffin beforehand, and then gently and horizontally transfer the block into the bottom of embedding box. After the paraffin solidify naturally, the solidified paraffin block can be stored at 4 °C.
NOTES: The pre-embedded sample can be used in cryosection by following the following steps. After the embryos are pre-embedded with agarose as described above, the samples are simply embedded in OCT Compound. Afterwards, cryosection can be carried out according to routine cryosection protocol. Subsequently, regular staining approaches can be used on the sections.
- Immerse the pre-embedded agar block in 35% ethanol, 50% ethanol, 75% ethanol, and 85% ethanol for 15 min each. Then dip 2 times in 95% ethanol and 100% ethanol.
4. Hematoxylin-eosin staining
- Prepare Drosophila embryo sections. Place prepared sections in an oven at 60 °C for 15 min and then submerge the sections in xylene 2 times for 10 min each. Submerge the sections once in 100% ethanol and xylene (1:1 ratio) for 2 min.
- Hydrate the sections in 100% ethanol I, 100% ethanol II, 95% ethanol, 85% ethanol, 75% ethanol, 50% ethanol, and in distilled water for 3 min.
- Stain the sections with hematoxylin solution for 2 min. Dip the slides in 1% acid alcohol solution (1 mL of hydrochloric acid, 100 mL of 70% ethanol) quickly and wash the slides in distilled water.
- Dip the slides in 50% ethanol, 75% ethanol, 85% ethanol, and 95% ethanol sequentially.
- Stain the sections with eosin solution for 1 min.
- Dehydrate the slides in 95% ethanol I, 95% ethanol II, 95% ethanol III, 100% ethanol I, 100% ethanol II and 100% ethanol III for 1 min each time.
- Clear the slides in 100% xylene I, 100% xylene II, and 100% xylene III for 5 min each time.
- Mount the slides with neutral balsam according to manufacturer's instructions.
5. Periodic acid-silver methenamine staining
- Deparaffinize and hydrate paraffin sections of Drosophila embryo to distilled water.
- Oxidize the sections in 1% periodic acid for 15 min at room temperature.
- Rinse the slides in distilled water 3 times for 1 min each.
- Incubate the slides in about 40 mL of freshly filtered silver methenamine working solution (3% methenamine, 5% silver nitrate, 5% sodium borate solution) for 1 h and 20 minutes at 60 °C. Then rinse the slides in distilled water for 1 min.
- Tone the slides in 0.2% gold chloride for 2 min and then rinse the slides in distilled water for 1 min.
- Place the slides in 3% sodium thiosulfate for 3 min and then rinse the slides in distilled water for 1 min.
- Counterstain the slides in hematoxylin for 30 s. Wash the slides in running tap water for 3-5 min.
- Dehydrate the slides in 70% ethanol, 80% ethanol, 95% ethanol I, 95% ethanol II, 95% ethanol III, 100% ethanol I, and 100% ethanol II for 5 min each.
- Clear the slides in 100% xylene I, 100% xylene II and 100% xylene III for 5 min each.
- Mount the slides in neutral balsam according to manufacturer's instructions.
6. Immunohistochemistry
- Deparaffinize and rehydrate paraffin sections of Drosophila embryo to PBS following the standard protocol19.
- Treat slides with 0.3% H2O2 in methanol for 10 min at room temperature and avoid light.
- Pre-heat the appropriate antigen retrieval buffer to approximate 100 °C in a container with the slides in the vegetable steamer. The container should also have a lid. Put the slides in the container. Close the lid of the steamer.
- Keep the container in the steamer for 20 minutes from this point. Note that the lid of the slide container should not be entirely tightened so water vapor can enter during the steaming process.
- Allow the slides return to room temperature before gently rinsing it in PBS-T (PBS with Tween-20, pH 7.2) 3 times for 5 min each, and then rinse the slides in PBS 3 times for 1 min each.
- Block the slides with 1% bovine serum albumin (BSA) in PBS for 60 min at room temperature.
- Incubate the slides with primary antibody (anti-FKBP12 at 1:1000) overnight at 4 °C or at room temperature for 4 h.
- Wash the slides in PBS-T 4 times for 5 min each.
- Apply the slides with HRP labeled polymer conjugated with secondary antibodies (anti-Rabbit, 1:5,000) for 90 min at room temperature. Do this in the dark to avoid photo bleaching.
- Wash the slides in PBS-T 3 times for 5 min each.
- Incubate the slides with 5% 3,3'-diaminobenzidine-tetrahydrochloride (DAB) for 2-10 min until the color of reaction products is appropriate under microscope.
- Counterstain the slides in hematoxylin for 30 s and wash the slides in distilled water 2 times for 5 min each.
- Dehydrate the slides in 50% ethanol, 70% ethanol, 90% ethanol, 95% ethanol, and 100% ethanol for 1 min each.
- Clear the slides in 100% xylene I, 100% xylene II and 100% xylene III for 5 min each.
- Mount the slides in neutral balsam according to manufacturer's instructions.
7. In-situ hybridization
NOTE: Drosophila embryo sections were prepared with 0.01% diethyl pyrocarbonate (DEPC) treated water. All accessories used for in situ hybridization apply DEPC overnight treatment in advance.
- Riboprobe preparation
- Linearize template DNA by cutting at a restriction enzyme site downstream from the cloned insert using a restriction enzyme that creates 5' overhangs. After linearization, purify the template DNA via phenol/chloroform extraction, and subsequent ethanol precipitation. Use standard protocols.
- Add 1 µg of purified template DNA to a RNase-free centrifuge tube. Place the centrifuge tube on ice. Add enough DEPC-treated water to the total volume 13 µL.
- Then add the following reagents in order: 2 µL of 10x NTP labeling mixture, 2 µL of 10x transcription buffer, 1 µL of protector RNase inhibitor, 2 µL of RNA Polymerase SP6 or RNA Polymerase T7.
- Mix the reaction by pipetting and centrifuge shortly. Incubate for 2 h at 37 °C.
NOTE: Longer incubations do not increase the yield of labeled RNA. A short spin (~800 rpm) applied on the reaction tube is critical necessary before continuing to the next step. This spin will allow all contents, which could generate from the incubation, to come to the bottom of the tube. - Add 2 µL of RNase-free DNase I to remove template DNA and incubate for 15 min at 37 °C.
- Add 2 µL of 200 mM EDTA (pH 8.0) to stop the reaction.
NOTE: DIG-labeled RNA probes can be stored at -15 to -25 °C in ethanol for one year.
- Pre-hybridization of Drosophila embryo section
- Place sections of Drosophila embryo in a 42 °C oven for 48 h.
- Deparaffinize the slides in 100% xylene for 10 min twice.
- Hydrate the slides by sequentially submerging in 100% ethanol, 95% ethanol, 90% ethanol, 70% ethanol, and 50% ethanol for 5 min.
- Incubate the slides in PBS for 5 min 3 times to remove the fixative solution/Bouin's solution from the tissue.
- Wash the slides with 100 mmol/L glycine/PBS solution 2 times for 5 min each.
- Wash the slides with PBS 2 times for 5 min each.
- Incubate the slides with 0.3% Triton X-100 in PBS for 15 min to permeabilize the membranes.
- Wash the slides with PBS 3 times for 5 min each.
- Incubate the slides in 15 µg/mL RNase-free proteinase K for 30 minutes at 37 °C.
- Incubate the slides with 4% paraformaldehyde in PBS for 3 min to stop the digestion.
- Wash the slides with PBS 2 times for 5 min each.
- Incubate the slides in 100 mM triethanolamine buffer (pH 8.0) with 0.25% (v/v) acetic anhydride 2 times for 5 min each.
- Add DEPC water into the slide moisture chamber. Add 20 µL of pre-hybridization solution (containing 100 µg/mL of salmon sperm DNA) on the slides. Put the slides into the slide moisture chamber. Incubate the slides at 42 °C for 4 h.
NOTE: It is necessary to ensure the tightness of the lid of the wet box and that the wet box is always kept in a horizontal to prevent volatilization and deviation from the tissue of the pre-hybridization solution and the followed hybridization solution.
- Hybridization of Drosophila embryo section
- Remove the pre-hybridization solution and wash the slides with 0.2x SSC (150 mM sodium chloride, 15 mM sodium citrate, pH 7.0) 3 times for 5 min each. Wipe off excess 0.2x SSC around the tissue samples.
- Apply 30 µL of probe hybridization solution (containing 2-6 ng of digoxin-labeled RNA probe by diluting in pre-hybridization solution) with a pipette. Incubate the slides in the slide moisture chamber at 42 °C for approximately 20 h.
NOTE: Be sure on the cap-tightness as the suggestion of the last note. Otherwise, the dry-up caused from the leakage of either pre-hybridization or pre-hybridization will generate dirty background including pseudo-positive in slides.
- Hybridization
- Remove the hybridization solution and wash the slides with 4x SSC solution 4 times for 15 min each at 37 °C.
- Wash the slides in 2x SSC solution with 20 µg/mL of RNase A for 30 min at 37 °C.
- Wash the slides in 1x SSC solution for 15 min at 37 °C.
- Wash the slides in 0.5x SSC solution for 15 min at 37 °C.
- Wash the slides in 0.1x SSC solution for 15 min at 37 °C.
- Wash the slides with PBS 3 times for 5 min each.
- Wash the slides in washing buffer (100 mM maleic acid, 150 mM NaCl, 0.3% (v/v) Tween 20, pH 7.5) for 1 min.
- Incubate the slides in 100 mL of freshly prepared blocking solution (2% sheep serum in maleic acid buffer without Tween 20) for 30 minutes.
- Incubate the slides in 20 mL of antibody solution for 45 minutes.
- Centrifuge the antibody for 5 min at 10,000 rpm in the original vial prior to each use, and pipet the necessary amount carefully from the surface. Dilute anti-digoxigenin-AP 1:5000 (150 mU/mL) in blocking solution.
- Wash the slides in 100 mL of washing buffer 2 times for 15 minutes each to remove unbound antibody conjugate.
- Equilibrate the slides in 20 mL of detection buffer (100 mM Tris-HCl, 100 mM NaCl, pH 9.5) for 2-5 min.
- Incubate the slides with 10 mL of freshly prepared color substrate solution in the slide moisture chamber. The reaction begins within a few minutes and is complete after 16 h. Do not shake or mix while color is developing.
- Wash the slides with 50 mL of distilled water for 5 min to stop the reaction.
- Dry the sample overnight in the dark.
- Dehydrate the slides in 70% ethanol, 80% ethanol, 90% ethanol, 95% ethanol, 100% ethanol I and 100% ethanol II for 3 min each.
- Clear the slides in 100% xylene I, 100% xylene II and 100% xylene III for 20 min each.
- Mount the slides with neutral balsam.
- Document images and analyze with the inverted microscopy and manufacturer software. Perform distribution analysis using ImageJ according to density of positive signal either along longitudinal axis or within differential zone of embryos.
NOTE: During the incubation with the freshly prepared color substrate solution in the step 7.4.12, the reaction color can be seen even with a bare eye. This observation on how color developing can be inspected under stereoscope. Once reaction color is reasonable and the step 7.4.13 can be carried out to stop the reaction.
Representative Results
The figures describe protocols used to overcome the challenge of attaching high lipid and chitin-containing chorion Drosophila embryos (Table 1) to the glass slide surface for examination and experimentation. Utilizing the chrome alum gelatin slide-coating method shown in Figure 1, we enhanced the attachment of Drosophila embryos onto the surface of slides while the embryo pre-embedding method shown in Figure 2 allows for efficient inspection of the dynamic distribution of protein and RNA DmFKBP12/Calstabin in all four early development stages (i.e., syncytial blastoderm, cellular blastoderm, early and late gastrula of the Drosophila embryos). Figure 3 shows the protein distribution of antibody detected FKBP12 / Calstabin at the syncytial and cellular blastoderm stages. From these histochemical results, it can be seen that FKBP12 expression is less polarized in syncytial blastoderm (Panels A to H), and then begins to localize within the basement membrane underneath the trophectoderm epithelium to form a gradient with more expression found in the anterior than the posterior poles (Panels I to S). Eventually, the DmFKBP12/Calstabin protein becomes restricted to certain architectural components of the primitive embryonic tissues with less extracellular distribution than that observed in early and late gastrula embryonic stages (Figure 4). Interestingly, the above-described dynamic protein distribution is not accompanied by corresponding mRNA levels of DmFKBP12 (Figure 5), indicating protein expression to be the result of a still unknown post-transcription molecular mechanism. The described method of CAG slide-coating and pre-embedding will allow us to further probe the importance of DmFKBP12 in calcium signaling and unearth this still unknown molecular mechanism while combining other techniques such as micro-injection RNA interference19.
In this study, we develop a chrome alum gelatin slide-coating and pre-embedding method for histology, histochemical and immune-histological analysis of Drosophila embryos at different development stages. Using this pre-embedding method, the embryos are effectively collected for both cryo- and paraffin sectioning that allows for the analysis of longitudinal cross sections. This allowed us to describe the evolving expression patterns for this protein during decisive developmental stages, linking expression of the protein to development of key physiologic systems such as the gut, the brain, and the connecting nervous system. By employing these methods, we were able to investigate the dynamic distributions of RyR regulating protein and mRNA FKBP12/Calstabin during the development of Drosophila embryos at definitive stages, including the syncytial and cellular blastoderm, and early and late gastrulation stages. Our data showed that the FKBP12/Calstabin protein and its RNA can be detected very early in the syncytial blastoderm. As the embryo develops, FKBP12 becomes more densely distributed in the basal cell layer underneath the epithelium of the blastoderm. The FKBP12/Calstabin protein then dynamically strengthens its expression and differentiates its distribution into embryonic muscle-containing tissues, including the stomodaeum, stomodaeum and posterior midgut rudiment. These distribution changes from the beginning of blastoderm to late gastrulation stages are also described in the developing neuronal system including neuroblasts, ventral nerve, supraesophageal ganglion and brain. While these dynamic changes in FKBP12/Calstabin hint at its importance in development, they are not reflected in mRNA levels of the protein, hinting at a yet unknown regulator of protein expression that remains to be identified.
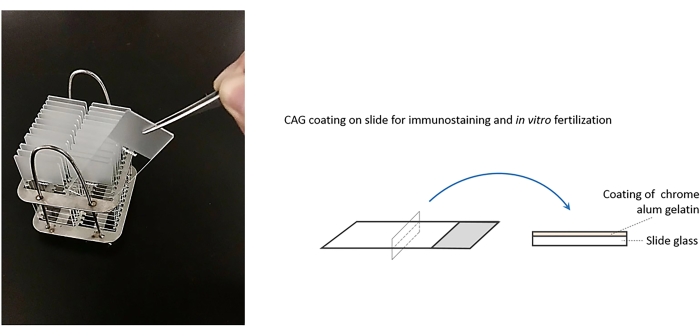
Figure 1: Sketch of chrome alum gelatin (CAG)coating method for paraffin-sections used in immunology and RNA in-situ hybridization to visualize the distribution of DmFKBP12 protein and mRNA in Drosophila embryos. Please click here to view a larger version of this figure.
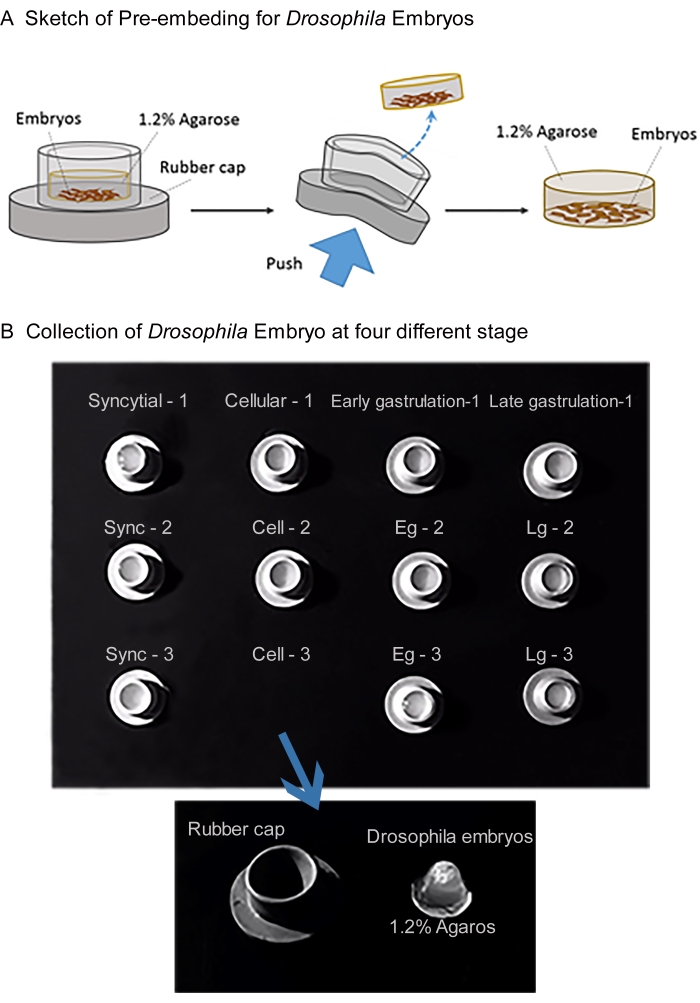
Figure 2: Sketch of pre-embedding method for expression profile of FKBP12 and DmFKBP12 in developing Drosophila embryos. Please click here to view a larger version of this figure.
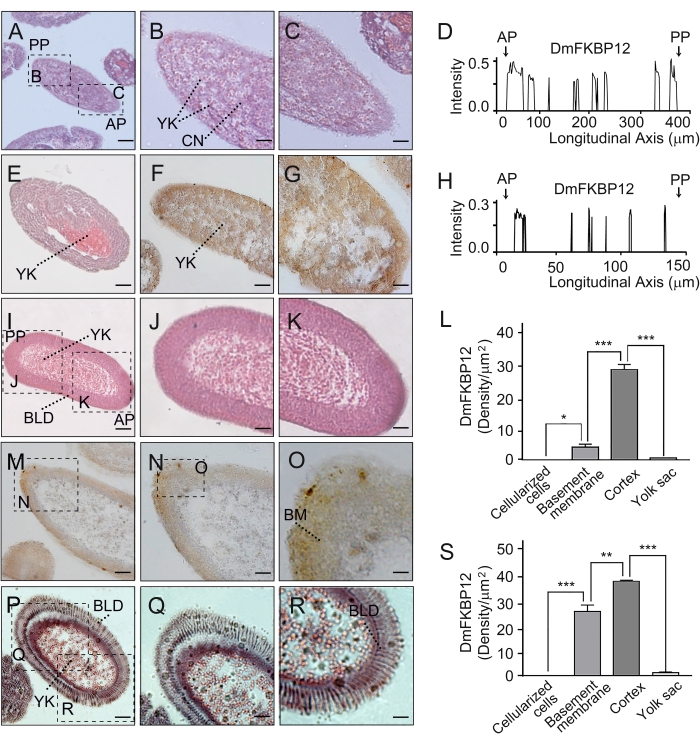
Figure 3: Expression profile of DmFKBP12 protein in syncytial and cellular blastoderms of the Drosophila embryo. (A-D) H&E staining of the syncytial blastoderm. Quantification of DmFKBP12 distribution in early syncytial blastoderm is presented in D. (E-G) Panels present early syncytial blastoderm in which nuclei are under division. Quantification of DmFKBP12 distribution in late syncytial blastoderm are presented in H. (I-L) H&E staining of the cellular blastoderm. The qualification of DmFKBP12 distribution in late cellular blastoderm is presented in L. (M-S) Panels exhibit the distribution of DmFKBP12 protein in the cellular blastoderm. O shows the larger views of M-N. (P-R) Panels present earlier cellular blastoderm shown in M-O. Quantifications of DmFKBP12 distribution in early cellular blastoderm is presented in S. Scale bar for A, E, I, M and P is 60 µm, for B, C, F, J, K, N, Q and R is 40 µm, and for G, O is 20 µm. AP, anterior pole of egg; YK, yolk; CN, cleavage nucleus; PP, posterior pole of egg; BLD, blastoderm, nuclei and cell; BM, Basement membrane. Data are expressed as mean± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001. The data quantifying was carried out with ImageJ. Firstly, the brown products were remained in the image along with removing all other colors. Secondly, the brown image was changed black-and-white and calculate its density for quantification consequently. Please click here to view a larger version of this figure.
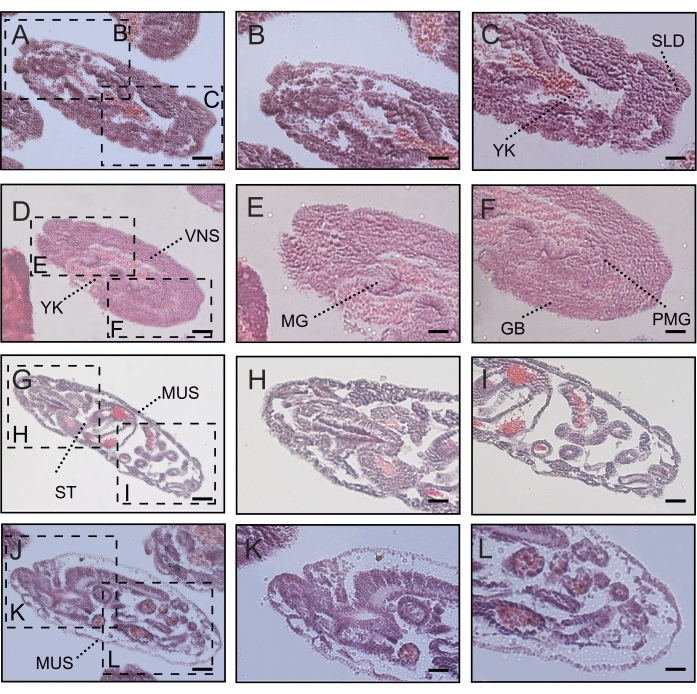
Figure 4: Expression profile of DmFKBP12 protein in early and late gastrulation stages of the Drosophila embryo. A, D and G show H&E staining of Drosophila embryo at the gastrulation stage. B, C, E and F display the distribution of DmFKBP12 at the early gastrulation stage. H, I, and J-L display the distribution of DmFKBP12 at the late gastrulation stage. Quantifications of DmFKBP12 distribution in early gastrulae are presented in M. Scale bar for A, D, E and G is 80 µm, for B, C, F, H, I and J-L is 40 µm. PP, posterior pole of egg; NBL, neuroblasts; ST, stomodaeum; YK, yolk; PMG, posterior midgut rudiment; BR, brain, supraesophageal ganglion; VNS, ventral nervous system; PV, proventriculus; MUS, muscle; MGC, midgut caecum; AMG, anterior midgut rudipighian; MMG, middle midgut; TR, trachea. Data are expressed as mean± SEM. ns, no significant; ** p < 0.01, *** p < 0.001. The data quantifying was carried out with ImageJ 1.50d by remaining product-brown and changing to black-and-white image, and following by density calculation of black-and-white as described in the previous figure legend. Please click here to view a larger version of this figure.
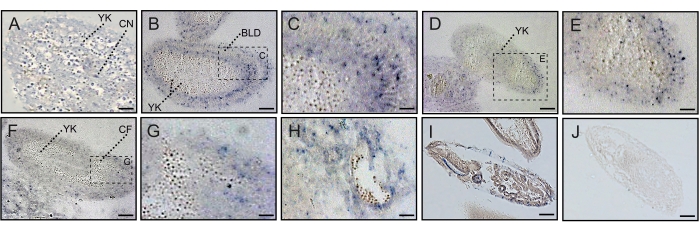
Figure 5: In-situ localization of DmFKBP12 mRNA during embryonic development of the Drosophila embryo. A shows in-situ localization of DmFKBP12 mRNA at the syncytial blastoderm stage when FKBP12 mRNA was expressed in the cytoplasm of the embryo. B and C show in-situ localization of DmFKBP12 mRNA at the cellular blastoderm stage when FKBP12 mRNA was expressed in the inner limits of blastoderm cells. D and E exhibit in-situ localization of FKBP12 mRNA at the early gastrulation stage. During this stage, it is mainly localized in the developing gut. F, G and H show in-situ localization of FKBP12 mRNA at the late gastrulation stage, when the mRNA is expressed in the muscle and gut. I is positive control of DmFKBP12 mRNA. J is negative control of DmFKBP12 mRNA. Scale bar for A, C, E, G and I is 5 µm, for B, D, F, I and J is 10 µm. YK, yolk; CN, cleavage nucleus; BLD, blastoderm, nuclei and cells; CF, anterior oblique cleft, cephalic furrow. Please click here to view a larger version of this figure.
| Embryonic stages | 0-2 hours (%) | 2-3 hours (%) | 3-12 hours (%) | 12-24 hours (%) |
| Syncytial blastoderm | 90.8 | 0 | 0 | 0 |
| Cellular blastoderm | 6.9 | 91.54 | 0 | 0 |
| Early gastrula | 2.3 | 4.23 | 91.1 | 0 |
| Late gastrula | 0 | 4.23 | 8.93 | 93.3 |
Table 1: Embryo collection of different stages in development of Drosophila melanogaster
Discussion
RyRs and IP3Rs mediated calcium signaling is a fundamental pathway in many physiological and pathological process of both vertebrate and invertebrate animals1,2,3,4. In humans, point mutations, such as CPVT-associated R4496C mutation, in the RyR2 gene lead to calcium leakage from the sarcoplasmic reticulum of cardiomyocyte, resulting in cardiac dysfunction. These mutations present as arrhythmia with a high risk of sudden cardiac death during physical activity, and is reproducible in mouse models engineered to carry this mutation. Under exercise conditions, the mice carrying this calcium-leak mutation experienced cardiac sudden death not demonstrated by its wildtype counter parts18,20,21,22. Overexpression of FKBP12, a RyRs regulator, in the heart, corresponded with high rates of potassium channels associated cardiac death in mouse models22,23, a finding that corresponded with the phenotype of FKBP12 knockout mice10,11. As mammalian RyRs including RyR1, RyR 2and RyR3 and their regulator FKBPs have been well studied for over three decades, most proceedings involved in cardiopathology, skeletal muscular dysfunction and brain function achievement after early development plus oncogenesis are the focus of research24,25.
Unlike in vertebrates, which have three isoforms of RyR, Drosophila flies have only one RyR1,2,3,5. Recently, we demonstrated the critical function of Drosophila RyR (DmRyR) in the early stages of Drosophila embryonic development. We unraveled DmRyR expression at both the translational and transcriptional level in Drosophila embryos during early development: the syncytial blastoderm stage, the blastoderm stage, and the early gastrula stages. In all four stages, DmFKBP12 mRNA was distributed evenly throughout the embryo and remained at the same level, while the protein expression of the gene was highly dynamic with expression profiles polarized anteriorly and then distributed in certain cell layers19. The methodological protocols we utilized in this publication were critical to the ability to obtain a high volume of representative results.
Paraffin sectioning is a basic and practical technique for the histological detection of protein expression for quantitative analysis. Paraffin sections are a foundational tool for studying protein expression in the adult insect. However, these techniques have been difficult to replicate in developing insects due to their lipid-rich embryo and chitin-rich chorion, which make embedding, preservation, and slide mounting difficult. The lipids found in embryos interfere with the forces allowing for attachment of embryonic sections onto the surface of the slide. While the chitin found in the chorion present physical barriers that protects the embryo from chemical and mechanical interference, it also restricts attachments of embryonic samples to glass slides. While these characteristics ensure a higher survival rate of insect larvae, it greatly hinders the ability to study their in-situ protein and mRNA expression. This problem has been troubleshot in many ways. The use of surface detergent such as Tween-20 often detaches Drosophila embryos and reduces the number of embryos available for study. This technique requires incubation for 20 hours to unveil mRNA for in-situ hybridization, but also detaches the samples from the slides almost entirely. The technique here of coating glass slides with chrome alum gelatin (CAG) before carrying out immunohistochemistry and anti-sense specific recognition for mRNA solves these technical barriers by enhancing the attachment between the embryo and the glass slide.
In addition to the CAG enhanced attachment, the described Drosophila embryo pre-embedding techniques allow the harvest of large numbers of precisely dated Drosophila embryos. This allowed us to report the dynamic expression of DmFKBP12/Calstabin at various stages of early embryonic development. The earliest stage we were able to study using this technique is the syncytial blastoderm stage, which showed less differentiated distribution and a generally lower level of expression. By studying embryos at the cellular blastoderm and gastrula stages, we found that the general level of DmFKBP12/Calstabin increased as embryos develop and begins to localize, first to the anterior poll, then to certain layers in the three-layer stage. The findings suggest that the FKBP12 involved in calcium signaling plays a critical role in early development and the differentiation of which plays an important role in Drosophila melanogaster development.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (#31771377/ 31571273/31371256), the Foreign Distinguished Scientist Program from the National Department of Education (#MS2014SXSF038), the National Department of Education Central Universities Research Fund (#GK201301001/201701005/GERP-17-45), and XZ is supported by Outstanding Doctoral Thesis Fund (#2019TS082 /2019TS079), Key Program of Shaanxi Provincial Education Department (#20JS138), the Natural Science Basic Research Program Youth Project of Shaanxi Provincial Science and Technology Department (#2020JQ-885).
Materials
| -20°C Refrigerator | Meiling Biology &Medical | DW-YL270 | Used for regent storage |
| -80°C Ultra low temperature refrigerator | Thermo | Forma 90 Series | Used for regent storage |
| Agar | Sigma-Aldrich | WXBB6360V | Preparation of grape juice agar plates |
| Anti-Digoxigenin-AP, Fab fragments | Roche | 11093274910 | For the detection of digoxigenin-labeled compound |
| Biochemical incubator | Shanghai Bluepard Instruments | LRH-250 | In-situ Hybridization |
| Bouin's solution | Sinopharm Chemical Reagent at Beijing | 69945460 | Drosophila Embryo Embedding |
| Centrifuge | Eppendorf | 540BH07808 | In-situ Hybridization |
| Centrifuge tube | Denville | C-2170 | Drosophila Embryo Collection |
| Chrome Alum | Sinopharm Chemical Reagent at Beijing | 10001018 | Coating Slides |
| Constant temperature water bath | Jintan Henfeng Instruments | KW-1000DC | Hematoxylin-Eosin Staining, Immunohistochemistry, In-situ Hybridization and Periodic Acid-Silver Methenamine Staining |
| Dako REAL EnVision Detection System | Dako | K5007 | In immunohistochemical reaction or in situ hybridization reaction, it binds to the primary antigen antibody, and the target is labeled by staining. |
| DEPC | Sigma-Aldrich | D5758 | In-situ Hybridization |
| DIG RNA Labeling Kit | Roche | 11093274910 | RNA labeling with diagoxigenin-UTP by in vitro transcription with SP6 and T7 RNA polymerase |
| Drosophila melanogaster | Bloomington Stock Center | BDSC_16799, BDSC_19894, BDSC_11664 | The stocks of Drosophila melanogaster mutant |
| Electric blast drying oven | Tianjin Taiste Instruments | 101-0AB | For coating slides and paraffin embedding |
| Eosin | Sigma-Aldrich | 230251 | Hematoxylin-Eosin Staining |
| Ethanol | Sinopharm Chemical Reagent at Beijing | 100092680 | Paraffin Embedding, Hematoxylin-Eosin Staining, Immunohistochemistry, In-situ Hybridization and Periodic Acid-Silver Methenamine Staining |
| Gelatin | Sinopharm Chemical Reagent at Beijing | 10010328 | Coating Slides |
| Gold chloride | Sigma-Aldrich | 379948 | Periodic Acid-Silver Methenamine Staining |
| Hematoxylin | Sigma-Aldrich | H3136 | Hematoxylin-Eosin Staining |
| High Pure PCR Product Purification Kit | Roche | 11732668001 | For purification of PCR products |
| Intelligent constant temperature and humidity box | Ningbo Jiangnan Instruments | HWS | For fly maintenance |
| LE Agarose | HyAgarose | 14190108029 | Pre-embedding |
| Methanol | Sinopharm Chemical Reagent at Beijing | 10014108 | Drosophila Embryo Collection |
| Microscope | ZEISS | Observer.A1 | Hematoxylin-Eosin Staining, Immunohistochemistry, In-situ Hybridization and Periodic Acid-Silver Methenamine Staining |
| Microscope Slides | MeVid Labware Manufacturing | P105-2001 | Coating Slides |
| Neutral Gum | Sinopharm Chemical Reagent at Beijing | 10004160 | Hematoxylin-Eosin Staining |
| N-heptane | Sinopharm Chemical Reagent at Beijing | 40026768 | Drosophila Embryo Collection |
| Paraffin slicer | Huahai science instrument | HH-2508III | In-situ Hybridization |
| Paraffin | Sinopharm Chemical Reagent at Beijing | 69019461 | Paraffin Embedding |
| pH/mV Meter | Sartorius | PB-10 | For determing the pH value of a solution |
| Silver nitrate | Sinopharm Chemical Reagent at Beijing | 10018461 | Periodic Acid-Silver Methenamine Staining |
| Ultrapure water meter | Thermo | AFXI-0501-P | In-situ Hybridization |
| Xylene | Sinopharm Chemical Reagent at Beijing | 10023418 | Paraffin Embedding |
References
- Weisleder, N., Ma, J. Altered Ca2+ sparks in aging skeletal and cardiac muscle. Ageing Research Reviews. 7 (3), 177-188 (2008).
- Cheng, H., Lederer, W. J. Calcium Sparks. Physiological Reviews. 88 (4), 1491-1545 (2008).
- Fan, J., et al. Ryanodine Receptors: Functional Structure and Their Regulatory Factors. Chinese Journal of Cell Biology. 37 (1), 6-15 (2015).
- Xu, X., Balk, S. P., Isaacs, W., Ma, J. Calcium signaling: an underlying link between cardiac disease and carcinogenesis. Cell & Bioscience. 8 (39), 1-2 (2018).
- Xu, X., Bhat, M. B., Nishi, M., Takeshima, H., Ma, J. Molecular cloning of cDNA encoding a Drosophila ryanodine receptor and functional studies of the carboxyl-terminal calcium Release Channel. Biophysical Journal. 78 (3), 1270-1281 (2000).
- George, G. K., et al. Comparative analysis of FKBP family protein: evaluation, structure, and function in mammals and Drosophila melanogaster. BMC Developmental Biology. 18 (1), 1-12 (2018).
- Zhou, X., et al. Syncytium calcium signaling and macrophage function in the heart. Cell & Bioscience. 8 (1), 1-9 (2018).
- Wang, L., et al. Calcium and CaSR/IP3R in prostate cancer development. Cell & Bioscience. 8 (1), 1-7 (2018).
- Xu, M., Seas, A., Kiyani, M., Ji, K. S., Bell, H. N. A temporal examination of calcium signaling in cancer- from tumorigenesis, to immune evasion, and metastasis. Cell & Bioscience. 8 (1), 1-9 (2018).
- Shou, W., et al. Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature. 391 (6666), 489-492 (1998).
- Xin, H., et al. Oestrogen protects FKBP12.6 null mice from cardiac hypertrophy. Nature. 416 (6878), 334-337 (2002).
- Zalk, R., et al. Structure of a mammalian ryanodine receptor. Nature. 517 (7532), 44-49 (2015).
- Ridgway, E. B., Gilkey, J. C., Jaffe, L. F. Free calcium increases explosively in activating medaka eggs. Proceedings of the National Academy of Sciences of the United States of America. 74 (2), 623-627 (1977).
- Gilkey, J. C., Jaffe, L. F., Ridgway, E. B., Reynolds, G. T. A free calcium wave traverses the activating egg of the medaka, Oryzias latipes. Journal of Cell Biology. 76 (2), 448-466 (1978).
- Kreko-Pierce, T., Azpurua, J., Mahoney, R. E., Eaton, B. A. Extension of health span and life span in Drosophila by S107 requires the calstabin homologue FK506-BP2. Journal of Biological Chemistry. 291 (50), 26045-26055 (2016).
- Sullivan, K. M., Scott, K., Zuker, C. S., Rubin, G. M. The ryanodine receptor is essential for larval development in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 97 (11), 5942-5947 (2000).
- Feng, R., et al. Dynamics expression of DmFKBP12/Calstabin during embryonic early development of Drosophila melanogaster. Cell & Bioscience. 9 (1), 1-16 (2019).
- Campos-Ortega, J. A., Hartenstein, V. . The embryonic development of Drosophila melanogaster. , 633-645 (1958).
- Xu, X., Dong, C., Vogel, B. Hemicentins Assemble on Diverse Epithelia in the Mouse. Journal of Histochemistry and Cytochemistry. 55 (2), 119-126 (2007).
- Wehrens, X. H., et al. FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell. 113 (7), 829-840 (2003).
- Wehrens, X. H., et al. Protection from cardiac arrhythmia through ryanodine receptor-stabilizing protein calstabin2. Science. 304 (5668), 292-296 (2004).
- Bellinger, A. M., et al. Remodeling of ryanodine receptor complex causes "leaky" channels: a molecular mechanism for decreased exercise capacity. Proceedings of the National Academy of Sciences of the United States of America. 105 (6), 2198-2202 (2008).
- Maruyama, M., et al. FKBP12 is a critical regulator of the heart rhythm and the cardiac voltage-gated sodium current in mice. Circulation Research. 108 (9), 1042-1052 (2011).
- Xu, X., et al. FKBP12 is the only FK506 binding protein mediating T-cell inhibition by the immunosuppressant FK506. Transplantation. 73 (11), 1835-1838 (2002).
- Zalk, R., Marks, A. R. Ca2+ release channels join the ‘resolution revolution’. Trends in Biochemical Sciences. 42 (7), 543-555 (2017).

