Automated, High-Throughput Detection of Bacterial Adherence to Host Cells
Summary
Detection of host-bacterial pathogen interactions based on phenotypic adherence using high-throughput fluorescence labeling imaging along with automated statistical analysis methods enables rapid evaluation of potential bacterial interactions with host cells.
Abstract
Identification of emerging bacterial pathogens is critical for human health and security. Bacterial adherence to host cells is an essential step in bacterial infections and constitutes a hallmark of potential threat. Therefore, examining the adherence of bacteria to host cells can be used as a component of bacterial threat assessment. A standard method for enumerating bacterial adherence to host cells is to co-incubate bacteria with host cells, harvest the adherent bacteria, plate the harvested cells on solid media, and then count the resultant colony forming units (CFU). Alternatively, bacterial adherence to host cells can be evaluated using immunofluorescence microscopy-based approaches. However, conventional strategies for implementing these approaches are time-consuming and inefficient. Here, a recently developed automated fluorescence microscopy-based imaging method is described. When combined with high-throughput image processing and statistical analysis, the method enables rapid quantification of bacteria that adhere to host cells. Two bacterial species, Gram-negative Pseudomonas aeruginosa and Gram-positive Listeria monocytogenes and corresponding negative controls, were tested to demonstrate the protocol. The results show that this approach rapidly and accurately enumerates adherent bacteria and significantly reduces experimental workloads and timelines.
Introduction
Bacterial adhesion is a process whereby bacteria attach to other cells or surfaces. Successful establishment of infection by bacterial pathogens requires adhesion to host cells, colonization of tissues, and in some cases, invasion of host cells1,2,3. Emerging infectious diseases constitute major public health threats, as evidenced by the recent COVID-19 pandemic4,5,6. Importantly, new or emerging pathogens may not be readily discerned using genomic-based approaches, especially in cases where the pathogen has been engineered to evade detection or does not contain genomic signatures that identify it as pathogenic. Therefore, the identification of potential pathogens using methods that directly assess hallmarks of pathogenicity, like bacterial adherence to host cells, can play a critical role in pathogen identification.
Bacterial adherence to host cells has been used to evaluate mechanisms of bacterial pathogenesis for decades1,7. Microscopic imaging8,9 and the enumeration of bacterial colony-forming unit (CFU)10,11,12,13 by post-infection plating are two well-developed laboratory methods for testing microbial adherence and/or infection of host cells14. Considering the micrometer scale size of bacterial cells, the enumeration of the adherent bacterial cells generally requires the use of advanced high-magnification microscopy techniques, as well as high-resolution imaging approaches, including electron microscopy, expansion microscopy (ExM)15,16, and three-dimensional imaging17. Alternatively, the enumeration of bacteria bound to or internalized within host cells can be performed by plating the dilution series of harvested bacteria on solid agar and counting the resultant CFUs10,12,13. This method is laborious and includes many manual steps, which introduces difficulties in establishing a standardized or automated procedure required for high-throughput analyses18,19. Therefore, the development of new methods for evaluating host cell attachment would address current limitations in the field.
One such method is described here that uses automated high throughput microscopy, combined with high throughput image processing and statistical analysis. To demonstrate the approach, experiments with several bacterial pathogens were performed, including Pseudomonas aeruginosa, an opportunistic Gram-negative bacterial pathogen of humans, animals, and plants14,20, which is frequently found to colonize the respiratory tract of patients with impaired host defense functions. This approach optimized the microscopic imaging process described in previous studies14,20. The imaging detection was simplified by fluorescence-labeled host cells and bacteria to rapidly track the proximity of them, which dramatically reduced the microscopy workload to get high-resolution images for distinguishing bacteria. In addition, the automated statistical analysis of images in counting host cells and bacteria replaced the hand-on experiment of bacterial CFU plating to estimate the ratio of adherent bacterial counts per host cell. To confirm the compatibility of this method, multiple bacterial strains and host cell types have also been tested, like Listeria monocytogenes, Staphylococcus aureus, Bacillus cereus, and Klebsiella pneumoniae, as well as human umbilical vein endothelial cells (HUVECs), and the results support the diversity and effectiveness of the method.
Protocol
1. A549 cell culture
- Maintain the A549 cell line in F-12K medium supplemented with 10% fetal bovine serum (FBS) and incubate at 37 °C, 5% CO2.
- Change the medium every 3-4 days and passage at 85%-95% confluency.
- Briefly, rinse the cells with 1 x phosphate-buffered saline (PBS, pH 7.4, unless otherwise indicated) and treat with 1 ml of 0.25% Trypsin-0.53 mM ethylenediaminetetraacetic acid (EDTA) solution (submerging the cell layer) for about 2 min at 37 °C.
- Add additional 6 mL of complete growth medium (F-12K medium + 10% FBS) to stop the protease activity. Then, plate the cells in a sterile polystyrene T-75 tissue culture flask at a subcultivation ratio of 1:3-1:8. The final volume of culture is 12 mL.
- Seed approximately 1 x 104 A549 cells (cell concentration: ~1 x 105 cells/mL) onto each well of a 96-well plate a day prior to the adherence assays.
2. Bacterial growth and staining
- Perform all bacterial work in a Biosafety Cabinet, Biosafety Level 2 laboratory.
- Inoculate all bacterial cultures, including P. aeruginosa (PAO1), Escherichia. coli, L. monocytogenes, and B. subtilis, etc., from the frozen glycerol stock and grow them in Tryptic Soy Broth (TSB, 3 mL) in a shaking incubator at 37 °C overnight maintained at 250 rpm.
- The next day, use a 1:100 dilution of the overnight culture to inoculate a subculture and grown them in TSB (1 mL) for 3 h to the exponential phase, measure the OD at 600 nm (OD600) to confirm. Ensure that the OD600 is in the range of 0.4-0.6.
- Prior to performing the bacteria-host adherence assay, plate serial dilutions of bacterial suspension onto TSB-agar plates, incubate them overnight at 37 °C, and then establish the bacterial CFUs from the number of colonies. For P. aeruginosa, an OD600 of 1.0 corresponds to 2 x 108 viable bacterial cells/mL, and for L. monocytogenes, an OD600 of 1.0 corresponds to 9 x 108 viable bacterial cells/mL.
- Harvest the bacterial cultures at exponential phase by centrifugation at 13,000 x g for 2 min at room temperature (RT), then wash them once using 1x PBS (1 mL). Resuspend the bacterial pellets in 1 mL of 1x PBS and determine the concentrations by measuring OD600 of bacterial suspensions. For example, P. aeruginosa with OD600 of 0.5 represents a concentration of 1 x 108 bacterial cells/mL.
- Stain the bacterial suspension using either a green or a red fluorescent dye at RT for 30 min with gentle rotation in the dark. For this, add 2 μL of the 500- fold concentrated stock staining dye into 1 mL of bacterial suspension to dilute the dye 1-fold. To wash off the staining dye, centrifuge the stained bacteria at 13,000 x g for 2 min and resuspend the pellet in 1 mL of 1x PBS for three times.
NOTE: If fluorescence- (GFP-, RFP-, mCherry-, etc.) tagged bacteria are used in the experiment, then skip this bacterial staining step. GFP- tagged P. aeruginosa and red fluorescent dye-stained L. monocytogenes were used in this protocol. - Collect the stained bacterial cells or GFP- tagged bacteria by centrifugation at 13,000 x g for 2 min. Resuspend in fresh F-12K medium (1 mL) and measure the OD600 of each culture. Subsequentially dilute the cultures to the desired concentrations based on the multiplicity of infection (MOI) and host cell concentration. The final volume used in this experiment is 500 μL.
NOTE: For example, if the host cell concentration counted using Trypan Blue staining is 1 x 105 cells/mL, the desired concentration of bacterial cells at an MOI of 100 will be 1 x 107 cells/mL. Concentration of P. aeruginosa at 0.5 OD600 is 1 x 108/mL. To obtain the desired concentration, dilute the P. aeruginosa culture 10-fold, add 50 μL of resuspended culture to 450 μL of fresh F-12K medium.
3. Bacterial adherence and host cell staining
- First, wash the seeded A549 cell monolayers three times with warm 1x PBS. For each wash, add 100 μL of 1x PBS to each well, gently pipette up and down three times, dispose of 1x PBS or wait for 10 s after addition and then vacuum to remove 1x PBS. To determine the kinetics of bacterial association, overlay the cells with 100 µL of desired concentrations of bacterial suspension with different MOIs (0, 1, 10, and 100). Spin down the bacteria at 200 x g for 10 min and incubate the infected A549 cells at 37 °C, 5% CO2 for an additional 1 h.
NOTE: In this experiment, each condition had a technical triplicate. - Remove the unbound bacteria by washing the monolayers five times with warm 1x PBS as described above. Add 100 µL of 4% formaldehyde (in 1x PBS) into each well of the 96-well plates to fix the cells. Let the plates sit on ice for 15 min and then wash off the fixation solution using 1x PBS three times.
- To stain the nuclei, add 50 ng/mL of 4′,6-diamidino-2-phenylindole (DAPI) and incubate it for 10 min at RT. After incubation, wash the wells three times using 1x PBS. Cover the infected A549 cells with 100 µL of 1x PBS to avoid drying. Process for the next step or store the plate at 4 °C for up to 2 days in the dark.
4. Automated fluorescence imaging, processing, and analysis
- To maintain the data integrity, randomly and manually pick five locations of each well to capture the images at 20x magnification. Capture the fluorescent images of A549 cells and bacteria under DAPI and GFP channels, respectively. Use PE-Cy5 channel for the bacteria stained by the red fluorescent dye.
- To have a better resolution, process all the images for background flattening and deconvolution. Set the parameters as 68 µm diameter of the rolling ball for smoothing and auto-measure the point spread function (PSF) of image deconvolution based on the objective.
- To count all qualified cells and bacteria, measure the fluorescence intensity of the host cells and bacteria and set the weakest fluorescence intensity of host cells and bacteria as the thresholds for cell count. Count all bacteria proximate to a host cell within 15 µm distance as the adherent bacteria as the diameter of A549 cell is 10.59-14.93 µm21.
NOTE: A default setting in the imaging system selectively counts the host cells with diameters ranging from 5-100 µm and bacteria ranging from 0.2-5 µm in size (width and length). - Based on the above-mentioned manual image processing results, apply the parameters of image smoothing, deconvolution, objects sizes, distance, and fluorescence intensities to the rest of the automated images. After the automated analysis, consider critical readouts, such as total host cell counts, cell sizes and shapes, total bacterial counts, and the average bacterial count per host cell, which was the most important indicator, to determine bacterial adherence.
- Data export and statistical analysis
- Export the analyzed results from all images to a spreadsheet (e.g., 'xlsx' format). The automated system generates two sets of results: 1) the average adherent bacterial count from each image; 2) the informative data of each single host cell, such as adherent bacterial count on a single cell, host size, and the average fluorescence intensity of host cell and bacteria, from the corresponding image.
- Calculate the mean number and standard deviation of adherent bacterial counts from all images to represent the bacterial adherence level compared to negative controls. In this method, E. coli and B. subtilis served as negative controls in testing Gram-negative and Gram-positive bacterial adherence, respectively. Perform two-way ANOVAs to test for significant variation between data points across treatment for three independent experiments.
Representative Results
To develop the fluorescence imaging-based bacterial adherence assay, P. aeruginosa strain PAO1 and its negative-adherence counterpart E. coli were used to test the protocol effectiveness, as the adherence of these bacteria to A549 cells had been reported14,20,22. First, GFP- labeled P. aeruginosa (PAO1) and GFP-labeled E. coli were co-incubated with a human immortalized epithelial cell line A549 at various MOIs, respectively. The results showed that PAO1 adhered to A549 cells in a dose-dependent fashion (Figure 1A,B); in the meantime, near null adherence of E. coli was also verified (Figure 1A,B). There were 50 images captured and analyzed at each MOI. Two-way ANOVAs were performed to test the significant variations from three independent experiments.
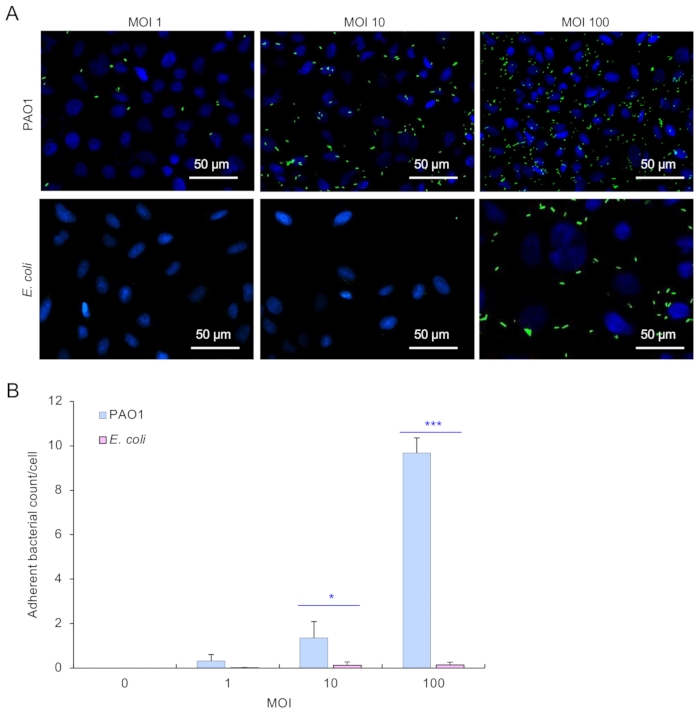
Figure 1: Gram-negative bacterial P. aeruginosa adhered to A549 cells within 1 h of co-incubation. (A) Microscopic images for an overview of bacterial adherence where the images were taken using 20x magnification. PAO1 and the negative-adherence control E. coli adhered to A549 cells at the indicated multiplicity of infections (MOIs). Bacteria were GFP-fluorescence tagged. A549 cell nuclei were stained by DAPI. The scale bar is 50 µm. (B) Quantification of adherent bacterial counts per A549 cell. For each tested bacterial strain (PAO1 and E. coli) in each MOI, a total of 50 images were applied to the analysis at each condition. Data are mean ± standard deviation (SD) from one representative of three independent experiments. The two-way ANOVA statistical analysis was performed. * p < 0.05, *** p < 0.001. Please click here to view a larger version of this figure.
Similar results were also observed when Gram-positive bacteria, L. monocytogens23,24 and its negative control B. subtilis, were used (Figure 2A,B). The adherence to A549 cells was significant in L. monocytogenes than B. subtilis (Figure 2A,B).
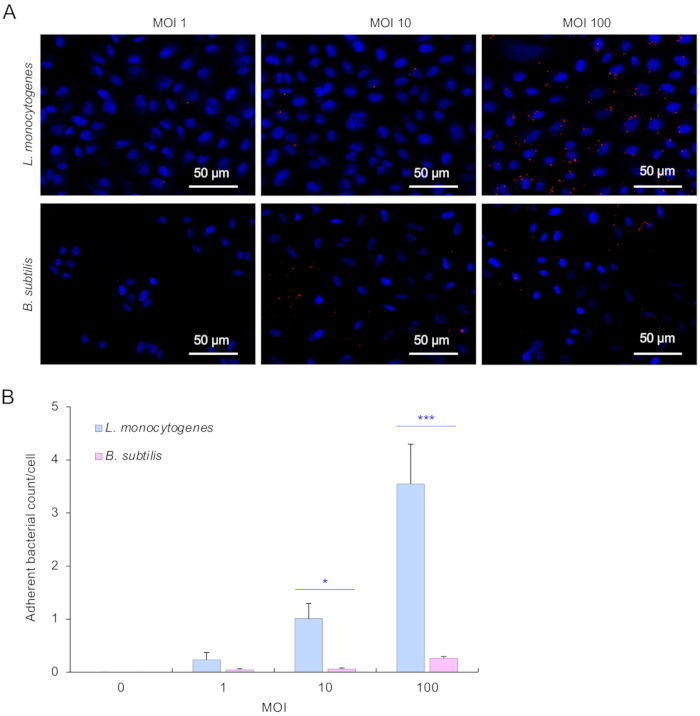
Figure 2: Gram-positive bacterial L. monocytogenes adhered to A549 cells within 1 h of co-incubation. (A) Microscopic images for an overview of L. monocytogenes, as well as the negative control B. subtilis adherence to A549 cells at different MOIs. Bacteria were stained using a red-fluorescent dye. A549 cell nuclei were stained with DAPI. The scale bar is 50 µm. (B) Quantification of adherent bacterial count per A549 cell. For each tested bacterial strain (L. monocytogenes and B. subtilis) in each MOI, a total of 50 images were applied to the analysis at each condition. Data are mean ± SD from a representative of three independent experiments. The two-way ANOVA statistical analysis was performed. ** p < 0.01, *** p < 0.001. Please click here to view a larger version of this figure.
A representative image of host-adherent P. aeruginosa segmentation and counts is shown in Figure 3 (Yellow masks: selected bacteria, red outline: single bacterial count). The settings for both host and bacteria targets are described in the Protocol section.
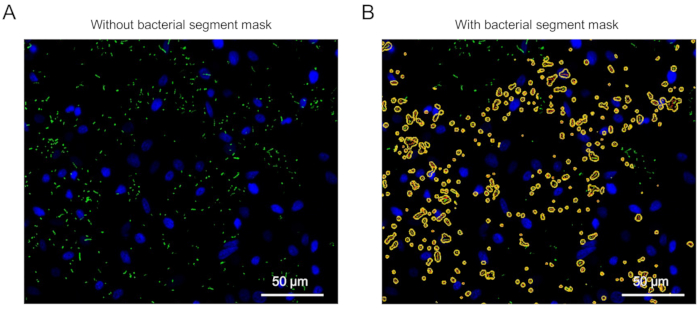
Figure 3: Example images of bacterial segmentation and counting in the automated analysis process. (A) Microscopic images of P. aeruginosa adhered to A549 at an MOI of 100 without bacterial segmentation and counting. (B) The same image after the statistical analysis of bacterial segmentation and counting. Yellow masks: selected bacteria, red outline: single bacterial count. Please click here to view a larger version of this figure.
The results of the automated analyses, including host cell sizes and areas, bacterial and host cell counts, as well as the average bacterial counts per host cell, representing the status of host cell health, bacteria adherence level, and bacterial cytotoxicity, are listed in Table 1 and Table 2. Table 1 represents the adherent bacterial counts at an average cellular level from different images, while Table 2 represents adherent bacterial counts analyzed on one image at a single A549 cellular level. These representative images were captured from A549 cells infected by PAO1 at an MOI of 100. The analyzed results of the initial image in Figure 3 were listed as Image 1 in Table 1 and Table 2.
Table 1: Statistical analysis results of 9 representative images at the cellular level. Please click here to download this Table.
In each image, host sum count and bacterial sum spots represent the total counts of host nuclei and total bacterial counts recognized by the system based on the parameters described above, respectively. In addition, the automated calculation of bacterial spots ratio represents the average bacterial count per host cell, derived from the bacteria sum spots/host sum count. Moreover, additional readings can also be included; for instance, the bacteria-adherent host counts illustrate the universal or specific phenomenon of host-bacteria adherence. When the bacteria-adherent host counts are much less than host sum counts, in contrast, the average bacterial count per host is relatively high, which represents that such adherence phenotype has a significant heterogeneity.
Table 2: Single cellular statistical analysis of a representative image. Please click here to download this Table.
Singular cell analysis provides more details about host cell and bacterial targets in each image, which is more useful when gathering other bacterial features and applying them to the automated computational analysis to develop a more powerful tool in evaluating the potential bacterial pathogenicity, like a Machine Learning model which is being studied. In this protocol, host size and area represent the diameters and areas of each stained host nuclei, and host fluorescence intensity represents the average intensity of the stained nuclei. Bacterial spot count and area represent the adherent bacterial targets to each host cell and their total areas. Bacterial fluorescence intensity represents the average intensity of all adherent bacteria.
It is not surprising that some virulent bacteria did not adhere to A549 while some bacteria with high levels of bacterial adherence do not necessarily correlate with pathogenicity. A different host cell type, HUVECs, was also tested to maximize the application of this method. The results showed the effective detection and the different adherence phenotypes of bacteria (Figure 4). Different host cells could increase the estimation of potential bacterial pathogens; for instance, pathogenic Serratia rubidaea and Streptococcus agalactiae were adherent to HUVEC but not to A549 cells (Figure 4). Moreover, the cytotoxic Enterohemorrhagic E. coli (EHEC) was non-adherent to either A549 or HUVEC (Figure 4). Therefore, it is critical to ensure that the method is suitable for multiple host cell types to respond to the specificity of host-bacteria interactions. Both A549 and HUVEC cells were co-incubated with bacteria at an MOI of 100 at 37 °C for 1 h.
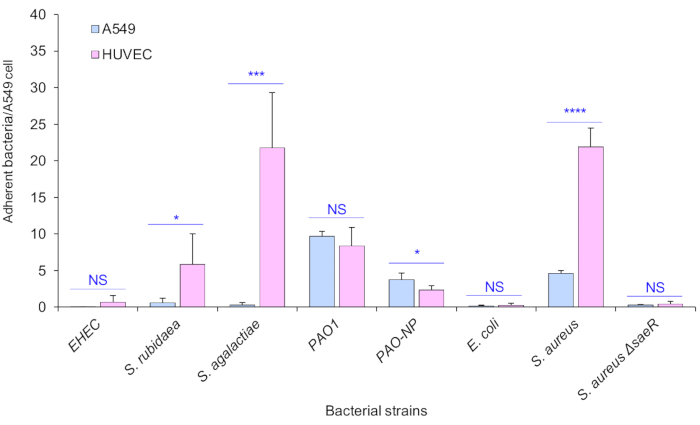
Figure 4: Specificity of bacterial adherence to host A549 and HUVEC cells. The bacterial strains, including S. rubidaea, S. agalactiae, cytotoxic Enterohemorrhagic E. coli (EHEC), PAO1, P. aeruginosa ΔpilA (PAO-NP), E. coli, S. aureus, and S. aureus ΔsaeR, were tested on both A549 and HUVEC cells at an MOI of 100 for 1 h of co-incubation at 37 °C, 5% CO2. Bacteria were stained with a red-fluorescent dye. Adherent bacterial counts were quantified from 45 images/bacterial strain in three independent experiments. Data are mean ± SD from one representative of three independent experiments. * p < 0.05, *** p < 0.001, **** p<0.0001. Please click here to view a larger version of this figure.
Discussion
The protocol describes an automated approach for enumerating bacterial attachment to host cells. The described approach has several attractive advantages over conventional methods. First, this approach enables the precise quantification of the number of microbial pathogen cells that are attached to individual host cells. Importantly, this quantification can be performed without the need for laborious bacterial harvesting, serial dilutions, plating on solid media, and determination of CFUs10,11,12. As such, the described technique reduces the overall workload required to quantify bacterial adherence. It should be appreciated that the advantages of the proposed approach are amplified when seeking to detect adherence phenotypes in large-scale screening experiments, including the identification of bacterial or host genes in mutant or CRISPR libraries, respectively, that regulate this process. Second, the direct evaluation of interactions between host and bacterial cells using fluorescence microscopy provides information for elucidating mechanisms of bacterial pathogenicity, including alterations in a host or bacterial cell survival, morphology, or dynamics. Finally, the automated statistical analysis performed on images collected in the analysis not only provides an overall quantification of the average bacterial association with host cells but also enables the evaluation of dynamic, single-cell, host-pathogen interactions.
Despite these advantages, the described methods have some important limitations. First, fluorescence staining of bacteria is required to enumerate their adherence to host cells. Thus, the fluorescent staining dye has to selectively stain bacteria than their host cell counterparts. In addition, the staining dye must not alter the attachment phenotype of the bacteria that carry it. Therefore, the optimization of bacterial fluorescence staining (e.g., concentration, staining duration) must be determined in advance of performing the described adherence studies. In this protocol, fluorescent staining of bacterial strains including P. aeruginosa, L. monocytogenes, E. coli, and B. subtilis for 30 min does not affect their adherence to host cells. Second, due to the specificity of different host-bacterium interactions, a single host cell type may not be sufficient to evaluate bacterial attachment to host cells. Therefore, multiple host cell types might be required to gain a complete understanding of the degree to which a given microbe can adhere to host cell surfaces. Third, bacterial segmentation was not fully efficient when bacteria propel aggregation during the co-incubation, e.g., S. aureus, or when bacteria formed clumps at a higher MOI (>100), e.g., P. aeruginosa. In this case, a higher magnification should be applied to capture the images, or more images should be used in the analysis to reduce the effect of bacterial aggregation.
Human lung epithelial cells (A549) and HUVECs were employed in the studies to demonstrate the plasticity of the approach with respect to host cell type, and both displayed high levels of susceptibility to bacterial adherence. The use of two host cell types also demonstrated that the described method could be widely applied to characterize adherent host-bacterium interactions rapidly.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We are thankful to Dr. Kaite Zlotkowski of Biotek Inc. for their technical support. We also thank Dr. Lori Burrows, McMaster University, for the generous gift of the Pseudomonas strains.This work was supported by the Department of Defense under contract number W911NF1920013 to PdF; the Defense Advanced Research Projects Agency (DARPA) and the Department of Interior under Contract No. 140D6319C0029 to PdF. The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
Materials
| 10x PBS | VWR | 45001-130 | |
| 4′,6-diamidino-2-phenylindole (DAPI) | Thermo Fisher | 62248 | Host cell staining dye |
| 96 well plate | Corning | 3882 | Half area well, flat clear bottom |
| A549 cells | ATCC | CCL 185 | Mammalian cell line |
| BactoView Live Red | Biotium | 40101 | Bacteria staning dye |
| Centrifuge | Eppendorf | 5810R | |
| CFSE cell division tracker | BioLegend | 423801 | |
| Cytation 5 | BioTek | Cytation 5 | Cell imaging multi-mode reader |
| E. coli | Laboratory stock | ||
| EGM bulletKit | Lonza | CC-3124 | HUVEC cell culture medium |
| EHEC | NIST collections | ||
| F-12k medium | ATCC | 302004 | A549 cell culture medium |
| Fetal bovine serum | Corning | 35-016-CV | |
| HUVEC | Laboratory stock | ||
| L. monocytogenes | NIST collections | ||
| OD600 DiluPhotometer | IMPLEN | ||
| P. aeruginosa | Dr. Lori Burrows laboratory stock | ||
| P. aeruginosa ΔpilA | Dr. Lori Burrows laboratory stock | ||
| S. agalactiae | NIST collections | ||
| S. aureus | BEI | NR-46543 | |
| S. aureus ΔsaeR | BEI | NR-48164 | |
| S. rubidaea | NIST collections | ||
| Typical soy broth | Growcells | MBPE-4040 |
References
- Pizarro-Cerda, J., Cossart, P. Bacterial adhesion and entry into host cells. Cell. 124, 715-727 (2006).
- Kipnis, E., Sawa, T., Wiener-Kronish, J. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Médecine et Maladies Infectieuses. 36 (2), 78-91 (2006).
- Josse, J., Laurent, F., Diot, A. Staphylococcal adhesion and host cell invasion: Fibronectin-binding and other mechanisms. Frontiers in Microbiology. 8, 2433 (2017).
- Cabibbo, G., Rizzo, G. E. M., Stornello, C., Craxì, A. SARS-CoV-2 infection in patients with a normal or abnormal liver. Journal of Viral Hepatitis. 28 (1), 4-11 (2021).
- Ortiz-Prado, E., et al. Clinical, molecular, and epidemiological characterization of the SARS-CoV-2 virus and the Coronavirus Disease 2019 (COVID-19), a comprehensive literature review. Diagnostic Microbiology and Infectious Disease. 98 (1), 115094 (2020).
- Chang, C. C., Senining, R., Kim, J., Goyal, R. An acute pulmonary coccidioidomycosis coinfection in a patient presenting with multifocal pneumonia with COVID-19. Journal of Investigative Medicine High Impact Case Reports. 8, (2020).
- Woo, V., et al. Microbiota inhibit epithelial pathogen adherence by epigenetically regulating C-type lectin expression. Frontiers in Immunology. 10, 928 (2019).
- Pandey, A., et al. Global reprogramming of host kinase signaling in response to fungal infection. Cell Host Microbe. 21 (5), 637-649 (2017).
- Ding, S., et al. Interactions between fungal hyaluronic acid and host CD44 promote internalization by recruiting host autophagy proteins to forming phagosomes. iScience. 24 (3), 102192 (2021).
- Qin, Q. M., et al. RNAi screen of endoplasmic reticulum-associated host factors reveals a role for IRE1alpha in supporting Brucella replication. PLoS Pathogens. 4 (7), 1000110 (2008).
- Qin, Q. M., et al. Functional analysis of host factors that mediate the intracellular lifestyle of Cryptococcus neoformans. PLoS Pathogens. 7 (6), 1002078 (2011).
- Qin, Q. M., et al. A tractable Drosophila cell system enables rapid identification of Acinetobacter baumannii host factors. Frontiers in Cellular and Infection Microbiology. 10, 240 (2020).
- Pandey, A., et al. Activation of host IRE1α-dependent signaling axis contributes the intracellular parasitism of Brucella melitensis. Frontiers in Cellular and Infection Microbiology. 8, 103 (2018).
- Chi, E., Mehl, T., Nunn, D., Lory, S. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infection and Immunity. 59 (3), 822-828 (1990).
- Götz, R., et al. Nanoscale imaging of bacterial infections by sphingolipid expansion microscopy. Nature Communications. 11, 6173 (2020).
- Lim, Y., et al. Mechanically resolved imaging of bacteria using expansion microscopy. PLOS Biology. 17 (10), 3000268 (2019).
- Bratton, B. P., Barton, B., Morgenstein, R. M. Three-dimensional Imaging of bacterial cells for accurate cellular representations and precise protein localization. Journal of Visualized Experiments. (152), e60350 (2019).
- Hoffmann, S., et al. High-throughput quantification of bacterial-cell interactions using virtual colony counts. Frontiers in Cellular and Infection Microbiology. 8, 43 (2018).
- Hazan, R., Que, Y. -. A., Maura, D., Rahme, L. G. A method for high throughput determination of viable bacteria cell counts in 96-well plates. BMC Microbiology. 12 (1), 259 (2012).
- Gellatly, S. L., Hancock, R. E. W. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathogens and Disease. 67, 159-173 (2013).
- Jiang, R. D., Shen, H., Piao, Y. J. The morphometrical analysis on the ultrastructure of A549 cells. Romanian Journal of Morphology and Embryology. 51 (4), 663-667 (2010).
- Farinha, M. A., et al. Alteration of the pilin adhesin of Pseudomonas aeruginosa PAO results in normal pilus biogenesis but a loss of adherence to human pneumocyte cells and decreased virulence in mice. Infection and Immunity. 62 (10), 4118-4123 (1994).
- Réglier-Poupet, H., Pellegrini, E., Charbit, A., Berche, P. Identification of LpeA, a PsaA-Like membrane protein that promotes cell entry by Listeria monocytogenes. Infection and immunity. 71 (1), 474-482 (2003).
- Ortega, F. E., et al. Adhesion to the host cell surface is sufficient to mediate Listeria monocytogenes entry into epithelial cells. Molecular Biology of the Cell. 28 (22), 2945-2957 (2017).

